Comparative Proteomic Analysis of Milk Fat Globule Membrane Proteins between Donkey Colostrum and Mature Milk
2022-05-09LIMohanZHANGXiuminSONGWanyingYUHaikunZHANGJuanYUEXiqingZHENGYan
LI Mohan, ZHANG Xiumin, SONG Wanying, YU Haikun, ZHANG Juan, YUE Xiqing,*, ZHENG Yan,*
(1.College of Food Science, Shenyang Agricultural University, Shenyang 110866, China; 2.Beijing Academy of Food Sciences,Beijing 100068, China; 3.College of Food Engineering, Harbin University of Commerce, Harbin 150076, China)
Abstract: To elucidate the differential milk fat globule membrane (MFGM) proteins between donkey colostrum (DC) and mature milk (DM), a comparative analysis was performed using proteomics.A total of 216 and 215 MFGM proteins were characterized in DC and DM, respectively.Among them, 15 differentially expressed and 25 specifically expressed MFGM proteins were identified.Bioinformatics analysis showed that the significantly differentially expressed MFGM proteins were mainly involved in cellular components including extracellular exosome, extracellular vesicle, and extracellular organelle compartments, and participated in biological processes such as external stimuli, cell proliferation, and blood vessel morphogenesis, and molecular functions such as metal ion binding, cation binding, and calcium ion binding.Additionally,these significantly differentially expressed MFGM proteins were mainly involved in metabolic pathways such as the complement and coagulation cascades and intestinal immune network for IgA production.Furthermore, some key protein factors with high connectivity, as determined by protein network interaction analysis, were identified as differently expressed MFGM proteins.This study provides a better understanding of the biological properties of donkey MFGM proteins and paves the way for future research of MFGM protein nutrition and for the development of formula milk powder.
Keywords: donkey colostrum; mature milk; milk fat globule membrane protein; proteomics
Bovine milk is one of the eight main allergic foods recognized by the Food and Agriculture Organization and World Health Organization, representing the earliest food allergen for infants[1].Recent data show that 2.69% of infants and young children in China are allergic to bovine milk protein[2].There are several types of bovine milk proteins,among which at least 25 proteins hold allergenic potential[3].Casein,β-lactoglobulin, andα-lactalbumin are considered the most important allergens in bovine milk, with casein representing approximately 80% of the total bovine milk protein[4].About 65% of patients with bovine milk allergy are allergic to casein[5], which is often characterized by eczema,diarrhea, and gastrointestinal bleeding[6-8].
Donkey milk has been suggested to be an ideal substitute for bovine milk because of its similarity in protein and lactose content with human milk[9-10].Moreover, donkey milk contains more whey proteins (about 35%–50%) than bovine milk (about 20%)[11], indicating its suitability as a potential substitute for bovine milk in infants with allergy to bovine milk protein[12-13].Donkey milk protein also has certain advantageous physiological functions, such as high tolerability[14],antimicrobial activity[15], and anticancer activity[16-17].
Compared with the characterization of bovine milk protein, that of donkey milk protein is at a relatively early stage.Chianese et al.[18]investigated donkey casein using a proteomic approach, based on one- and two-dimensional electrophoresis, stained with either Coomassie Brilliant Blue or specific polyclonal antibodies, and structural mass spectrometry (MS) analysis, and finally observed eleven components ofκ-casein, six phosphorylated components forβ- andαS1-casein, and three main phosphorylated components forαS2-casein.Zhang Xinhao et al.[19]characterized donkey whey proteins of different milk yields of Dezhou donkey using a label-free based comparative proteomics approach,and identified 216 whey proteins, 19 of which showed significant differences in high-milk-yield samples.Li Weixuan et al.[20]identified and quantified 288 and 287 donkey whey proteins in donkey colostrum (DC) and mature milk (DM), respectively.Li Weixuan et al.[21]characterized a total of 947 milk fat globule membrane (MFGM) proteins in DC and DM, including 902 and 913 MFGM proteins in DC and DM, respectively.Despite the revelations of these previous studies, research on the characterization of donkey whey and MFGM proteins is still very limited.
The formation of MFGM is closely related to the milk fat secretion process in breast milk.During the release of milk fat globules from breast cells to the milk, they are wrapped by three layers of phospholipids as they pass through the cell membrane.This triple phospholipid layer is collectively called the MFGM[22-23].This special secretion process of MFGM can collect and transfer membrane-bound proteins, carbohydrates, and lipids from the mother cell surface into the breast milk[19].MFGM is a heterogeneous membrane that can be used as an emulsifier to prevent milk fat globule fusion and degradation[24].MFGM protein accounts for approximately 5% of the total milk protein, but MFGM protein contains a large number of proteins with various functions[22,25].Previously, major MFGM proteins,including mucin-1, fatty acid synthase, xanthine oxidase,butyrophilin, and lactadherin, were also identified via sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis[26].However, a comparative analysis of differential MFGM proteins in DC and DM is still very limited.
Thus, the aim of this study was to characterize and compare MFGM proteins from DC and DM using labelfree high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS)-based proteomics, and investigate their interactions and related metabolic pathways.The collected data will contribute to a deeper understanding of the MFGM protein composition and provide a theoretical basis for the optimization of dairy product formulations based on donkey milk.
1 Materials and Methods
1.1 Materials
The donkey milk samples used in this study were the same as those described in previous studies[27-28].Briefly, DC(0–5 days postpartum) and DM (1–6 months postpartum)samples were collected from 45 Dezhou donkeys (Equus asinus) in Dalian, China, using milk sample devices (Waikato Milking Systems, Horotiu, New Zealand).The donkeys were aged 2-3 years at the first delivery time, weighed 250-350 kg,ate a constant amount of feed (mainly alfalfa), and lived in the same environment during the study duration.Each category of samples was randomly divided into three groups(each group contained 15 milk samples), and each group was pooled before analysis.The experiments were repeated three times to exclude individual differences.All samples were stored at -80 ℃ until analysis.
1.2 Instruments and equipment
ultrafiltration (Millipore Sigma, Burlington, MA, USA),EASY-nLC 1000 HPLC system (Thermo Fisher Scientific,Waltham, MA, USA); Q-Exactive mass spectrometer(Thermo Fisher Scientific); the upper C18column (Thermo Scientific Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18), the analytical column (Thermo Scientific EASY column,10 cm × 75 μm, 3 μm, C18-A2).
1.3 Methods
1.3.1 Preparation of MFGM proteins
The extraction of MFGM proteins from DC and DM was performed as described previously[21].Briefly, 4 mL of milk samples were centrifuged to separate the milk fat (10 000 ×g,15 min, 4 ℃).The upper layer containing the MFGM was collected and washed by adding 10 mL of phosphate-buffered saline solution.The sample was dispersed with ultrasound at 80 W for 15 s, repeated three times.The mixture was then centrifuged (10 000 ×g, 60 min, 4 ℃) and the supernatant was collected.Acetone was added to the supernatant and the proteins were precipitated for 12 h.The mixture was centrifuged (15 000 ×g, 30 min, 4 ℃) and the supernatant was discarded.Lysis solution was added to the pelleted protein sample and the mixture was dispersed with ultrasound at 80 W for 15 s, repeated 10 times.
1.3.2 Trypsin digestion of MFGM proteins
MFGM proteins (600 μg) were added to a mixture solution consisting of 4% SDS, 100 mmol/L Tris-HCl (pH 8.0), and 100 mmol/L dithiothreitol.Then, the mixture was heated at 90 ℃ for 10 min.The samples were placed statically and cooled to room temperature.The sample was then loaded onto a 10 kDa ultrafiltration containing 200 μL UT buffer (8 mol/L urea and 150 mmol/L Tris-HCl; pH 8.0).The sample mixture was centrifuged (14 000 ×g, 30 min,4 ℃) and washed with UT buffer.Iodoacetamide (50 mmol/L,final concentration in UT buffer) was added to the ultrafilter and mixed with the sample for 1 min.Subsequently, the sample mixture was transferred to a dark environment and incubated at room temperature for 30 min.Incubated mixture samples were centrifuged at 14 000 ×gfor 15 min at 4 ℃and washed with 100 μL UT buffer.This step was repeated twice.The filter was then pre-washed by adding dissolution buffer (UT buffer and 25 mmol/L NH4HCO3).The proteins were digested at 37 ℃ for 24 h by adding 40 μL of trypsin to the filter.Digested samples were centrifuged at 14 000 ×gfor 15 min at 4 ℃, and the peptide segments were collected for analysis.The absorbance of the peptides was then measured at 280 nm using a UV spectrophotometer.
1.3.3 HPLC-MS/MS analysis
Sample peptide fragments were separated using the EASY-nLC 1000 HPLC system and the proteins were identified with a Q-Exactive mass spectrometer.The separation system consisted of the mobile phase A of 0.1%formic acid and 99.9% water, and the mobile phase B of 84% acetonitrile, 0.1% formic acid, and 15.9% water.The chromatography column was equilibrated for 30 min using 95% mobile phase A.The mixture of peptide segments was loaded into the upper column (nanoViper C18) using an automatic sampler.Subsequently, the samples were separated using an analytical column (C18-A2) at 300 nL/min.The mobile phase B was used to separate the mixture of peptide segments, with a separation time of 120 min.When the separation time was 0 to 110 min, the linear gradient of mobile phase B ranged from 0% to 55%.The gradient of mobile phase B rose to 100% when the separation time was extended to 115 min.Thereafter, the liquid-phase gradient of the mobile phase B remained at 100% until the separation time reached 120 min.MS analysis was performed in the positive ion mode.The analysis time was set to 120 min,and the scanning range of parent ions wasm/z300–1 800.The first-order MS resolution of Q-Exactive was set to 70 000 full width at half maximum (FWHM) atm/z200,and 1 × 106was set to the automatic gain control target.The maximum isolation time (IT) was 50 ms, and the dynamic exclusion operation was conducted at 60 s.MS/MS data were obtained from the first 20 fragment maps withcharge ≥ 2 collected from each full scan.The MS2activation type was set to higher-energy collisional dissociation, and an isolation window with a mass/charge ratio ofm/z2 was selected.Second-order MS was conducted at a spectrum of 17 500 FWHM andm/z200.The normalized collision energy was 30 eV, and the underfill ratio was 0.1%.
1.3.4 Analysis of MS raw data and protein identification
MaxQuant software[29](version 1.5.3.17) was used to analyze the raw MS data and identify the proteins.Trypsin was used as a lyase and two maximum missed cleavages were allowed for database search.Peptide mass tolerance and MS/MS tolerance were set at 2.0 × 10-5and 0.1 Da, respectively.Formamide methyl (C) was used for fixed modifications, and oxidation (M) was used for variable modifications.Reversed versions of the target database were used as the database pattern to calculate the false discovery rate (FDR).Screening criteria for proteins and peptides identified with > 99%confidence at FDR ≤ 1%.
1.3.5 Bioinformatics analysis
Gene Ontology (GO, http://geneontology.org/)annotation[30], Kyoto Encyclopedia of Genes and Genomes(KEGG, https://www.genome.jp/kegg/) pathway analysis[31],and the online Database for Annotation, Visualization and Integrated Discovery[32](DAVID, http://david.abcc.ncifcrf.gov/home.jsp) were used to analyze the functional categories and related metabolic pathways of the identified MFGM proteins in DC and DM.The differently expressed proteins were also analyzed using the STRING software (https://string-db.org) to obtain a protein-protein interaction (PPI)network map.
2 Results and Analysis
2.1 Characterization of MFGM proteins in DC and DM
In total, 216 and 215 MFGM proteins were characterized in DC and DM, respectively, with 203 MFGM proteins(89.0%) being present in both groups (Fig.1A and 1B).Thirteen MFGM proteins (5.7%) were found only in DC(F7AAA8, K9K2M0, Q3ZCH5, Q8SPP7, A0A0A1E6K2,F1MPP2, E1BGJ0, F1MW68, Q9MXD5, F7B847, F7DGX8,F7E0P3, and H9GZN7), while 12 MFGM proteins (5.3%)were unique to DM (F6PZ29, K9K4A2, H9GZZ3, G3MYD7,F1N2N5, G3X807, F6VWX6, O97567, F6QIV8, F6XP08,F7D7M7, and Q9N2C4).
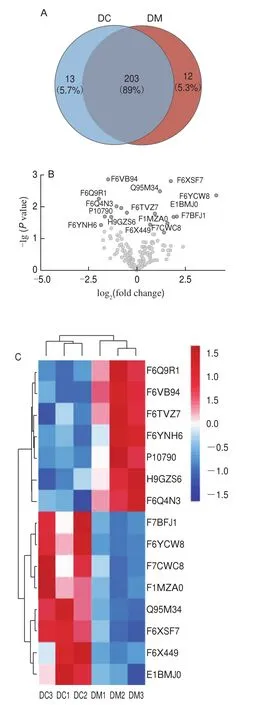
Fig.1 Significantly differentially expressed MFGM proteins between DC and DM
2.2 Identification of significantly different MFGM proteins between DC and DM
The differences found in MFGM protein content in donkey milk during different lactation periods laid thefoundation for screening for altered MFGM proteins.Thus, a threshold was set to screen for differently expressed MFGM proteins between DC and DM.In this study, aP-value <0.05 and a fold change > 1.50 or < 0.667 were indicative of differentially expressed proteins.As shown in Fig.1C, 15 differentially expressed MFGM proteins (eight upregulated and seven downregulated) were identified.In total, 40 significantly different MFGM proteins were selected for subsequent analysis, including 15 differentially expressed and 25 specifically expressed proteins (Table 1).

Table 1 Significantly differentially expressed MFGM proteins between DC and DM
2.3 GO analysis of significantly different MFGM proteins between DC and DM
The 40 significantly different MFGM proteins were classified into three sub-components (Fig.2), including biological processes, cellular components, and molecular functions by GO annotation.The cellular component subgroup, which contained the most significantly different MFGM proteins, included extracellular exosome,extracellular vesicle, extracellular organelle, membranebounded vesicle, extracellular region part, extracellular region, and extracellular space.The most common biological processes were response to external stimuli, cell proliferation,blood vessel morphogenesis, blood vessel development,vasculature development, positive regulation of cell proliferation, regulation of response to external stimulus, and positive regulation of protein phosphorylation.Furthermore,regarding molecular function the identified proteins were mainly involved in metal ion binding, cation binding, calcium ion binding, peptide binding, and amide binding.
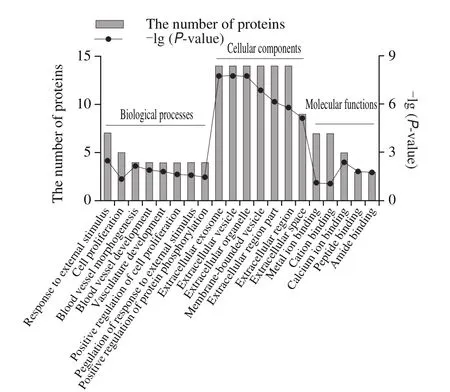
Fig.2 GO annotation of the 40 significantly differential MFGM proteins between DC and DM
2.4 Metabolic pathway analysis of significantly different MFGM proteins between DC and DM
To investigate whether the metabolism of MFGM proteins in donkey milk was affected by the lactation stage,metabolic pathway analysis was performed based on the 40 identified significantly different MFGM proteins.The pathway effects andP-values were calculated from the pathway topology analysis.The first five major metabolic pathways (Fig.3) included the complement and coagulationcascades, which were the most relevant pathways, followed by the intestinal immune network for IgA production,Staphylococcus aureusinfection, glutathione metabolism, and antigen processing and presentation.
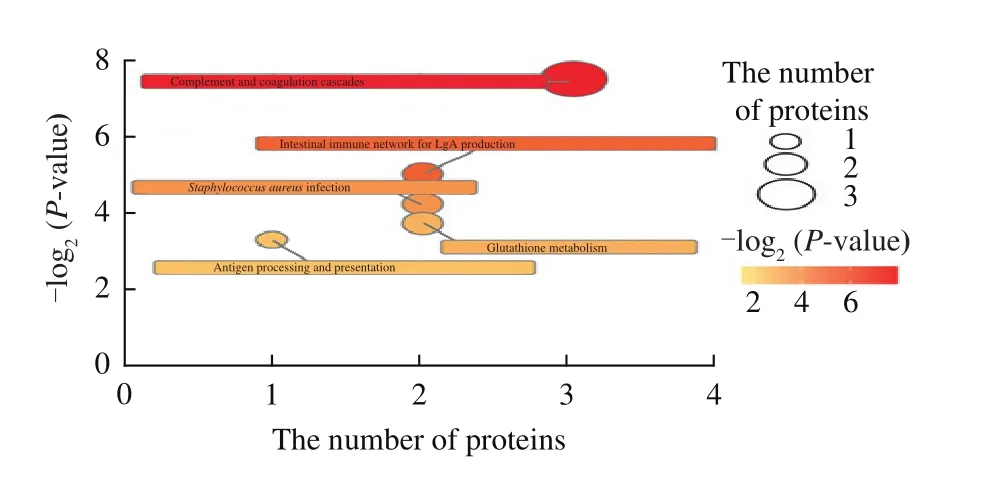
Fig.3 Metabolomic pathways of the 40 significantly differential MFGM proteins between DC and DM
2.5 PPI network of the significantly different MFGM proteins in DC and DM
The PPI network of the 40 significantly different MFGM proteins was found to include 17 MFGM proteins and 18 interactions (Fig.4).Apolipoprotein B (APOB; F6YCW8),which interacts with seven other proteins, was found to represent a node with the most interactions, followed by the fatty acid-binding protein (P10790) with five interactions, and prothrombin (F7BFJ1) and cathepsin Z (F1MW68) with three interactions each.
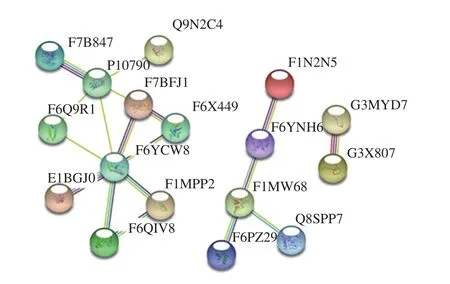
Fig.4 Protein-protein interaction network of the 40 significantly differential MFGM proteins between DC and DM
3 Discussion
A variety of studies have shown that the lactation period has a great impact on the composition of milk,including lipids[33-34], proteins[35-37], and amino acids[38-39].In this study, the alterations of MFGM proteins in DC and DM were analyzed, revealing eight upregulated and seven downregulated proteins in DC compared with DM.Among them, APOB was the protein with the highest (17.15) fold change value.APOB is the primary apolipoprotein of chylomicrons, very-low-density, intermediate-density, and low-density lipoprotein particles, which are responsible for carrying fat molecules, including cholesterol, throughout the body[40].Studies showed that APOB is the primary organizing protein component of lipoprotein particles and is absolutely required for the formation of these particles[41].Furthermore, APOB on low-density lipoprotein particles acts as a ligand for low-density lipoprotein receptors in various cells in the body[42].In our previous studies, APOB was also identified in human and bovine MFGM proteins,and was also found to be more concentrated in the colostrum compared with mature milk among different species[43-44],revealing its important role in the neonatal stage of mammals.Additionally, some fragments of immunoglobulins were found to be highly expressed or specifically expressed in DC.The concentration of immunoglobulin gamma 1 heavy chain constant region (Q95M34) in DC was 2.28-fold higher than that in DM, and immunoglobulin lambda light chain variable region (A0A0A1E6K2) was specifically expressed in DC.Consistent with these results, Turini et al.also found that the concentration of IgG in DC was the highest just after delivery, which then gradually decreased by 0.024 4(lg (mg/mL)) per hour[45].These results provide a basis for the study of functional components in DC.In addition, 25 specifically expressed proteins were identified, including 13 DC-specific and 12 DM-specific proteins.Peptidoglycan recognition protein 1 (Q8SPP7) is one of the proteins specifically expressed in DC.In mammals, peptidoglycan recognition proteins either directly kill bacteria by interacting with their cell wall or outer membrane, or by hydrolyzing peptidoglycans[46-47].They also modulate inflammation and the microbiome, and interact with host receptors[48-49].These proteins specifically expressed in DC may provide certain physiological functions for mammalian newborns, and their activities and mechanisms need to be further studied.
The functions that these significantly different MFGM proteins may perform and the biological processes they may participate in were also analyzed using GO annotation.It is worth noting that most identified MFGM proteins were found to share the cellular component.Of these, the extracellular vesicle, extracellular exosome, and extracellular organelle were the cellular components in which most MFGM proteinsmost significantly participated.Extracellular vesicles are lipid bilayer-delimited particles that are naturally released from a cell, and carry proteins, nucleic acids, lipids, metabolites, and organelles as cargo from the parent cell[50-51].Studies have shown that milk contains heterogeneous cell populations,including milk-secretory cells, myoepithelial cells, and a hierarchy of progenitor and stem cells[52].Therefore, these significantly different MFGM proteins are likely to be secreted by breast myoepithelial cells, among others, and participate in the formation of extracellular vesicles that are then transported to the milk.However, whether these proteins contribute to the formation of extracellular vesicles, or how they participate in the formation of extracellular vesicles,remains to be explored.
In addition, the metabolic pathway and PPI network analyses of these significantly different MFGM proteins were also performed.The complement and coagulation cascades were the most significantly affected metabolic pathways during different lactation periods, which was also found to be significantly different in whey proteins within different lactation periods in our[20,53]and other previous studies[54-55].The PPI network of these MFGM proteins can broaden our view of the dynamic alterations in donkey milk MFGM proteins, and can also support and provide a theoretical basis for clarifying the causes of nutrient alterations in donkey milk during lactation.In particular, APOB was found to be the most interactive protein in donkey milk, interacting with seven other proteins, followed by fatty acid-binding protein(namely glutamic-oxaloacetic transaminase 2 (GOT2)),prothrombin, and cathepsin Z.Studies showed that GOT2 plays a key role in amino acid metabolism and metabolite exchange between mitochondria and cytosol, and can also facilitate cellular uptake of long-chain free fatty acids[56-57].The PPI network analysis of the differently expressed MFGM proteins in DC and DM would be helpful to understand the underlying mechanism of protein alterations during different lactation stages, which might also reflect the special needs of mammal newborn growth.
4 Conclusions
Herein, the donkey MFGM proteins present in different lactation stages were identified based on labelfree quantitative proteomics.In total, 216 and 215 MFGM proteins were characterized in the DC and DM, respectively.Among them, some fragments of immunoglobulins were found to be highly expressed or specifically expressed in DCs.In particular, the concentration of the immunoglobulin gamma 1 heavy chain constant region in DC was 2.28-fold higher than in DM, and the immunoglobulin lambda light chain variable region was specifically expressed in DC.Moreover, 40 significantly different MFGM proteins and their related GO annotations, metabolic pathways, and interaction networks were also investigated.These results could serve as a basis for future investigations on the biological functions of donkey milk MFGM proteins, and could also pave the way for the development of a more suitable commercial milk formula for infants.
