基于活体微生物揭示蚯蚓对污泥耐药基因转归的影响
2022-07-19魏枫沂徐俊杰李建辉董夕琳
魏枫沂,徐俊杰,陈 进,李建辉,黄 魁,2*,董夕琳,夏 慧
基于活体微生物揭示蚯蚓对污泥耐药基因转归的影响
魏枫沂1,徐俊杰1,陈 进1,李建辉1,黄 魁1,2*,董夕琳3,夏 慧1
(1.兰州交通大学环境与市政工程学院,甘肃 兰州 730070;2.甘肃省黄河水环境重点实验室,甘肃 兰州 730070;3.长春水务集团有限公司,吉林 长春 130000)
为削减污泥蚯蚓堆肥产物中耐药基因(ARGs),以无蚯蚓组为对照,采用叠氮溴化丙锭对蚯蚓堆肥样品进行预处理,探究蚯蚓对污泥中活微生物种群及其ARGs的影响.结果显示,与对照组相比,蚯蚓堆肥产物中有机物矿化度与降解量分别显著提升82.5%与5.2%(<0.05).并且接种蚯蚓使其产物中放线菌门的丰度显著增加了65.6%(<0.05),而厚壁菌门和拟杆菌门的丰度分别显著降低了74.7%和34.6%(<0.05).相较于对照组,蚯蚓堆肥致使1、2、F和M基因丰度分别显著减少了66.5%、82.8%、72.8%和77.6%(<0.05),但B的丰度显著增加了5.7倍(<0.05).蚯蚓堆肥产物中I1基因丰度比对照组显著降低了67.2%(<0.05),蚯蚓处理后ARGs总绝对丰度为4.19×1013copies/g,ARGs总去除率为82.6%,比对照组高45.4%.研究表明,蚯蚓可通过改变活细菌种群结构,减少ARGs潜在活体宿主的丰度,进而降低其传播扩散的潜在风险.
堆肥;微生物种群;抗性基因;蚯蚓粪;污泥资源化
随着我国城市污水处理量的逐年增加,剩余污泥产量也与日俱增[1].然而,当前多数污水厂剩余污泥稳定化处理能力不足,污泥处理处置形势非常严峻[2].此外,污泥成分极为复杂,既含有碳、氮、磷等资源性物质,也含有重金属、持久性有机污染物、ARGs、微塑料等污染性物质[3].其中ARGs是一种新型的生物污染物,在环境中具有较强的扩散传播能力[4].研究表明,剩余污泥中ARGs种类多样,总丰度高达1015copies/g[5].同时,污泥中多元的微生物亦为ARGs的水平转移起到促进作用,增加人畜共患抗生素抗性致病细菌的传播风险[5-6].因此,如何消减污泥处理过程中的ARGs,已成为污泥处理处置所要解决的关键问题[7].
蚯蚓堆肥通过虫体与微生物的相互作用完成有机物的生物降解与稳定,具有工艺简单、能耗低、蚓粪肥效高等优点[8-9],被认为是一种生态环境友好的绿色技术.由于污泥来源及性质复杂多样,堆肥处理后的蚯蚓粪中亦含有大量的ARGs.虽有研究表明蚯蚓堆肥可减少污泥中ARGs总丰度,但各研究结果存在争议性[10-11].例如相关研究发现蚯蚓堆肥能有效降低污泥中磺胺类、四环素类抗性基因和整合子的丰度[12-13],陈景阳等[15]也证实蚯蚓堆肥可以显著降低整合子的丰度,控制ARGs的传播.但另有研究报道污泥蚯蚓粪中2、G及C的丰度在堆肥后显著增加[14-15].前人的研究均基于总DNA,未区分堆肥体内的活死菌可能是造成以上结果存在差异的原因之一.虽然死菌编码的ARGs仍有传播潜力,但环境中ARGs只有在活体微生物内才能进行代谢和转录等生命活动[16].因此,活体微生物体内ARGs的丰度及转移机制更为重要.然而目前关于ARGs的研究极少关注活体微生物.
叠氮溴化丙锭(PMA)可在强光下进入膜受损细胞,形成不能PCR扩增的修饰DNA[17],已广泛应用于细菌、病毒等各种微生物的活性检测[18].因此,本研究采用PMA对堆肥样品进行预处理,结合荧光定量PCR和高通量测序技术,分析蚯蚓处理污泥过程中活体微生物及其ARGs的影响,旨在为削减污泥蚯蚓粪中ARGs提供科学依据.
1 材料和方法
1.1 供试材料
选用赤子爱胜蚓()作为堆肥蚓种,实验前蚯蚓用脱水污泥饲养驯化.堆肥实验反应器选用带盖的矩形塑料箱(60cm×40cm×30cm).供试污泥取自兰州市安宁区七里河污水厂脱泥车间(含水率73%),供试污泥的理化性质见表1.
1.2 实验方法
将10kg新鲜脱水污泥投加至反应器中,块状污泥稍加分散,堆体厚度为10cm.之后接种1200条单体重约0.5g活跃的赤子爱胜蚓作为蚯蚓处理开始堆肥实验,以不添加蚯蚓的污泥作为对照处理.每个处理设3个重复,实验在室温(20~25℃)下进行30d.为保持水分、湿度和有氧条件,在各反应器上覆盖带孔保鲜膜,每隔一周手动翻堆减少污泥颗粒积压团聚,翻堆后喷洒少量自来水.堆肥20d后,蚯蚓处理组中污泥全部转化为颗粒状蚯蚓粪,将蚯蚓从堆肥反应器中取出,堆肥产物继续腐熟10d.随后从各反应器中取样,一式两份.一份自然风干后研磨,过60目筛,置于4℃冰箱中保存,用于理化性质分析;另一份新鲜样品直接经过PMA处理后提取DNA,并置于-20℃冰箱中冷冻保存,用于DNA相关分析.
1.3 测定方法
1.3.1 理化性质分析 采用灼烧减量法测定样品有机质(HJ 761-2015)[19].将风干研磨样品与去离子水混匀(干样:水=1:50;质量浓度),磁力搅拌30min后测定混合液的pH值(雷磁,PHS-3C,上海)和电导率(雷磁,DDS-307,上海).将上述混合液稀释10倍后,经0.45µm薄滤膜抽滤,采用碳氮分析仪(耶拿,MULTI N/C2100,德国)测定溶解性有机碳.硝酸盐氮采用紫外分光光度法(HJ/T 346-2007)[20],氨氮采用纳氏试剂分光光度法(HJ 535-2009)[21],总氮采用碱性过硫酸钾消解紫外分光光度法(HJ 636-2012)[22],总磷采用钼酸铵分光光度法(GB 11893-89)[23]测定.具体理化测试参照黄魁等[24]方法进行.
1.3.2 PMA处理与DNA提取 对采集样品的PMA预处理参考Van Frankenhuyzen等[25]的方法进行,简述如下.将1g新鲜样品加入200mL无菌超纯水和2mL磷酸缓冲盐溶液(0.01mol/L,pH=7.4)中,充分混匀后以300r/min磁力搅拌30min.随后取2mL混匀液,加入5µL PMA(25µmol/L),将其充分均匀后,在4℃下静置10min.随后将溶液在发光二极管光解装置(Takara,EM200,日本)中光解20min.每隔5min将离心管取出摇匀一次,以确保PMA能充分与死菌DNA结合.取上述经PMA处理后的样品用DNeasy®Power Soil®Kit(QIAGEN,德国)试剂盒提取DNA,并用1%琼脂糖凝胶电泳检测其浓度.
1.3.3 荧光定量PCR 采用荧光定量PCR仪(Takara,TP700,日本)对细菌16S rDNA(V3~V4区)、四环素类抗性基因(M)、大环内酯类抗性基因(B、F)、磺胺类抗性基因(1、2)以及移动遗传元件(MGEs)整合酶基因(I1)进行定量.引物序列及PCR扩增条件参照文献[14]的方法进行.所用引物均购置于生工生物工程(上海)股份有限公司.25μL的定量PCR反应体系为:TB Green II(Takara,日本)12.5μL,20μmoL上下游引物各0.5μL, DNA模板1μL,DNA-free超纯水10.5μL.利用TB Green II与双链 DNA结合发出的强烈荧光信号来监测整个扩增过程,扩增效率控制在90%~110%.然后通过Ct值(扩增循环次数)和标准曲线对样品中DNA的起始浓度进行定量检测.其中绘制标准曲线的标准品为携带目的基因的质粒(Takara,pMD20-T,大连),详细制备过程见文献[26].
1.3.4 PCR和高通量测序 采用带有Barcode碱基信息的细菌通用引物341F(5'-CCTACGGGAGGC- AGCAG-3')和806R(5'-GGACTACVSGGGTATCT- AAT-3')对16S rDNA的V3~V4区进行扩增.PCR扩增使用Phusion® High-Fidelity PCR Master Mix with GC Buffer(New England Biolabs)的高效高保真酶进行,其反应条件为:98℃预变性30s;30个循环包括98℃变性15s,58℃退火15s,72℃延伸15s;72℃终延伸1min.所得扩增产物使用2%浓度琼脂糖凝胶进行电泳检测,并利用Thermo Scientific公司GeneJET 胶回收试剂盒对其进行纯化.使用Illumina公司TruSeq DNA PCR-Free Library Preparation Kit建库试剂盒进行文库构建,经过Qubit定量和文库检测合格后,使用NovaSeq 6000平台进行上机测序(诺禾致源生物信息科技有限公司,北京).所得序列使用FLASH(v1.2.7)进行拼接,而后使用QIIME(v1.9.1)进行质控过滤.Tags序列通过(https://github.com/ torognes/vsearch/)与物种注释数据库进行比对检测嵌合体序列,去掉嵌合体,得到有效Clean reads.使用Uparse(v7.0.1001)对序列进行聚类,随后通过MUSCLE 3.8.31与Silva 138数据库(http://www.arb- silva.de/)比对分类,最终得到有效的测序数据.测序结果已上传至NCBI数据库,序列号为SAMN21235626~21235634.
1.4 统计方法
使用Statistica 10.0软件对样品的理化性质、ARGs数量在各组之间的差异进行单因素方差分析(One-way ANOVA)和相关性分析,显著性水平为0.05.ARGs的丰度图和活微生物细菌群落丰度堆积图使用OriginPro 2018(version 9.5)绘制.用HemI 1.0软件绘制热图.用Canoco 4.5软件对环境因子、微生物和ARGs之间的关系进行冗余分析.
2 结果与讨论
2.1 堆肥前后理化性质的变化
电导率和有机质的变化常用来表征蚯蚓堆肥过程中有机物降解与转化的程度.由表1可知,实验结束后,蚯蚓堆肥组的电导率较对照组相比显著增加了0.82倍(<0.05),而有机质下降了1.40%.电导率的增加可能是由于蚯蚓提升了污泥中有机物的矿化作用,释放出矿物盐和无机离子等[27].在蚯蚓与微生物的共同作用下,有机质的降解速率提升.溶解性有机碳(DOC)可以作为判断堆肥腐熟的指标,相关研究[28]表明堆肥产物中DOC含量为4g/kg时即可认为腐熟.相比对照组,蚯蚓组DOC减少了10.42%.这可能是由于蚯蚓的取食、破碎等刺激作用促进了微生物量的增长[29],加速了堆肥基质中有机质的分解,进而加快了蚯蚓对DOC的利用[30],产物蚯蚓粪的稳定化程度较好.
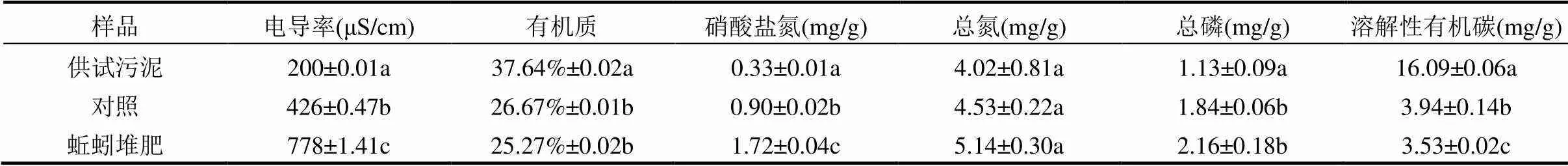
表1 供试污泥及不同处理堆肥产物的理化性质
注:同列指标后字母不同表明其两两之间具有显著性差异(<0.05),同行字母之间没有比较意义,下同.
表1结果显示,堆肥结束后两处理组硝酸盐氮的含量均显著增加(<0.05),且蚯蚓组比对照组显著提升了4.26倍(<0.05),表明蚯蚓能提升堆肥过程中的硝化作用.吴颖等[31]研究发现蚯蚓堆肥可显著增加氨氧化古菌和氨氧化细菌的数量.同时,蚯蚓活动增加了污泥内部的孔隙率,为硝化细菌提供充足的氧气,促进硝化反应的进行,从而提高了有机物的转化速率.对总氮而言,污泥蚯蚓堆肥产物比对照组增加了13.47%,但二者并不显著.这可能是在蚯蚓活动过程中,蚯蚓排泄物及其体壁分泌的黏液所致[32].本文蚯蚓堆肥组总磷相比于对照组增加了17.39%,可能是因为微生物渗出有机酸、磷酸酶的活化导致的有机磷矿化[33].Busato等[34]人认为磷能富集在蚯蚓的粪便中,并向可利用的形态转化.以上结果表明,蚯蚓堆肥可显著提高有机物降解转化的速率,使堆肥产物更加稳定,其产物蚯蚓粪含大量营养元素,具有很大的农用潜力.
2.2 堆肥前后活细菌群落的变化
由表2可知,与对照组相比,蚯蚓堆肥组活细菌群落的Shannon指数和Simpson指数分别增加了1.25%和0.34%.可见,蚯蚓能增加堆肥产物中活细菌的丰富度和均匀度,促进微生物的生长繁殖.此前黄魁等[24]基于总DNA的污泥蚯蚓堆肥实验中,蚯蚓堆肥细菌群落的Shannon指数为7.40,相较于无蚯蚓组减小了2.63%.蚯蚓堆肥活细菌多样性的增加,可能是由于蚯蚓的摄食和掘穴活动增加了体系孔隙率,促进了好氧微生物的生长,同时蚯蚓黏液和蚓粪中富含的多种可生物利用的营养组分[35],也可以刺激某些微生物的生长.

表2 供试污泥与堆肥产物中微生物群落的Shannon和Simpson指数
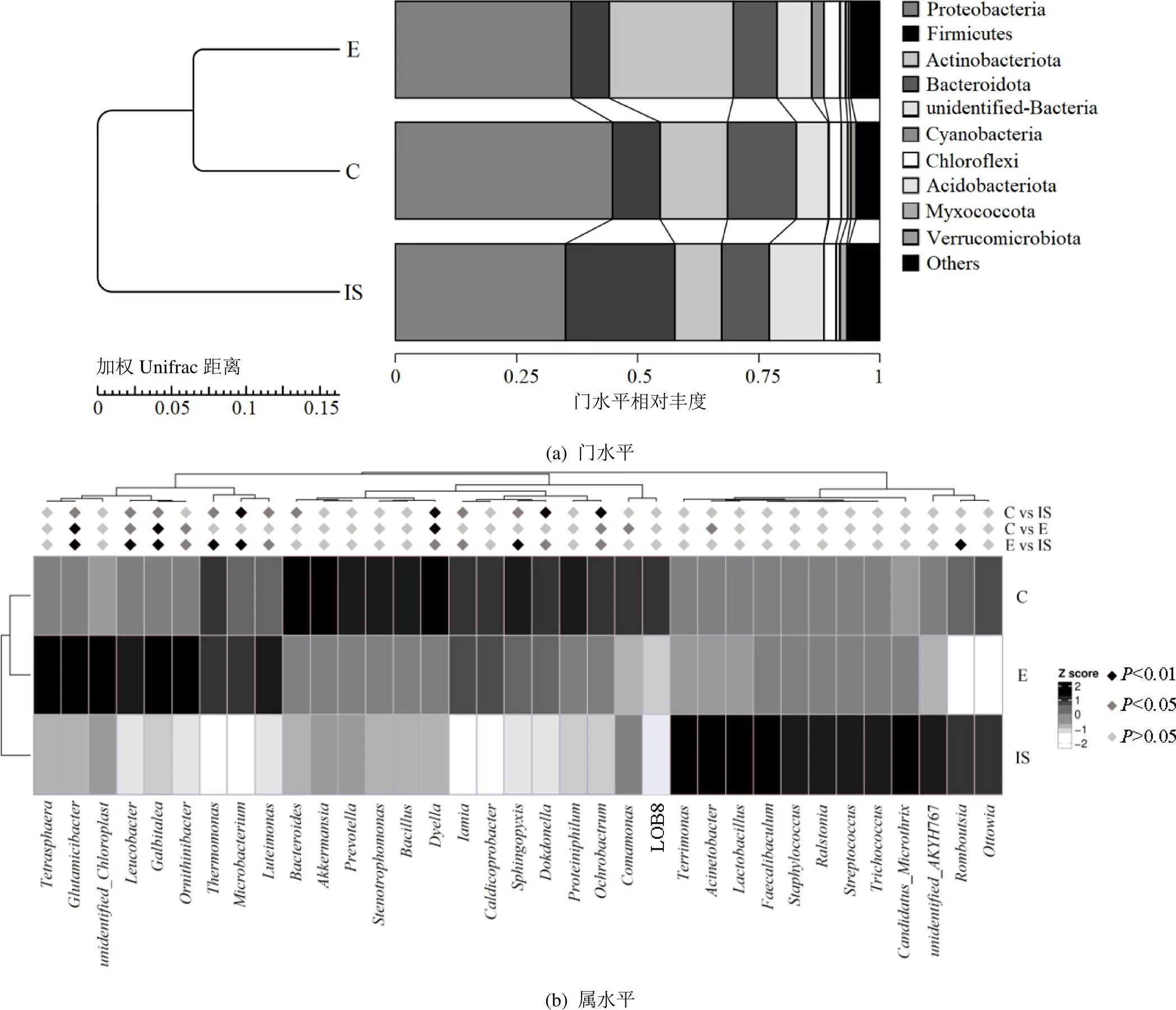
图1 供试污泥与不同处理堆肥产物中活细菌门和属水平相对丰度
由图1(a)可知,变形菌门(32.6%)、厚壁菌门(19.4%)、拟杆菌门(8.6%)与放线菌门(6.2%)是原始污泥活细菌的优势菌门.堆肥结束后,对照产物中活细菌的优势菌门依次为变形菌门(43.7%)、拟杆菌门(14.2%)和放线菌门(12.8%).而蚯蚓堆肥产物活细菌种群中变形菌门(35.2%)占比最大,其次是放线菌门(21.2%)和拟杆菌门(5.6%).上述结果表明,与对照组相比,蚯蚓堆肥后活细菌种群在门水平结构上发生了显著改变,其中放线菌门丰度增加了65.6%,而厚壁菌门和拟杆菌门丰度分别减少了74.6%和34.6%.先前针对总细菌DNA的研究[24]结果显示,接种蚯蚓致使堆肥产物中变形菌门与厚壁菌门减少,而拟杆菌门和放线菌门显著增加(<0.05).放线菌门常被认为是堆肥腐熟的指示菌门[36],蚯蚓堆肥产物中较高的活放线菌丰度表明蚯蚓堆肥可产生更稳定的污泥蚯蚓粪.拟杆菌门能将污泥中有机碳、有机氮化合物转化为相对稳定的产物[13],经过蚯蚓肠道转运会降低拟杆菌门的丰度[37].也可能是本文采用了PMA预处理对活死细菌进行区别,而活细菌中的拟杆菌门丰度较小.
图1(b)为活细菌属水平复杂热图,对照组中活体微生物(6.1%)、(3.8%)、(2.7%)、(1.9%)和(1.8%)等菌属丰度占比较高,而蚯蚓堆肥产品中优势活体菌属为(6.9%)、(4.1%)、(3.8%)、(1.9%)和(1.7%).结果显示,与对照组相比,蚯蚓组中和的丰度分别显著增加了2.6%和1.9%(<0.05).其中贝氏谷氨酸杆菌()属于农业益生菌[38],进一步说明蚯蚓污泥堆肥产物的农用潜力高.放线菌属()占比的增加很可能与堆肥过程中抗生素类物质有 关.
2.3 堆肥前后ARGs和MGEs的丰度变化
两处理组堆肥前后各ARGs绝对丰度的变化如图2所示,对照组和蚯蚓组堆肥后1、F和M的丰度变化呈相近的下降趋势.相较于供试污泥,对照组和蚯蚓组中1、F和M的丰度分别显著降低了46.3%~82.1%、48.3%~86.0%和66.1%~ 92.4%(<0.05).其中蚯蚓堆肥对1、F和M的去除率均显著高于对照组(<0.05),见图2(a、d、e).陈景阳等[15]的研究证实,蚯蚓可以在污泥蚯蚓堆肥过程中去除部分四环素抗性基因.先前的研究结果表明,与对照处理相比,蚯蚓堆肥产物中1和F分别降低了24.6%和69.4%[11].这表明蚯蚓堆肥可以减少污泥活细菌中1、F和M的丰度,ARGs的选择性减少可能归因于蚯蚓的肠道消化过程[26].此外,与原始污泥相比,堆肥后对照组中2的丰度显著增加5.5倍(<0.05). Qian等[39]在对牛粪进行堆肥后也发现其中2丰度增加了21.4~30.8倍.相比而言,蚯蚓堆肥产物中2的丰度比对照组显著降低了82.8%(<0.05),说明蚯蚓能降低污泥中2的丰度,见图2(b).对B来说,其丰度在两处理组中均低于原始污泥.但与对照组相比,蚯蚓堆肥产物中B的丰度显著增加了5.74倍(<0.05).在此前好氧堆肥实验中发现,污泥中B丰度可减少23.9%~99.3%[40].由此推断,在堆肥体内添加蚯蚓能增加B基因的丰度.同时B和F丰度的不同变化趋势可能与其抗性机制不同有关.
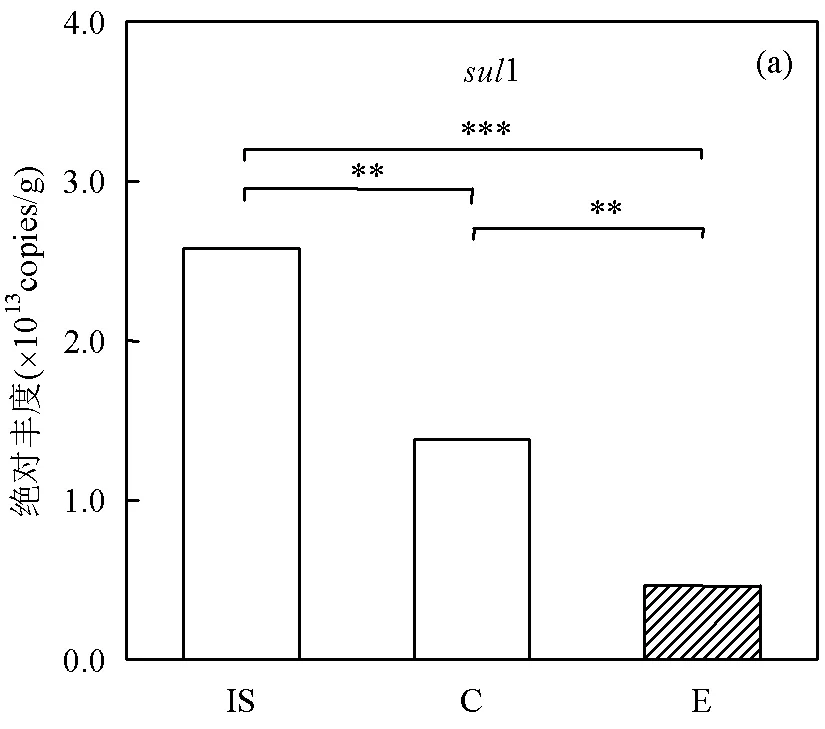
IS代表供试污泥,C代表对照,E代表蚯蚓堆肥,下同
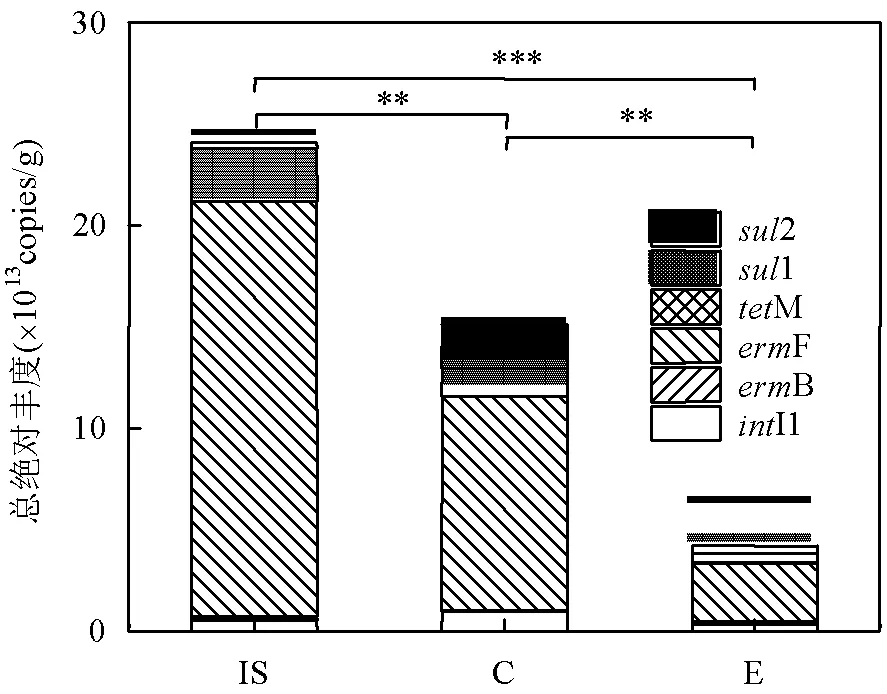
图3 蚯蚓堆肥前后ARGs的总绝对丰度
大量可移动遗传元件(质粒、整合子、转座子)在细菌群落ARGs的水平转移中发挥着重要作用[41],因此本文分析了一类整合子I1在堆肥过程中的丰度变化,见图2(f).与原始污泥相比,I1的绝对丰度在对照组中增加了91.1%,但在蚯蚓组中显著减少了37.3%(<0.05).该结果进一步表明蚯蚓能够降低堆肥产物中ARGs水平转移的风险.Cui等[26]研究新鲜蚯蚓排泄粪便发现,经PMA处理后蚯蚓粪便中I1的绝对丰度较原始污泥降低了82.1%,进而证实蚯蚓肠道短消化对I1的丰度具有显著削减作用.而本实验蚯蚓堆肥产物中I1丰度的减少率低于其在蚯蚓粪便当中,可能是由于经过肠道短消化后,I1的丰度在堆肥产物活微生物体内再次增加.另外,两组中2与I1的丰度变化具有显著正相关性(<0.05),表明2的增多可能与堆肥产物中I1的丰度有关[42].
如图3所示,堆肥结束后对照组与蚯蚓组中代表性ARGs的总丰度均显著降低(<0.05).两处理组对ARGs总去除率分别为37.2%和82.6%.与对照组相比,蚯蚓堆肥后的污泥活体微生物中ARGs的总丰度显著减少了72.3%(<0.05).可见,蚯蚓堆肥可显著降低堆肥产物中ARGs的绝对丰度[43],减轻污泥蚯蚓粪后续农用的潜在生物风险.先前针对总细菌DNA的研究[11]结果显示,蚯蚓堆肥减少了整合子的类型和丰度,堆肥后污泥中的ARGs总丰度比对照组降低了41.5%.Cui等[26]研究发现蚯蚓肠道中的厌氧环境可能会使部分携带ARGs的优势需氧细菌难以生存,蚯蚓可在一定程度上降低堆肥产物中ARGs的绝对丰度.说明蚯蚓活动对活体微生物的群落变化影响更大.同时这种微生物的群落变化和可移动遗传元件的减少可能是蚯蚓堆肥过程中ARGs丰度减少的主要原因.在蚯蚓组样品中绝对丰度占比最高的ARGs为F(2.88×1013copies/g),丰度最低的为M(3.30×108copies/g).此前的研究[44]显示蚯蚓堆肥产物中2丰度最高,其次为X.说明活体微生物中ARGs多样性与之前总DNA中有差别.
2.4 环境因子、微生物和ARGs之间的关系
图4显示活细菌群落及环境因子对堆肥过程中ARGs丰度变化的贡献,其中活细菌群落前10个菌门可以解释驱动ARGs变化的62.1%,表明ARGs的丰度受活细菌群落的影响较大.其中蚯蚓堆肥样品中高含量的EC及NO3-,表明蚯蚓可以促进污泥中有机物的降解与矿化.同时,EC对F丰度的增加有显著的积极影响(<0.05).以上结果表明本文所研究的理化性质会导致ARGs的丰度发生改变,控制这些关键的环境因子有助于去除堆肥体中的ARGs.
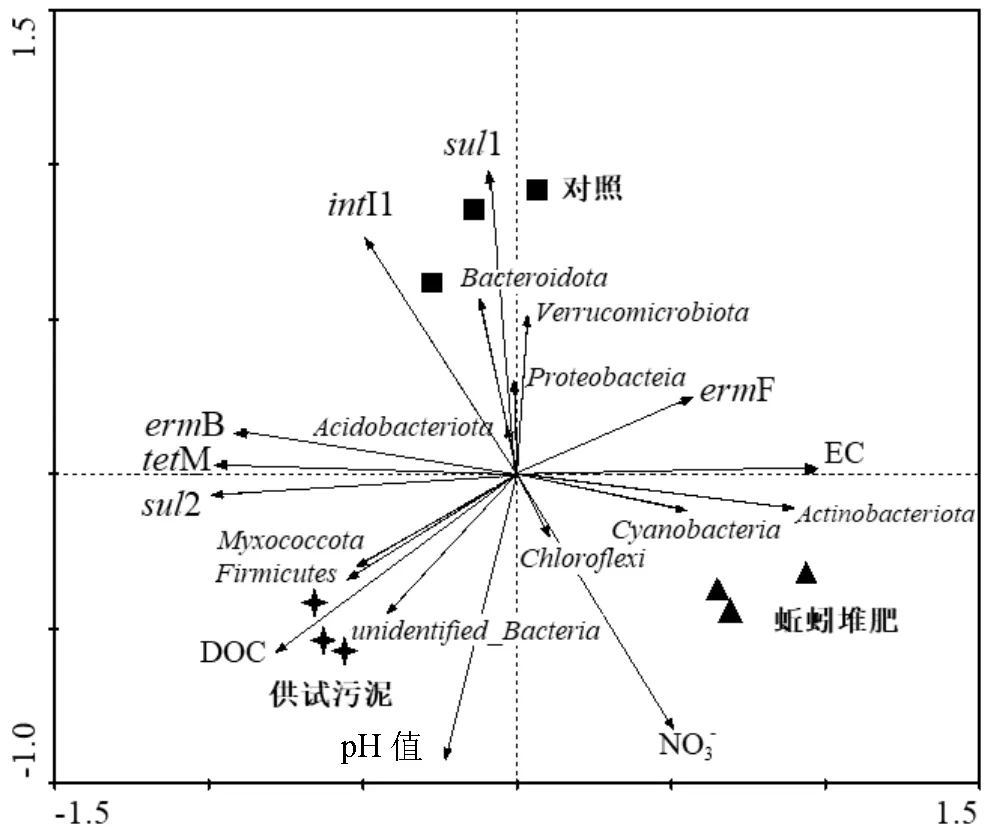
图4 ARGs、活细菌种群和环境因子的冗余分析
先前研究表明放线菌门是抗生素的主要生产者,可以携带和传播ARGs[45].本文蚯蚓堆肥前后Actinobacteria(放线菌门)丰度显著增加了2.8倍,但蚯蚓堆肥产物中ARGs的总绝对丰度却显著降低.造成这一结果的原因可能是放线菌门产生的抗生素可以杀死污泥中的ARGs,使大部分ARGs存在于游离态DNA中.本文研究的是堆肥体系中活体微生物的菌群结构,而此前的研究并未区分活死微生物,导致宿主体内可表达的ARGs定量不准确.此外,I1与Proteobacteria(变形菌门)和Bacteroidetes (拟杆菌门)存在显著正相关性(<0.05),这一结果显示蚯蚓堆肥过程中Proteobacteria和Bacteroidetes可能是I1的潜在携带菌.进一步相关分析表明I1菌属水平的宿主为和,该宿主在堆肥产物中较高的丰度可能会促进ARGs的水平传播[46].以上相关性结果表明,在蚯蚓组中,变形菌门的和是B的潜在宿主,M、F和1的共同潜在宿主属于厚壁菌门,推断它们之间可能存在共生互惠关系.可见,蚯蚓能通过控制堆肥产物中Proteobacteria和Bacteroidetes的丰度来降低参与ARGs水平转移的I1基因的增殖,减小ARGs传播扩散的潜在风险.
冗余分析结果显示pH值、DOC、EC及NO3-等环境因子可以调控ARGs的丰度,且活细菌群落变化对堆肥过程中ARGs的变化有一定的影响[14].因此,环境因子对ARGs丰度的影响主要取决于它们对其潜在宿主细菌种群结构的影响[11-47].本文结果显示,蚯蚓可通过改变污泥中环境因子及活体微生物种群结构,进而对蚯蚓堆肥产物中ARGs的分布和丰度产生影响[48].
3 结论
3.1 污泥蚯蚓堆肥使活体细菌群落的丰富度和均匀度显著增加,污泥蚯蚓粪中变形菌门(35.2%)、放线菌门(21.2%)、拟杆菌门(5.6%)为优势菌门.
3.2 蚯蚓堆肥后的污泥活细菌中总ARGs和I1的绝对丰度分别显著减少了72.3%和37.3% (< 0.05),污泥蚯蚓堆肥能减轻ARGs在环境中的传播风险.
3.3 蚯蚓通过改变堆体环境,影响活细菌群落演替,减少活微生物中ARGs潜在宿主菌的丰度,是削减污泥蚯蚓粪中ARGs的主要原因.
[1] 戴晓虎.我国污泥处理处置现状及发展趋势 [J]. 科学, 2020,72 (6):30-34.
Dai X H. Applications and Perspectives of Sludge Treatment and Disposal in China [J]. Science, 2020,72(6):30-34.
[2] 安 叶,张义斌,黎 攀,等.我国市政生活污泥处置现状及经验总结 [J]. 给水排水, 2021,57(S1):94-98.
An Y, Zhang Y B, Li P, et al. Current situation and experience summary of municipal sewage sludge treatment and disposal in China [J].Water & Wastewater Engineering, 2021,57(S1):94-98.
[3] 戴晓虎,张 辰,章林伟,等.碳中和背景下污泥处理处置与资源化发展方向思考 [J]. 给水排水, 2021,57(3):1-5.
Dai X H, Zhang C, Zhang L W, et al. Thoughts on the development direction of sludge treatment and resource recovery under the background of carbon neutrality [J]. Water & Wastewater Engineering, 2021,57(3):1-5.
[4] 罗 义,周启星.抗生素抗性基因(ARGs)——一种新型环境污染物 [J]. 环境科学学报, 2008,(8):1499-1505.
Luo Y, Zhou Q X. Antibiotic resistance genes(ARGs) as emerging pollutants [J]. Acta Scientiae Circumstantiae, 2008,(8):1499-1505.
[5] 彭兰生,关孟欣,黄 魁,等.蚯蚓摄食污泥对其肠道功能区微生物种群及耐药基因的影响 [J]. 中国环境科学, 2022,42(1):465-473.
Peng L S, Guan M X, Huang K, et al. Effects of excess sludge fed by earthworms on microbial community and antibiotic resistance genes in their intestinal functional area [J]. China Environmental Science, 2022, 42(1):465-473.
[6] 杨凤霞,毛大庆,罗 义,等.环境中抗生素抗性基因的水平传播扩散 [J]. 应用生态学报, 2013,24(10):2993-3002.
Yang F X, Mao D Q, Luo Y, et al. Horizontal transfer of antibiotic resistance genes in the environment [J]. Chinese Journal of Applied Ecology, 2013,24(10):2993-3002.
[7] 薛重华,孔祥娟,王 胜,等.我国城镇污泥处理处置产业化现状、发展及激励政策需求 [J]. 净水技术, 2018,37(12):33-39.
Xue C H, Kong X J, Wang S, et al. Industrialization status, development analysis and incentive policy demands of municipal sludge treatment and disposal industry in china [J]. Water Purification Technology, 2018,37(12):41-47.
[8] 王亚利,杨 光,熊才耘,等.蔬菜废弃物蚯蚓堆肥对鸡毛菜生长的影响 [J]. 农业环境科学学报, 2017,36(10):2129-2135.
Wang Y L, Yang G, Xiong C Y, et al.Effect of vegetable waste vermicompost on the growth of Brassica chinensis [J]. Journal of Agro-Environment Science, 2017,36(10):2129-2135.
[9] Hait S, Tare V. Optimizing vermistabilization of waste activated sludge using vermicompost as bulking material [J]. Waste Management, 2011,31(3):502-511.
[10] Li Z H, Yuan L, Shao W, et al. Evaluating the interaction of soil microorganisms and gut of soil fauna on the fate and spread of antibiotic resistance genes in digested sludge-amended soil ecosystem [J]. Journal of Hazardous Materials, 2021,420:126672.
[11] Huang K, Xia H, Zhang Y, et al. Elimination of antibiotic resistance genes and human pathogenic bacteria by earthworms during vermicomposting of dewatered sludge by metagenomic analysis [J]. Bioresource Technology, 2020,297:122451.
[12] 夏 慧,陈学民,黄 魁.宏基因组学揭示蚯蚓对污泥中抗生素抗性基因的影响 [J]. 兰州交通大学学报, 2019,38(3):80-84.
Xia H, Chen X M, Huang K. Effects of earthworms on antibiotic resistance genes during vermicomposting of dewatered sludge by metagenomic analysis [J]. Journal of Lanzhou Jiaotong University, 2019,38(3):80-84.
[13] Huang K, Xia H, Wu Y, et al. Effects of earthworms on the fate of tetracycline and fluoroquinolone resistance genes of sewage sludge during vermicomposting [J]. Bioresource technology, 2018,259:32-39.
[14] 关孟欣,彭兰生,陈景阳,等.玉米芯生物炭对污泥蚯蚓粪中微生物种群及ARGs的影响 [J]. 中国环境科学, 2021,41(6):2744-2751.
Guan M X, Peng L S, Chen J Y, et al. Effects of corncob biochar on the fate of microbial communities and antibiotics resistance genes during vermicomposting of dewatered sludge [J]. China Environmental Science, 2021,41(6):2744-2751.
[15] 陈景阳,夏 慧,黄 魁,等.四环素对污泥蚯蚓粪中微生物种群和抗性基因的影响 [J]. 环境科学, 2019,40(7):3263-3269.
Chen J Y, Xia H, Huang K, et al. Effects of tetracycline on microbial communities and antibiotic resistance genes of vermicompost from dewatered sludge [J]. Environmental Science, 2019,40(7):3263-3269.
[16] Miller J H, Novak J T, Knocke W R, et al. Survival of antibiotic resistant bacteria and horizontal gene transfer control antibiotic resistance gene content in anaerobic digesters [J]. Frontiers in Microbiology, 2016,7:263.
[17] Nocker A, Cheung C Y, Camper A K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells [J]. Journal of Microbiological Methods, 2006,67(2):310-320.
[18] 陶怡君,谌志筠,何秋水.叠氮溴化丙锭结合qPCR检测与区分活菌和死菌的研究进展 [J]. 微生物学免疫学进展, 2020,48(6):63-68.
Tao Y J, Zhan Z Y, Hei Q S. Advances in detection and differentiation of live and dead bacteria by PMA- qPCR technology [J]. Progress in Microbiology and Immunology, 2020,48(6):63-68.
[19] HJ 761-2015 固体废物有机质的测定灼烧减量法 [S].
HJ 761-2015 Solid waste-Determination of organic matter-Ignition loss method [S].
[20] HJ/T 346-2007 水质硝酸盐氮的测定紫外分光光度法(试行) [S].
HJ/T 346-2007 Water quality-Determination of nitrate-nitrogen- Ultraviolet spectrophotometry [S].
[21] HJ 535-2009 水质氨氮的测定纳式试剂分光光度法 [S].
HJ 535-2009 Water quality-Determination of ammonia nitrogen Nessler’s reagent spectrophotometry [S].
[22] HJ 636-2012 水质总氮的测定碱性过硫酸钾消解紫外分光光度法 [S].
HJ 636-2012 Water quality-Determination of total nitrogen- Alkaline potassium persulfate digestion UV spectrophotometric method [S].
[23] GB 11893-89 水质总磷的测定钼酸铵分光光度法 [S].
GB 11893-89 Water quality-Determination of total phosphorus- Ammonium molybdate spectrophotometric method [S].
[24] 黄 魁,夏 慧,陈景阳,等.蚯蚓对城市污泥蚯蚓堆肥过程中微生物特征变化的影响 [J]. 环境科学学报, 2018,38(8):3146-3152.
Huang K, Xia H, ChenJ Y, et al. Effects of earthworms on changes of microbial feature during vermicomposting of municipal sludge [J]. Acta Scientiae Circumstantiae, 2018,38(8):3146-3152.
[25] Van Frankenhuyzen J K, Trevors J T, Lee H, et al. Molecular pathogen detection in biosolids with a focus on quantitative PCR using propidium monoazide for viable cell enumeration [J]. Journal of Microbiological Methods, 2011,87(3):263-272.
[26] Cui G, Bhat S A, Li W, et al. Gut digestion of earthworms significantly attenuates cell-free and-associated antibiotic resistance genes in excess activated sludge by affecting bacterial profiles [J]. Science of the Total Environment, 2019,691:644-653.
[27] Hait S, Tare V. Optimizing vermistabilization of waste activated sludge using vermicompost as bulking material [J]. Waste Management, 2011,31(3):502-511.
[28] Khan N, Clark I, Sánchez-Monedero M A, et al. Maturity indices in co-composting of chicken manure and sawdust with biochar [J]. Bioresource Technology, 2014,168:245-251.
[29] Aira M, Monroy F, Domínguez J. Earthworms strongly modify microbial biomass and activity triggering enzymatic activities during vermicomposting independently of the application rates of pig slurry [J]. Science of the total Environment, 2007,385(1-3):252-261.
[30] Nigussie A, Bruun S, de Neergaard A, et al. Earthworms change the quantity and composition of dissolved organic carbon and reduce greenhouse gas emissions during composting [J]. Waste Management, 2017,62:43-51.
[31] 吴 颖,黄 魁,夏 慧,等.污泥四环素含量对蚯蚓堆肥中氨氧化菌群的影响 [J]. 环境科学, 2019,40(6):2954-2960.
Wu Y, Huang K, Xia H, et al. Effects of different concentrations of tetracycline in sludge on ammonia oxidizers during vermicomposting [J]. Environmental Science, 2019,40(6):2954-2960.
[32] Tripathi G, Bhardwaj P. Decomposition of kitchen waste amended with cow manure using an epigeic species () and an anecic species () [J]. Bioresource Technology, 2004, 92(2):215-218.
[33] Gaume A, Mächler F, Frossard E. Aluminum resistance in two cultivars of Zea mays L.:root exudation of organic acids and influence of phosphorus nutrition [J]. Plant and Soil, 2001,234(1):73-81.
[34] Busato J G, Lima L S, Aguiar N O, et al. Changes in labile phosphorus forms during maturation of vermicompost enriched with phosphorus-solubilizing and diazotrophic bacteria [J]. Bioresource Technology, 2012,110:390-395.
[35] Zhao L, Wang Y, Yang J, et al. Earthworm–microorganism interactions: a strategy to stabilize domestic wastewater sludge [J]. Water Research, 2010,44(8):2572-2582.
[36] Gerzova L, Babak V, Sedlar K, et al. Characterization of antibiotic resistance gene abundance and microbiota composition in feces of organic and conventional pigs from four EU countries [J]. PLoS One, 2015,10(7):e0132892.
[37] Wang N, Wang W, Jiang Y, et al. Variations in bacterial taxonomic profiles and potential functions in response to the gut transit of earthworms () feeding on cow manure [J]. Science of the Total Environment, 2021,787:147392.
[38] Ji J, Yuan D, Jin C, et al. Enhancement of growth and salt tolerance of rice seedlings (Oryza sativa L.) by regulating ethylene production with a novel halotolerant PGPR strain Glutamicibacter sp. YD01containing ACC deaminase activity [J]. Acta Physiologiae Plantarum, 2020, 42(4):1-17.
[39] Qian X, Sun W, Gu J, et al. Variable effects of oxytetracycline on antibiotic resistance gene abundance and the bacterial community during aerobic composting of cow manure [J]. Journal of Hazardous materials, 2016,315:61-69.
[40] Wei H, Ma J, Su Y, et al. Effect of nutritional energy regulation on the fate of antibiotic resistance genes during composting of sewage sludge [J]. Bioresource Technology, 2020,297:122513.
[41] 安新丽,苏建强.活性污泥抗生素抗性基因研究进展 [J]. 微生物学通报, 2019,46(8):2069-2079.
An X L, Su J Q. Resistome in activated sludge: current knowledge and future directions [J]. Microbiology China, 2019,46(8):2069-2079.
[42] Duan M, Li H, Gu J, et al. Effects of biochar on reducing the abundance of oxytetracycline, antibiotic resistance genes, and human pathogenic bacteria in soil and lettuce [J]. Environmental Pollution, 2017,224:787-795.
[43] Gómez-Brandón M, Aira M, Lores M, et al. Epigeic earthworms exert a bottleneck effect on microbial communities through gut associated processes [J]. PloS One, 2011,6(9):e24786.
[44] 李建辉,张莹莹,黄 魁,等.宏基因组学解析蚯蚓粪中微生物种群及耐药基因的组成 [J]. 中国环境科学, 2020,40(12):5375-5382.
Li J H, Zhang Y Y, Huang K, et al. Composition of microbial community and antibiotic resistance genes in vermicomposts revealed by metagenomic analysis [J]. China Environmental Science, 2020, 40(12):5375-5382.
[45] Zhang J, Lu T, Shen P, et al. The role of substrate types and substrate microbial community on the fate of antibiotic resistance genes during anaerobic digestion [J]. Chemosphere, 2019,229:461-470.
[46] Shen Q, Tang J, Wang X, et al. Fate of antibiotic resistance genes and metal resistance genes during the thermophilic fermentation of solid and liquid swine manures in an ectopic fermentation system [J]. Ecotoxicology and Environmental Safety, 2021,213:111981.
[47] Zhang R, Gu J, Wang X, et al. Contributions of the microbial community and environmental variables to antibiotic resistance genes during co-composting with swine manure and cotton stalks [J]. Journal of Hazardous Materials, 2018,358:82-91.
[48] Sun W, Qian X, Gu J, et al. Mechanisms and effects of arsanilic acid on antibiotic resistance genes and microbial communities during pig manure digestion [J]. Bioresource Technology, 2017,234:217-223
Effects of earthworms on the antibiotic resistance genes of vermicompost from dewatered sludge revealed by active microbes.
WEI Feng-yi1, XU Jun-jie1, CHEN Jin1, LI Jian-hui1, HUANG Kui1,2*, DONG Xi-lin3, XIA Hui1
(1.School of Environmental and Municipal Engineering, Lanzhou Jiaotong University, Lanzhou 730070, China;2.Key Laboratory of Yellow River Water Environment in Gansu Province, Lanzhou 730070, China;3.Changchun Water Group Co. Ltd, Changchun 130000, China)., 2022,42(7):3425~3433
To eliminate the abundances of ARGs in sludge vermicompost, this study aimed to reveal the underlying effects of earthworms on the active bacterial community structure and their ARGs involved in vermicomposting systems for sludge recycling. For this, vermicomposting with and without earthworms was set up in parallel. Moreover, the dyeing pretreatment for samples with propidium monoazide (PMA) was adopted to screen out the DNA of active bacteria. The results showed that the electrical conductivity of sludge vermicompost was significantly increased by 82.5% (<0.05), the degradation rate of organic matter was increased by 5.2% (<0.05). Compared with the control treatment, the abundance of Actinobacteria significantly increased by 65.6% (<0.05), while the abundance of Firmicutes and Bacteroidetes significantly decreased by 74.7% and 34.6%, respectively. Meanwhile, vermicomposting led to the selected ARGs abundances ofM,1,2,B andF significantly decreased by 66.5%, 82.8%, 72.8% and 77.6% (<0.05), while the abundance ofB significantly increased by 5.7times (<0.05) in active bacteria, compared to the counterpart. The abundance ofI1gene in vermicompost products significantly reduced by 67.2% compared with the control treatment. The total absolute abundance of ARGs was 4.19×1013copies/g, and the total removal rate of ARGs was 82.6%, 45.4% higher than that of the counterpart. This study suggests that earthworms can reduce the abundance of dominant hosts of ARGs by modifying the active microbial community structure of sludge, thus reducing the associated dissemination risks of the spread of ARGs.
composting;microbial community;resistance genes;vermicompost;sludge recycling
X171.5
A
1000-6923(2022)07-3425-09
魏枫沂(1997-),女,甘肃白银人,兰州交通大学硕士研究生, 主要研究方向为生物污染物归趋与控制.发表论文1篇.
2021-12-06
国家自然科学基金资助项目(51868036,52000095);兰州交通大学百人计划;甘肃省青年博士基金资助项目(2021-QB051);甘肃省科技计划(20JR2RA002);甘肃省优秀研究生“创新之星”项目(2021CXZX-629)
* 责任作者, 教授, huangkui@mail.lzjtu.cn
