小麦旗叶叶绿素含量的QTL定位及验证
2023-01-16赵佳佳武棒棒温宏伟张树伟郑兴卫
杨 斌 乔 玲 赵佳佳 武棒棒 温宏伟 张树伟 郑兴卫 郑 军
小麦旗叶叶绿素含量的QTL定位及验证
杨 斌**乔 玲**赵佳佳 武棒棒 温宏伟 张树伟 郑兴卫 郑 军*
山西农业大学小麦研究所/ 省部共建有机旱作农业国家重点实验室(筹), 山西临汾 041000
旗叶是小麦主要的光合器官, 叶绿素既是旗叶最主要的光合色素, 也是品种选育中重要的表型指标, 因此挖掘和利用旗叶叶绿素含量有关的主效基因/位点, 对于培育高产稳产小麦新品种意义重大。以旗叶叶绿素含量差异较大双亲构建的双单倍体群体(DH群体)为材料, 利用小麦90K SNP芯片对5个环境旗叶叶绿素含量进行QTL分析, 共定位到20个旗叶叶绿素含量有关的遗传位点, 表型贡献率为4.10%~27.16%; 其中3个QTL (、和)能在多个环境条件下检测到;的遗传效应最高, 该位点与2D染色体上已报道的其他叶绿素位点不同, 初步确定是1个新的主效QTL。并进一步将紧密连锁的SNP标记开发为KASP标记, 通过在含有共同亲本金麦919的RIL群体中验证其效应, 发现在多个环境条件下具有有利等位基因的家系叶绿素含量显著或极显著高于其他家系。对、和所在功能区段进行基因注释, 筛选到12个与叶绿素相关的候选基因, 其中3个基因参与镁等金属离子的结合过程, 5个基因参与调控叶绿体结构组成, 4个基因参与调控光合作用过程中相关电子链的传递活性。本文研究结果为叶绿素调控的遗传机制提供了有价值的信息, 并为高光效分子标记辅助育种提供依据与参考。
小麦; 叶绿素含量; SNP标记; QTL定位
小麦(L.)是我国主要粮食作物, 小麦高产与稳产对我国粮食安全意义重大。在遭受逆境胁迫时, 叶绿素含量较高的品种可以保持较强的光合能力, 有助于产量稳定形成[1-8]。小麦旗叶叶绿素含量与产量紧密相关, 是品种选育中重要的表型指标[9]。叶绿素的合成与降解由多基因调控, 易受到环境的影响, 对叶绿素含量进行QTL分析, 发掘稳定主效QTL, 不仅有利于完善叶绿素调控的遗传机制[10-12], 亦有助于分子标记辅助选育小麦高光效品种。利用不同遗传群体对苗期、抽穗期以及灌浆期的叶绿素含量进QTL分析已有报道[3,13-18], 目前已获得了上百个控制叶绿素含量的QTL, 分布于21条染色体上[4,19-27], 但检测到的位点普遍效应较低, 易受环境条件影响, 且多在个别环境下进行, 主效QTL缺少多个环境中验证。此外, 早期研究采用SSR标记构建遗传连锁图谱, 标记间平均遗传距离大于10 cM, 鉴定的位点数量较少, 主效QTL检出率较低。因此, 利用高密度SNP遗传连锁图谱, 在多年多点的不同环境中检测旗叶叶绿素含量QTL, 并在不同遗传群体中进行验证, 可发掘稳定主效QTL。
迄今, 尚无小麦旗叶叶绿素含量相关QTL精细定位的报道, 仅有利用早衰突变体对持绿主效位点进行精细定位的研究[7,28-29], 如Li等[28]利用早衰突变体M114构建的F2群体进行了BSR-Seq分析, 将基因定位在2BS染色体上, 遗传距离为1.5 cM; Wang等[30]利用早衰突变体LF2099构建的F2群体, 将基因定位在2BL上2BIP09~2BIP14的标记区间。通过同源克隆已获得、和等调控叶绿素的相关基因。编码细胞分裂素氧化酶的位于3A染色体, 在4个环境下Jing411与Hongmangchun 21构建的RIL群体中,对叶绿素含量的贡献率为8.9%~20.1%[31]; 编码2-Cys过氧还蛋白BAS1的基因位于2B染色体, RIL群体(Jing 411×Hongmangchun 21)在3个环境中,对叶绿素含量的贡献率为9.0%~19.2%[32]; 在7A染色体上, 编码脱镁叶绿酸水解酶的基因与干旱胁迫环境下的旗叶叶绿素含量紧密相关[33]。但这3个基因并不在已定位的QTL区间内, 且均未进行转基因验证。目前叶绿素含量有关的稳定主效QTL报道较少, 因此利用遗传群体和自然群体挖掘定位叶绿素含量有关的主效QTL, 对于认识小麦叶绿素调控机制很有价值。本文以DH群体(晋春7号×金麦919)为材料, 利用90K芯片在5个环境中对小麦旗叶叶绿素含量进行QTL分析, 旨在发掘调控旗叶叶绿素含量的稳定主效QTL, 为基因克隆和分子育种提供有价值的信息。
1 材料与方法
1.1 试验材料
供试材料为2个遗传群体, 其中DH群体为晋春7号和金麦919构建的152个家系用于QTL定位, 母本晋春7号的叶绿素含量高于父本金麦919。RIL群体为尧麦(DH118)和金麦919构建的165个家系用于QTL效应的验证。
1.2 田间试验
供试材料于2018—2019、2019—2020和2020— 2021年种植于山西农业大学小麦研究所尧都区试验地(36°08′N, 111°52′E, YD)和韩村试验站(36°25′N, 111°67′E, HC), 简称为E1 (18YD)、E2 (18HC)、E3 (19YD)、E4 (19HC)、E5 (20YD)和E6 (20HC); DH群体种植在除E6环境外的其余5个环境中。试验采用随机区组设计, 3次重复, 每个家系种植2行, 行长1.5 m, 行距0.3 m, 每行21粒。播种后韩村试验站全生育期只依靠自然降水, 2018—2019、2019—2020和2020—2021年度小麦整个生育期内降水量分别为132、154和147 mm; 尧都区试验地分别在越冬前、拔节期和灌浆期正常灌溉, 其他同常规管理方法。
1.3 表型数据统计
在亲本与各家系中随机选取抽穗期一致、发育正常的10株小麦进行标记, 开花后10 d使用SPAD- 502 (Konica-Minolta, 日本)叶绿素仪对旗叶叶绿素含量进行测定, 计算10株测定结果的平均值[18]。采用SPSS 21.0进行检验、相关性和方差分析; 使用SAS计算不同环境下旗叶叶绿素含量的最佳线性无偏预测(best linear unbiased prediction, BLUP)和广义遗传力(2)。
1.4 遗传连锁图谱构建与QTL分析
采用改良CTAB法[34]提取基因组DNA, 利用Illumina SNP Genotyping技术测试平台(北京博奥生物有限公司)微珠芯片技术(Bead Array)对DH群体进行90K SNP标记分型, 根据Li等[35]的方法采用IciMapping V4.0 (https://www.isbreeding.net/)进行高密度遗传连锁图谱的构建。
利用WinQTLCart 2.5 (https://brcwebportal.cos. ncsu.edu/qtlcart/WQTLCart.htm)复合区间作图法对叶绿素含量进行QTL定位, LOD阈值为2.5。将间距小于1 cM或共用侧翼标记的QTL视为同一个位点, 参照McCouch等[36]方法对QTL命名。将定位到的QTL侧翼标记与小麦多组学网站数据库(http:// wheatomics.sdau.edu.cn/)中的中国春参考基因组序列进行比对[37]。
1.5 KASP标记开发与验证
基于QTL定位结果, 针对主效位点的连锁SNP标记开发KASP标记(PolyMarker, http://polymarker. tgac.ac.uk/), 设计2条SNP特异性引物KASP- Excalibur_c18324_390F(5′-GTACTAGTGTTTCTAG AGCTGTCAT-3′), KASP-Excalibur_c18324_390H(5′- GTACTAGTGTTTCTAGAGCTGTCAC-3′)和1条通用引物KASP-Excalibur_c18324_390R(5′-AAGCGT ACAATGGAGAGAAGT-3′)。KASP引物由北京嘉程生物科技有限公司合成。利用开发的KASP标记对RIL群体的165个家系进行检测, 验证主效QTL的效应。
1.6 候选基因预测
利用小麦多组学网站(http://wheatomics.sdau. edu.cn/)的JBrowse平台获得QTL目标区间内的基因。使用GO (gene ontology)数据库对这些区域的基因进行功能注释和富集分析, 筛选出与叶绿素相关的基因。利用小麦多组学网站分析小麦和水稻的同源基因(http://wheat.cau.edu.cn/TGT/)。
2 结果与分析
2.1 DH群体叶绿素含量表型分析
对5个环境下的双亲和DH群体旗叶叶绿素含量进行方差分析, 结果表明, 晋春7号的旗叶叶绿素含量为46.17~58.59, 金麦919为44.43~57.79, 晋春7号均高于金麦919, 其中在E1、E4和E5环境中差异显著(<0.05) (表1)。DH群体叶绿素含量的变异范围为48.42~56.90, 峰度与偏度绝对值大多小于1, 表明各环境中叶绿素含量均呈现连续性正态分布, 适用于QTL分析。叶绿素含量的2为0.58, 说明叶绿素含量主要受遗传因素影响。
2.2 遗传连锁图谱
利用小麦Wheat 90K SNP芯片数据, 经过滤、筛选和去冗余后构建了高密度遗传连锁图谱(图1), 该图谱总长2456.98 cM, 包含3470个SNP标记, 标记间平均遗传距离为0.71 cM, 覆盖小麦21条染色体。分布在A、B和D染色体组的标记数目分别为1346、1410和714个, 连锁长度分别为739.52、949.54和767.92 cM, 平均遗传距离分别为0.55、0.67和1.08 cM。
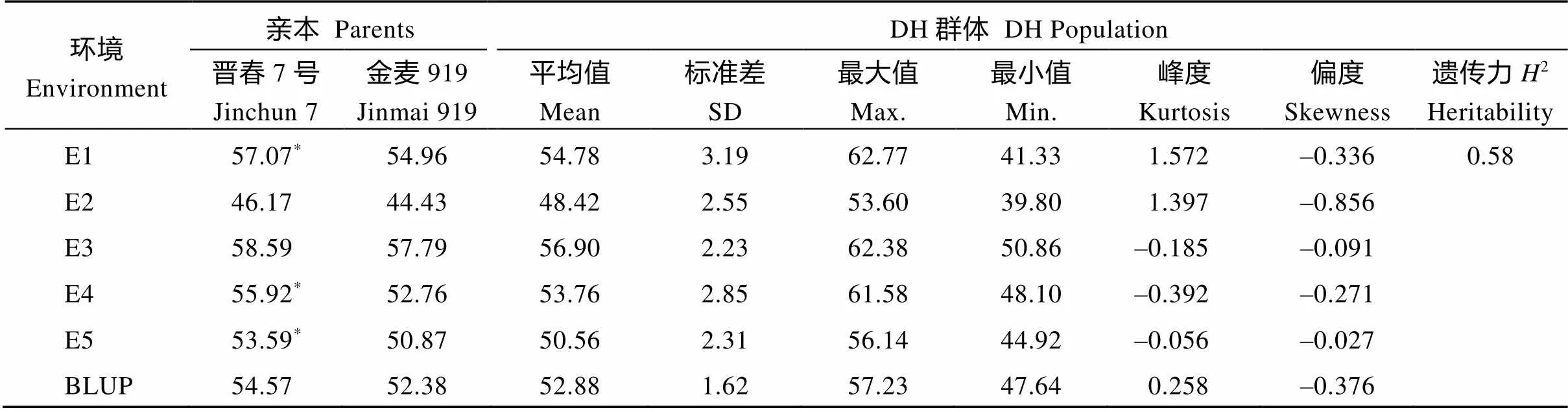
表1 晋春7号×金麦919亲本与DH群体的旗叶叶绿素含量变异特征

图1DH群体的遗传连锁图谱
2.3 旗叶叶绿素含量QTL分析
对DH群体的旗叶叶绿素含量进行QTL分析, 共检测到20个控制叶绿素含量的QTL, 分布于2A、2B、2D、3A、3B、4A、4B、4D、6A和6B染色体上, LOD值介于2.55~13.60之间, 可解释4.10%~27.16%的表型变异率(表2), 其中6个QTL加性效应来自于晋春7号, 14个QTL加性效应来源于金麦919 (表2)。和能在多个环境中检测到, 可解释4.10%~26.07%的表型变异率。其中位于2DS染色体BS00012148_51~Excalibur_c18324_390标记区间的对表型的贡献率大于10%, 加性效应来自于金麦919, 是1个稳定主效旗叶叶绿素含量相关的QTL。
2.4 稳定QTL位点的聚合效应
对检测到的3个稳定QTL、4D和的加性效应分析表明, 随着有利等位基因数量的增多相关株系旗叶叶绿素含量增高(图2-A), 具有3个有利等位基因的家系的叶绿素含量极显著高于缺失这些等位基因的家系(<0.01)。DH群体中仅具有有利等位基因的家系的旗叶叶绿素含量高于仅具有和等位基因的家系, 表明对叶绿素含量的遗传效应最高。此外, 根据连锁的SNP标记将DH群体划分为两类, 携带有金麦919 (正向增加叶绿素含量)等位变异的家系与携带有晋春7号等位变异的家系在各环境中差异均极显著(<0.01), 金麦919等位变异可增加0.49-3.41的叶绿素含量(图2-B)。

图2 Qchl.saw-2D.1、Qchl.saw-4D.2和Qchl.saw-6A的加性效应(A)、Qchl.saw-2D.1在DH群体中的效应(B)
2.5 Qchl.saw-2D.1标记开发与验证
利用不同的遗传群体在2D、4D和6A染色体上已分别检测到10个、1个和12个旗叶叶绿素含量的QTL (表3), 根据中国春参考基因组序列v1.0, 发现在2D染色体上检测到的[16]和[38]与的标记区间相邻; 4D染色体上检测到的[39]与的标记区间重合;与Hassan等[40]在6A染色体上检测到的QTL相邻。已有研究在2D染色体上定位到的10个叶绿素QTL位于23.03~570.40 Mb区间内(表3), 而本文检测的定位在2DS的3.77~7.97 Mb区间内, 物理间距为4.20 Mb (图3-A, B), 此区段内尚无叶绿素含量QTL/基因的报道, 可以初步确定是1个新的叶绿素含量有关的主效QTL。
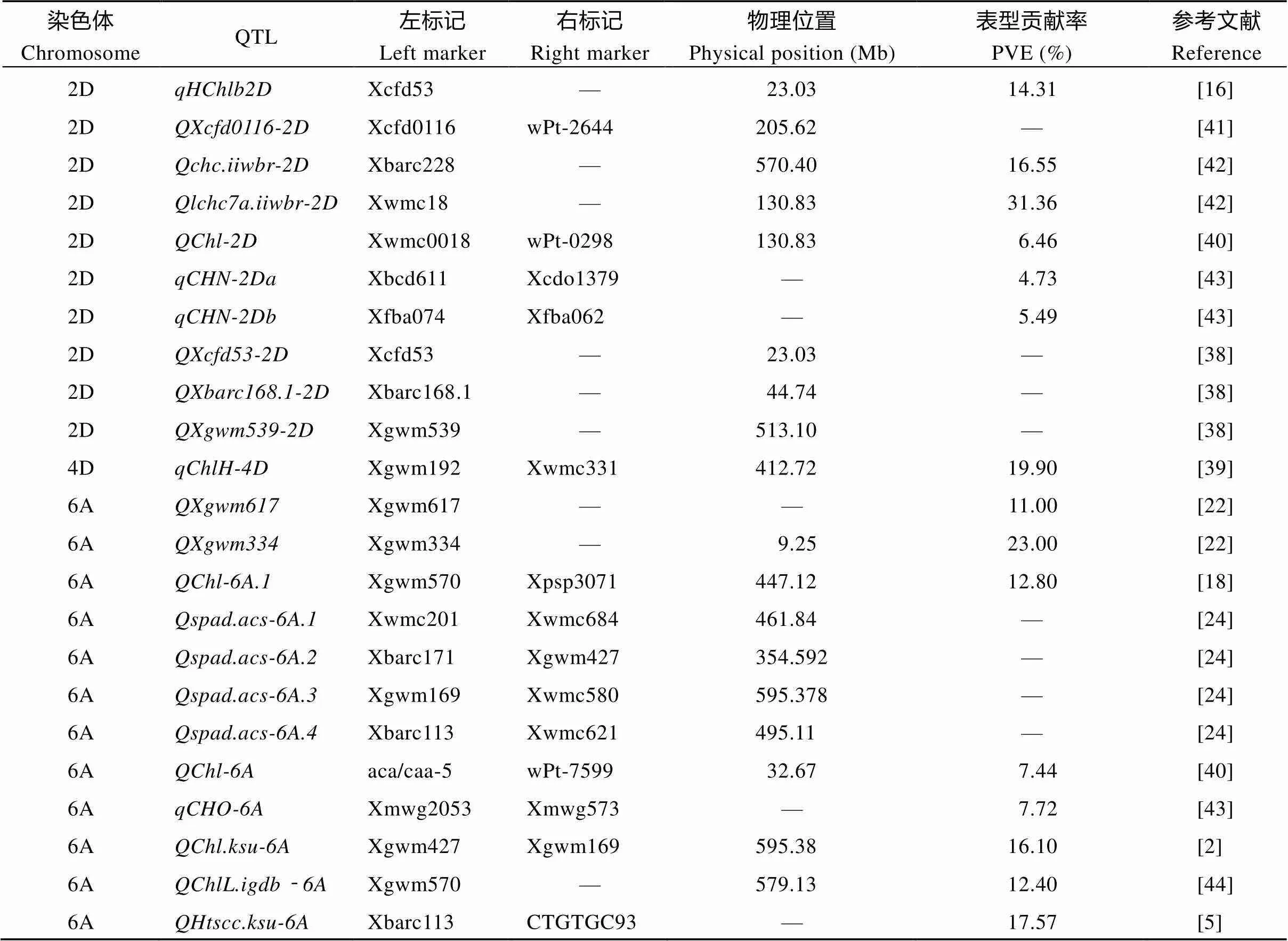
表3 前人研究在2D、4D和6A染色体上检测到的叶绿素QTL
为了验证的遗传效应, 利用其连锁SNP标记Excalibur_c18324_390设计KASP标记, 命名为。在RIL群体(尧麦33 (DH118)×金麦919)中进行QTL验证(图3-C), 结果表明, 携带有金麦919等位变异有68个家系, 携带有尧麦33 (DH118)的等位变异家系有97个, 组间在E2和E6环境中差异显著(<0.05), 在E1和E3环境中差异极显著(<0.01)。金麦919等位变异可增加0.35~2.37的叶绿素含量(图3-C), 说明具有较强的增加旗叶叶绿素的作用。
2.6 Qchl.saw-2D.1、Qchl.saw-4D.2和Qchl.saw- 6A的候选基因预测
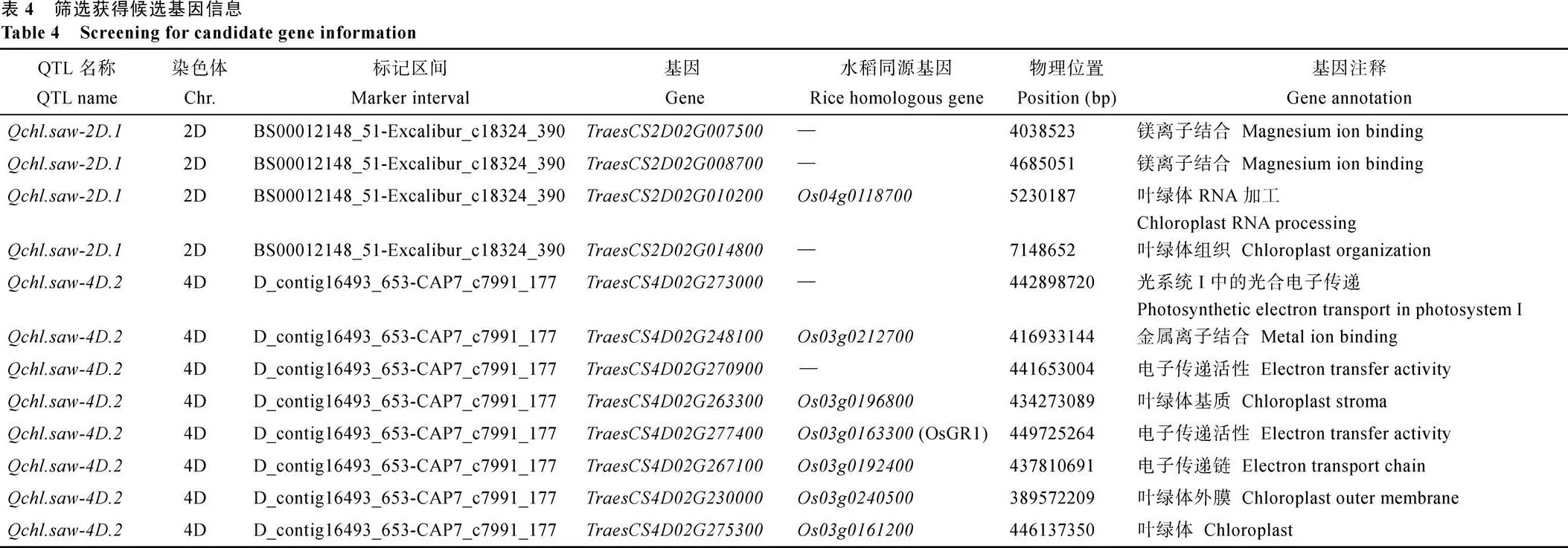
对、和区间内基因进行功能注释, 共筛选到12个叶绿素相关的候选基因, 其中2D染色体上4个, 4D染色体上8个(表4), 候选基因中与水稻同源的基因有7个。经过GO富集发现、和通过参与镁等金属离子结合, 影响小麦光合作用过程中相关氧化还原反应;、、、和参与叶绿体RNA加工过程和叶绿体基质、外膜等结构组成;、、和在光合作用过程参与相关电子链的电子传递活性。
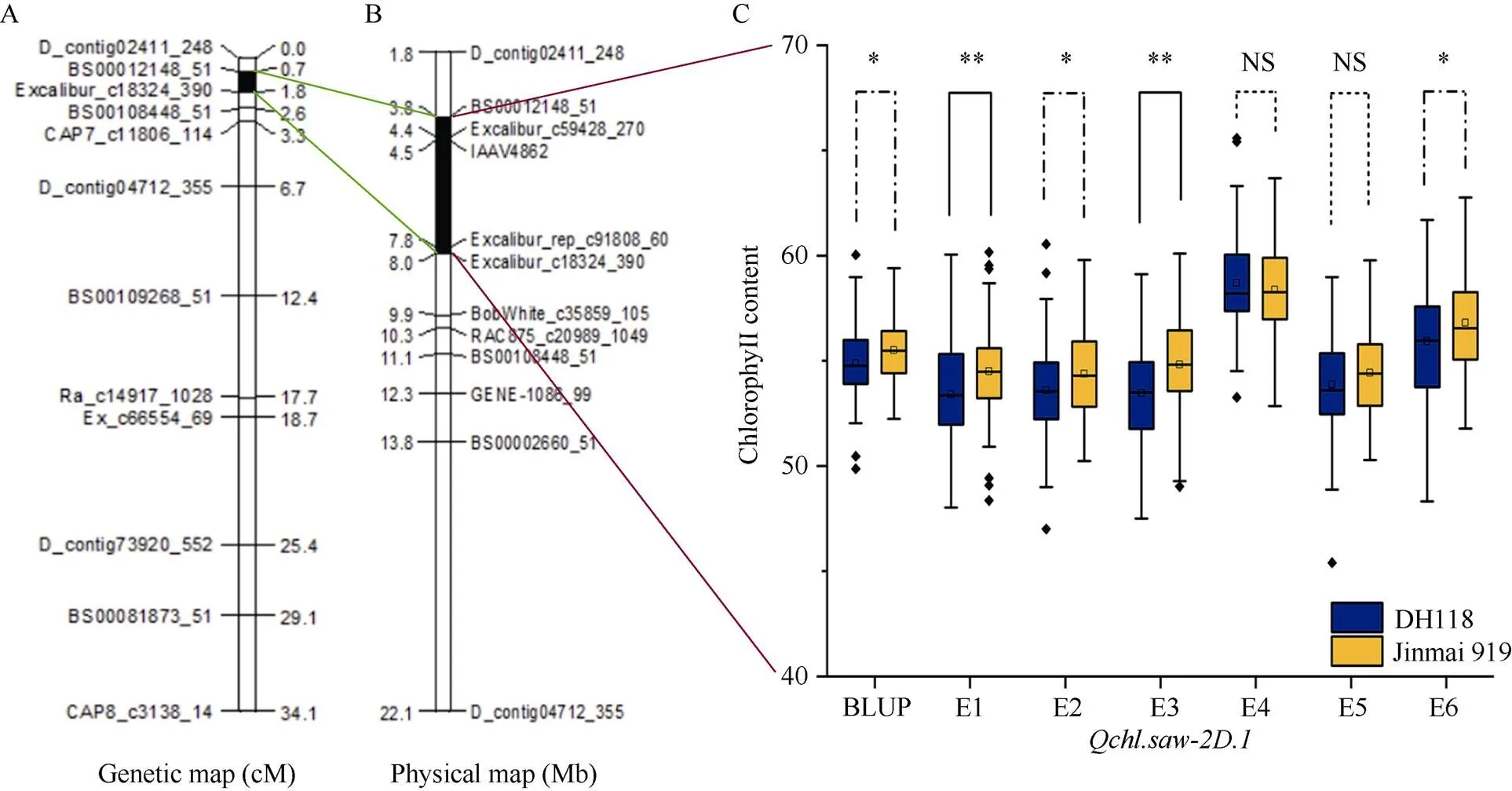
图3 Qchl.saw-2D.1遗传图谱(A)、物理图谱(B)及Qchl-2D.1在RIL群体中的效应分析(C)
3 讨论
3.1 Qchl.saw-2D.1是新的旗叶叶绿素含量有关的主效QTL
旗叶叶绿素含量受多基因调控, 易受环境因素影响, 发掘叶绿素含量有关的稳定主效QTL是应用于分子标记辅助育种的前提。目前已检测到上百个控制叶绿素含量的QTL[7,45], 但共定位的QTL较少。造成这种现象的原因: 1) 叶绿素检测方法不同, 有些研究使用分光光度计测定[19], 有些采用叶绿素仪测定[42], 表型值的差异必然导致定位结果不同; 2) 控制数量性状的基因表达具有时空特异性, 调控叶片叶绿素含量的QTL在各发育时期具有差异[24], 现有研究分为苗期、抽穗期以及灌浆中后期, 检测时期的不同造成鉴定出的QTL具有差异; 3) 分子标记的类型不同, 造成不同研究结果无法直接进行比较; 4) 检测到的QTL多数效应值较低, 定位结果易受环境因素干扰。因此, 能够在不同研究中或不同环境下被多次检测到, 且具有较高表型贡献率的主效QTL, 可能更具应用于分子标记辅助育种的潜力[24]。
以中国春物理图谱v1.0为参考, 发现本文检测到的大多数QTL与前人报道的结果不同, 这是因为这些QTL多为效应值较低的微效QTL, 容易受到环境条件或遗传背景的影响; 本文定位到3个QTL与已有研究位置相同, 在2D染色体上20.63 Mb至22.14 Mb区间, 本文与已有研究共同检测到叶绿素含量的QTL[16,38]; 这个区间内还检测到许多产量相关性状QTL, 如控制千粒重的[46]和[47]以及同时控制千粒重和穗长的[48]。大量研究已从表型的角度证实了叶绿素含量与产量性状高度相关[4,19], 本文定位到的叶绿素QTL与已报道的千粒重等产量相关性状QTL位置重合, 说明该区段是调控多种重要农艺性状的热点区间, 具有深入挖掘的潜在价值。在4D染色体上386.44~451.30 Mb区间, 本文与Li等[39]共同检测到叶绿素含量的主效QTL。在6A染色体29.56~32.88 Mb区间, 本文与Hassan等[40]共同定位到叶绿素含量的QTL。可见、和能够在不同遗传背景及不同环境条件下被多次检测到, 是稳定的QTL。目前小麦中已克隆的3个叶绿素相关基因、和分别位于2B、3A和7A染色体上, 在2D染色体上尚无叶绿素相关基因克隆的报道; 本文在2D染色体3.77~7.97 Mb区间检测到叶绿素含量的稳定主效QTL, 该区间内尚未见到叶绿素QTL的报道, 表明是1个新的主效叶绿素含量QTL。
3.2 主效QTL区段中与叶绿素相关的候选基因
叶绿素的生物合成和生物反应过程受基因调控,筛选调控基因并进行功能分析有助于深入了解叶绿素调控机制[49]。对3个稳定QTL (、和)的功能区间进行基因功能分析, 筛选到12个叶绿素相关功能的候选基因,第一类是5个叶绿体组成的相关基因,参与调控叶绿体基因组转录的初级RNA分子转化为一个或多个成熟的RNA分子, 促进叶绿体的合成。参与调控叶绿体基质的合成, 叶绿体基质作为光合作用暗反应相关酶的储存场所, 含量越多, 光合作用越强。叶绿体外膜作为信号蛋白和离子的转运通道, 对叶绿体相关蛋白的运转起着至关重要的作用[50],调控叶绿体外膜的组成, 水稻中同源基因调控叶绿体外膜转运子和GTPase亚基toc34 (叶绿体34转位酶)[51],和也涉及叶绿体组成的调控。第二类是7个参与调控光合作用的相关基因, 涉及镁等金属离子结合和光合电子传递过程。叶绿体在光合作用过程中离不开镁离子等螯合酶, 它们在调控昼夜环境下光反应和暗反应的转化过程起着重要作用[52]。、和通过调控镁等金属离子结合, 调节小麦叶绿体光合作用过程中相关氧化还原反应,有利于小麦的生物量积累和抵御胁迫环境的能力。和调控光系统I反应中心的电子传递过程, 而光合电子传递链的调控是植株正常生长和存活的关键, 还影响叶绿体内的能量调节和氧化还原平衡[53]。水稻的同源基因,的同源基因也参与调控电子传递链的传递过程, 在水稻中基因则主要调控叶绿体中的谷胱甘肽还原酶活性[54]。后续工作我们将构建次级分离群体, 筛选重组株, 重点对进行精细定位。
4 结论
利用DH群体在2D、4D和6A染色体上定位到3个稳定的旗叶叶绿素含量QTL、和, 其中是新的主效位点, 基于其紧密连锁标记开发了可供育种利用的KASP标记, 并在RIL群体中证实其具有较强的增加旗叶叶绿素的作用。对3个稳定QTL的物理区间进行了功能注释, 筛选到12个叶绿素相关的候选基因。
[1] Kumar S, Sehgal S K, Kumar U, Prasad P V V, Joshi A K, Gill B S. Genomic characterization of drought tolerance-related traits in spring wheat., 2012, 186: 265–276.
[2] Ilyas M, Ilyas N, Arshad M, Kazi A G, Kazi A M, Waheed A. QTL mapping of wheat doubled haploids for chlorophyll content and chlorophyll fluorescence kinetics under drought stress imposed at anthesis stage., 2014, 46: 1889–1897.
[3] Vijayalakshmi K, Fritz A K, Paulsen G M, Bai G, Pandravada S, Gill B S. Modeling and mapping QTL for senescence-related traits in winter wheat under high temperature., 2010, 26: 163–175.
[4] Talukder S K, Babar M A, Vijayalakshmi K, Poland J, Prasad P V V, Bowden R, Fritz A. Mapping QTL for the traits associated with heat tolerance in wheat (L.)., 2014, 15: 97.
[5] Awlachew Z T, Singh R, Kaur S, Bains N S, Chhuneja P. Transfer and mapping of the heat tolerance component traits ofin tetraploid wheat., 2016, 36: 78–92.
[6] Gupta P K, Balyan H S, Sharma S, Kumar R. Genetics of yield, abiotic stress tolerance and biofortification in wheat (L.)., 2020, 133: 1569–1602.
[7] Bhoite R, Si P, Siddique K H M, Yan G. Comparative transcriptome analyses for metribuzin tolerance provide insights into key genes and mechanisms restoring photosynthetic efficiency in bread wheat (L.)., 2021, 113: 910–918.
[8] Borjigin C, Schilling R K, Jewell N, Brien C, Sanchez-Ferrero J C, Eckermann P J, Watson-Haigh N S, Berger B, Pearson A S, Roy S J. Identifying the genetic control of salinity tolerance in the bread wheat landrace Mocho de Espiga Branca., 2021, 48: 1148–1160.
[9] Avenson T J, Cruz J A, Kanazawa A, Kramer D M. Regulating the proton budget of higher plant photosynthesis., 2005, 102: 9709–9713.
[10] Thomas H, Ougham H. The stay-green trait., 2014, 65: 3889–3900.
[11] Verma V, Foulkes M J, Worland A J, Sylvester-Bradley R, Caligari P D S, Snape J W. Mapping quantitative trait loci for flag leaf senescence as a yield determinant in winter wheat under optimal and drought-stressed environments., 2004, 135: 255–263.
[12] Rasheed A, Takumi S, Hassan M A, Imtiaz M, Ali M, Morgunov A I, Mahmood T, He Z. Appraisal of wheat genomics for gene discovery and breeding applications: a special emphasis on advances in Asia., 2020, 133: 1503–1520.
[13] Quarrie S, Pekic Quarrie S, Radosevic R, Rancic D, Kaminska A, Barnes J D, Leverington M, Ceoloni C, Dodig D. Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes., 2006, 57: 2627–2637.
[14] Guo P, Baum M, Varshney R K, Graner A, Grando S, Ceccarelli S. QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought., 2008, 163: 203–214.
[15] Czyczylo-Mysza I, Marcińska I, Skrzypek E, Chrupek M, Grzesiak S, Hura T, Stojałowski S, Myśków B, Milczarski P, Quarrie S. Mapping QTLs for yield components and chlorophyll a fluorescence parameters in wheat under three levels of water availability., 2011, 9: 291–295.
[16] Zhang K, Fang Z, Liang Y, Tian J. Genetic dissection of chlorophyll content at different growth stages in common wheat., 2009, 88: 183–189.
[17] Yu M, Mao S L, Chen G Y, Liu Y X, Li W, Wei Y M, Liu C J, Zheng Y L. QTLs for waterlogging tolerance at germination and seedling stages in population of recombinant inbred lines derived from a cross between synthetic and cultivated wheat genotypes., 2014, 13: 31–39.
[18] Yang B, Yan X, Wang H Y, Li X X, Ma H X, Wang S G, Sun D Z, Jing R L. Dynamic QTL analysis of chlorophyll content during grain filling stage in winter wheat (L.)., 2016, 33: 77–85.
[19] Zhang K, Zhang Y, Chen G, Tian J. Genetic analysis of grain yield and leaf chlorophyll content in common wheat., 2009, 37: 499–511.
[20] Kumar U, Joshi A K, Kumari M, Paliwal R, Kumar S, Roeder M S. Identification of QTLs for stay green trait in wheat (L.) in the ‘Chirya 3’ × ‘Sonalika’ population., 2010, 174: 437–445.
[21] Saleh M S, Al-Doss A A, Elshafei A A, Moustafa K A, Al-Qurainy F H, Barakat M N. Identification of new TRAP markers linked to chlorophyll content, leaf senescence, and cell membrane stability in water-stressed wheat., 2014, 58: 64–70.
[22] Barakat M N, Saleh M, Al-Doss A A, Moustafa K A, Elshafei A A, Al-Qurainy F H. Identification of new SSR markers linked to leaf chlorophyll content, flag leaf senescence and cell membrane stability traits in wheat under water stressed condition., 2015, 66: 93–102.
[23] Li X M, He Z H, Xiao Y G, Xia X C, Trethowan R, Wang H J, Chen X M. QTL mapping for leaf senescence-related traits in common wheat under limited and full irrigation., 2015, 203: 569–582.
[24] Yang D, Li M, Liu Y, Chang L, Cheng H, Chen J, Chai S. Identification of quantitative trait loci and water environmental interactions for developmental behaviors of leaf greenness in wheat., 2016, 7: 273–288.
[25] Shi S, Azam F I, Li H, Chang X, Li B, Jing R. Mapping QTL for stay-green and agronomic traits in wheat under diverse water regimes., 2017, 213: 246–264.
[26] Yan X, Wang S, Yang B, Zhang W, Cao Y, Shi Y, Sun D, Jing R. QTL mapping for flag leaf-related traits and genetic effect ofon flag leaf width using two related introgression line populations in wheat., 2020, 15: e0229912.
[27] 郑军, 李晓华, 赵佳佳, 尚保华, 曹勇, 马小飞, 张晓军, 乔玲, 乔麟轶, 郑兴卫, 张建诚. 山西省小麦育成品种遗传多样性分析. 植物遗传资源学报, 2018, 19: 619–626.
Zheng J, Li X H, Zhao J J, Shang B H, Cao Y, Ma X F, Zhang X J, Qiao L, Qiao L Y, Zheng X W, Zhang J C. Genetic diversity analysis of wheat cultivars in Shanxi province., 2018, 19: 619–626 (in Chinese with English abstract).
[28] Li M, Li B, Guo G, Chen Y, Xie J, Lu P, Wu Q, Zhang D, Zhang H, Yang J, Zhang P, Zhang Y, Liu Z. Mapping a leaf senescence geneby BSR-Seq in common wheat., 2018, 6: 236–243.
[29] Wang N, Xie Y, Li Y, Wu S, Li S, Guo Y, Wang C. High-resolution mapping of the novel early leaf senescence genein common wheat.(Basel), 2020, 9: 698.
[30] Wang N, Xie Y Z, Li Y Z, Wu S N, Wei H S, Wang C S. Molecular mapping of a novel early leaf-senescence genein common wheat by SNP genotyping arrays., 2020, 71: 356–367.
[31] Chang C, Lu J, Zhang H P, Ma C X, Sun G. Copy number variation of cytokinin oxidase geneassociated with grain weight and chlorophyll content of flag leaf in common wheat., 2015, 10: e0145970.
[32] Zhu X F, Zhang H P, Hu M J, Wu Z Y, Jiang H, Cao J J, Xia X C, Ma C X, Chang C. Cloning and characterization of-gene associated with flag leaf chlorophyll content and thousand-grain weight and development of a gene-specific marker in wheat., 2016, 36: 142–153.
[33] Wang H, Wang S, Chang X, Hao C, Sun D, Jing R. Identification of-haplotypes and development of a molecular marker associated with important agronomic traits in common wheat., 2019, 19: 296.
[34] 乔玲, 刘成, 郑兴卫, 赵佳佳, 尚保华, 马小飞, 乔麟轶, 盖红梅, 姬虎太, 刘建军, 张建诚, 郑军. 小麦骨干亲本临汾5064单元型区段的遗传解析. 作物学报, 2018, 44: 931–937.
Qiao L, Liu C, Zheng X W, Zhao J J, Shang B H, Ma X F, Qiao L Y, Ge H M, Ji H T, Liu J J, Zhang J C, Zheng J. Genetic analysis of haplotype-blocks from wheat founder parent Linfen 5064., 2018, 44: 931–937 (in Chinese with English abstract).
[35] Li F, Wen W, Liu J, Zhai S, Cao X, Liu C, Cheng D, Guo J, Zi Y, Han R, Wang X, Liu A, Song J, Liu J, Li H, Xia X. Genome-wide linkage mapping for canopy activity related traits using three RIL populations in bread wheat., 2021, 217: 67–82.
[36] McCouch S R, Chen X L, Panaud O, Temnykh S, Xu Y B, Cho Y G, Huang N, Ishii T, Blair M. Microsatellite marker development, mapping and applications in rice genetics and breeding., 1997, 35: 89–99.
[37] 胡文静, 李东升, 裔新, 张春梅, 张勇. 小麦穗部性状和株高的QTL定位及育种标记开发和验证. 作物学报, 2022, 48: 1346–1356.
Hu W J, Li D S, Yi X, Zhang C M, Zhang Y. Molecular mapping and validation of quantitative trait loci for spike-related traits and plant height in wheat., 2022, 48:1346–1356 (in Chinese with English abstract).
[38] 李玮瑜, 张斌, 张嘉楠, 昌小平, 李润植, 景蕊莲. 利用关联分析发掘小麦自然群体旗叶叶绿素含量的优异等位变异. 作物学报, 2012, 38: 962–970.
Li W Y, Zhang B, Zhang J N, Chang X P, Li R Z, Jing R L. Exploring elite alleles for chlorophyll content of flag leaf in natural population of wheat by association analysis., 2012, 38: 962–970 (in Chinese with English abstract).
[39] Li H, Tong Y, Li B, Jing R, Lu C, Li Z. Genetic analysis of tolerance to photo-oxidative stress induced by high light in winter wheat (L.)., 2010, 37: 399–412.
[40] Hassan F S C, Solouki M, Fakheri B A, Nezhad N M, Masoudi B. Mapping QTLs for physiological and biochemical traits related to grain yield under control and terminal heat stress conditions in bread wheat (L.)., 2018, 24: 1231–1243.
[41] Tahmasebi S, Heidari B, Pakniyat H, McIntyre C L. Mapping QTLs associated with agronomic and physiological traits under terminal drought and heat stress conditions in wheat (L.)., 2017, 60: 26–45.
[42] Bhusal N, Sharma P, Sareen S, Sarial A K. Mapping QTLs for chlorophyll content and chlorophyll fluorescence in wheat under heat stress., 2018, 62: 721–731.
[43] 曹卫东, 贾继增, 金继运. 不同供氮水平下小麦苗期叶绿素含量的QTL及互作研究. 植物营养与肥料学报, 2004, 10: 473–478.
Cao W D, Jia J Z, Jin J Y. Identification and interaction analysis of QTL for chlorophyll content in wheat seedlings., 2004, 10: 473–478 (in Chinese with English abstract).
[44] Li H, Wang G, Zheng Q, Li B, Jing R, Li Z. Genetic analysis of biomass and photosynthetic parameters in wheat grown in different light intensities., 2014, 56: 594–604.
[45] Gupta P K, Balyan H S, Gahlaut V. QTL analysis for drought tolerance in wheat: present status and future possibilities.(Basel), 2017, 7: 5.
[46] Huang X Q, Cloutier S, Lycar L, Radovanovic N, Humphreys D G, Noll J S, Somers D J, Brown P D. Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (L.)., 2006, 113: 753–766.
[47] Guan P F, Lu L H, Jia L J, Kabir M R, Zhang J B, Lan T Y, Zhao Y, Xin M M, Hu Z R, Yao Y Y, Ni Z F, Sun Q X, Peng H R. Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (L.)., 2018, 9: 529–546.
[48] Wu X Y, Cheng R R, Xue S L, Kong Z X, Wan H S, Li G Q, Huang Y L, Jia H Y, Jia J Z, Zhang L X, Ma Z Q. Precise mapping of a quantitative trait locus interval for spike length and grain weight in bread wheat (L.)., 2014, 33: 129–138.
[49] 姜雪, 龙武华, 李祖军, 唐会会, 吴朝昕, 张习春, 吴娴, 朱速松. 水稻叶绿素合成途径关键基因自然变异分析. 分子植物育种, 2022, [2022-04-15]. http://kns.cnki.net/kcms/detail/46. 1068.S.20220118.1547.004.html.
Jiang X, Long W H, Li Z J, Tang H H, Wu C X, Zhang X C, Wu X, Zhu S S. Variation analysis of chlorophyll biosynthesis key genes in natural populations of rice (L.)., 2022, [2022-04-15]. http://kns.cnki.net/kcms/detail/46.1068. S.20220118.1547.004.html (in Chinese with English abstract).
[50] 杨小龙, 李漾漾, 刘玉凤, 齐明芳, 李天来. 植物叶绿体蛋白质周转的研究进展及潜在应用. 植物生理学报, 2019, 55: 577–586.
Yang X L, Li Y Y, Liu Y F, Qi M F, Li T L. Recent advances in chloroplast protein turnover and potential applications., 2019, 55: 577–586 (in Chinese with English abstract).
[51] Zhou J, Wang Z G, Huang Z W, Lu C, Han Z, Zhang J F, Jiang H M, Ge C L, Yang J C. Expression of sulfur uptake assimilation-related genes in response to cadmium, bensulfuron-methyl and their co-contamination in rice roots., 2014, 26: 650–661.
[52] 罗莎, 罗韬, 彭鹏, 李艳萍, 黎小刚. 镁离子螯合酶活性调控及其功能. 植物生理学报, 2015, 51: 806–812.
Luo S, Luo T, Peng P, Li Y P, Li X G. Activity regulation and functions of Mg chelatase., 2015, 51: 806–812 (in Chinese with English abstract).
[53] 薛娴, 许会敏, 吴鸿洋, 沈应柏, 肖建伟, 万迎朗. 植物光合作用循环电子传递的研究进展. 植物生理学报, 2017, 53: 145–158.
Xue X, Xu H M, Wu H Y, Shen Y B, Xiao J W, Wan Y L. Research progress of cyclic electron transport in plant photosynthesis., 2017, 53: 145–158 (in Chinese with English abstract).
[54] Hong C Y, Chao Y Y, Yang M Y, Cheng S Y, Cho S C, Kao C H. NaCl-induced expression of glutathione reductase in roots of rice (L.) seedlings is mediated through hydrogen peroxide but not abscisic acid., 2009, 320: 103–115.
QTL mapping and validation of chlorophyll content of flag leaves in wheat (L.)
YANG Bin**, QIAO Ling**, ZHAO Jia-Jia, WU Bang-Bang, WEN Hong-Wei, ZHANG Shu-Wei, ZHENG Xing-Wei, and ZHENG Jun*
Institute of Wheat Research, Shanxi Agricultural University / State Key Laboratory of Sustainable Dryland Agriculture (in preparation), Linfen 041000, Shanxi, China
Flag leaf is the main photosynthetic organ in wheat. The chlorophyll content is not only the major photosynthetic pigment in flag leaf but also an important phenotypic indicator in crop breeding. Therefore, the identification of major loci/genes related to chlorophyll content in the flag leaf play an important role in breeding wheat varieties with higher grain yields and stability. In this study, we constructed a double haploid (DH) population from a cross of two cultivars with significant difference in chlorophyll content, and the chlorophyll contents of DH lines were detected under five environments. A total of 20 QTLs associated with chlorophyll content were detected using Wheat 90K single-nucleotide polymorphism (SNP) array, with contributions to phenotypic variation explained (PVE) from 4.10% to 27.16%. Three QTLs (,andwere identified under multiple environmental conditions, in whichwith the strongest genetic effect was different from previous studies and identified as a novel major QTL. Furthermore,was validated by a tightly linked kompetitive allele specific PCR (KASP) marker in a recombinant inbred line (RIL) population containing the co-parent Jinmai 919. Those lines with the favorable allele ofrevealed significantly higher chlorophyll content than other lines under multiple environments. Moreover, a total of 12 candidate genes controlling chlorophyll content were identified in the three QTL regions. Based on gene annotation, three genes were involved in the binding process of metal iron, such as magnesium. Five genes were regulated the structural composition of chloroplasts, and four genes were engaged in the regulation of electron transfer activities during the photosynthetic process. In conclusion, this study will broaden the understanding of the genetic mechanism and provide a molecular basis for the marker-assisted breeding (MAS) of chlorophyll content in the flag leaf of wheat.
wheat; chlorophyll content; SNP marker; QTL mapping
10.3724/SP.J.1006.2023.21018
本研究由中央引导地方科技发展资金项目(YDZJSX2022A033), 山西农业大学育种工程(YZGC013)和山西省基础研究计划项目(202103021223156)资助。
This study was supported by the Central Guidance on Science & Technology Development of Shanxi (YDZJSX2022A033), the Shanxi Agricultural University Breeding Project (YZGC013), and the Basic Research Program of Shanxi Province (202103021223156).
郑军, E-mail: sxnkyzj@126.com
同等贡献(Contributed equally to this work)
杨斌, E-mail: sxxmsyb83@126.com; 乔玲, E-mail: qiaolingsmile@163.com
2022-02-25;
2022-06-07;
2022-07-07.
URL: https://kns.cnki.net/kcms/detail/11.1809.S.20220706.1444.004.html
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
