GmNMHC5 may promote nodulation via interaction with GmGAI in soybean
2022-02-19WentingWngZhiliWngWenshengHouLiChenBingjunJingWenyLijunBiWenwenSongCilongXuTinfuHnYongjunFengCunxingWu
Wenting Wng, Zhili Wng, Wensheng Hou, Li Chen, Bingjun Jing, Weny M, Lijun Bi,Wenwen Song, Cilong Xu, Tinfu Hn,*, Yongjun Feng,*, Cunxing Wu,*
a School of Life Science, Beijing Institute of Technology, Beijing 100081, China
b MARA Key Laboratory of Soybean Biology (Beijing), Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
Keywords:Glycine max GmNMHC5 GmGAI Nodulation GA pathway
ABSTRACT Soybean(Glycine max[L.]Merr.)is a food and oil crop whose growth and yield are influenced by root and nodule development.In the present study,GmNMHC5 was found to promote the formation of nodules in overexpressing mutants.In contrast, the number of nodules in Gmnmhc5 edited with CRISPR/Cas9 decreased sharply.In 35S:GmNMHC5 mutants, expression levels of genes involved in nodulation were significantly up-regulated.Both in vitro and in vivo biochemical analyses showed that GmNMHC5 directly interacted with GmGAI (a DELLA protein), and the content of gibberellin 3 (GA3) in overexpressing mutants was lower than that in the wild type.These results revealed that GmNMHC5 participates in the classical GA signaling pathway,and may regulate the content of GA3 to match the optimal concentration required for nodule formation,thereby promoting nodulation by directly interacting with GmGAI.A model illustrating the mechanism by which GmNMHC5 promotes soybean nodulation is presented.
1.Introduction
MADS-box family proteins are plant transcriptional regulators[1] with similar secondary structures, including an N-terminal MADS-box domain that binds to DNA.The MADS-box domain is followed by the K-box and the C-terminus involved in proteinprotein interactions, which differ widely among members [2].Members of this family have a wide range of functions,in particular in the regulation of flowering time and the development of various vegetative organs [3,4].The AGL17 subfamily of MADSbox functions in the regulation of plant vegetative growth.Among the MADS-box family, nmhC5, which is orthologous to rootexpressed AGL17 subfamily proteins in Arabidopsis,forms homodimers and performs its functions by binding to a CArG consensus sequence [5].GmNMH7, a MADS-box transcription factor, inhibited soybean root and nodule development [6].GmNMHC5was originally cloned from soybean (Glycinemax [L.] Merr.) root nodules and confirmed to promote the growth of soybean lateral roots and root nodules in a soybean root transformation system [7].Recent study [8] has shown thatGmNMHC5also accelerates flowering and maturity in soybean.
Leguminous plants have the ability to form nitrogen-fixing symbiotic nodules with rhizobia [9], and nodules strongly influence yield.The physiological process of nodulation is precisely regulated by a metabolic network in which nodule factors influence the development of symbiotic nodules[10].Root nodule formation begins with the interaction between host plant and rhizobia,which symbiotic relationship is highly specific [11].The process can be divided into two phases:pre-infection and infection.In the preinfection phase, the infection of roots by rhizobia is triggered by the perception by the plant of specific bacterial signals, called Nod factors(NFs)[12,13].Among the early nodulin genes,ENOD40is considered to be a key gene, and its expression strongly affects the number of nodules [14,15].The perception of NFs is mediated by nodule factor receptor(NFR)which encoded by the nodule factor receptor gene[16].The physical interactions between NFR and NFs promote the deformation of root hairs,the formation of infection threads,and the division of cortical and pericycle cells to form nodulation primordia.The NFR genes have been identified asGmNFR1α andGmNFR1β [17] andGmNFR5α andGmNFR5β [18] in soybean.
Several hormones have been reported to regulate nodulation.Ethylene and ABA were unfavorable for nodule formation [19].Cytokinins, in contrast, were necessary for initiation of nodule organogenesis [20].Gibberellins (GAs) played a dual role in nodulation.On the one hand,GA was dependent on DELLA proteins and negatively regulatedM.truncatulaandL.japonicusrhizobial infection [21-23].Exogenous application of biologically active GA3,inhibited the formation of nodules, and this inhibition was counteracted by the application of a biosynthesis inhibitor of GA [24].In legume-rhizobia symbiosis, GA could regulate the number of nodule from the already-incorporated rhizobia to prevent delayed infection by other rhizobia [25].These results indicate that higher GA signaling levels specifically inhibit the nodulation signaling pathway.On the other hand, GA promoted nodule organogenesis but inhibited the infection stages of nodulation [26].The number of nodules was reduced in severely GA-deficientnamutants, supporting the promotion by GA of nodule formation [27].Recently[28], addition of GA3at low concentration (10-9mol L-1) in pea was shown to increase nodule formation,whereas at 10-6mol L-1or higher the process was inhibited.An optimal GA concentration was key to successful nodulation.
GmGAI, a member of the DELLA protein family, modulated GA response.GA destabilized the DELLA protein, which repressed GA signaling [29].The mechanism of this process has been revealed:GA binds to the receptor protein GID1[30]and then interacts with a DELLA protein,leading to its targeted degradation by an SCF-26S proteasome and thereby removing its inhibition of the GA activity[31,32].Given that DELLA induces the expression of the upstream GA biosynthesis geneGA3ox,DELLA may be involved in the maintenance of GA homeostasis through this feedback regulation mechanism [33,34].
In the present study,the role ofGmNMHC5in nodulation of soybean was determined by observing the nodulation phenotypes in overexpressing mutants and theGmnmhc5edited with CRISPR/Cas9,as well as detecting the expression level of nodulation genes.The interacting protein of GmNMHC5 were identified by yeast two-hybrid assay, bimolecular fluorescence complementation and pull-down.By analyzing the regulatory pathway involved in the GmNMHC5 interacting protein,the expression levels of related genes in GA synthesis pathway and the content of GA3in plants were detected.Finally, a model illustrating the mechanism by which GmNMHC5 promotes soybean nodulation is presented.
2.Materials and methods
2.1.Plant materials and growth conditions
The soybean cultivar Jack was denoted as the wild type (WT)and used as a control group.The T3 progeny of the homozygous linesGmNMHC5-#5,GmNMHC5-#25, andGmNMHC5-#32 were overexpressing mutants,and the genotypes were examined to confirm thatGmNMHC5was stably overexpressed when inherited from the T2 generation (data not shown).CRISPR/Cas9 was used to createGmnmhc5mutants from the WT Jack, yielding three edited lines(Gmnmhc5-#5-1,Gmnmhc5-#5-2,andGmnmhc5-#5-3).A mutation(including 112 bp insertion of 6 bp mutation)at the target site of GmNMHC5-SP1 was identified, which produces premature translation stop codon (PTC), resulting in the inability of GmNMHC5 to be translated and loss of function[8].All seeds were separately grown in a controlled culture room under long-day(LD,16 h light/8 h dark)photoperiodic conditions at 27°C with 50%relative humidity.
2.2.Nodule phenotype statistics and gene expression detection
Bradyrhizobium japonicumUSDA 110 was used to induce root nodules.Fourteen-day-old seedlings were inoculated with 1 mL of USDA 110 culture (A600= 0.6) for each plant.Nodules were counted on the 14th day after injection of rhizobia.Root materials for real-time quantitative PCR (qPCR) were selected at 48 h after injection of rhizobia to measure the expression levels ofGmNMHC5,GmGAIand those genes which involved in nodule development.TheActingene was used as a reference.Primers are listed in Table S1.qPCR was performed with an ABI 7900 thermocycler (Applied Biosystems, Foster City, CA, USA) with Takara SYBR Premix Ex Taq (Takara, Kyoto, Japan).Statistical analyses were performed with Microsoft Excel(Microsoft Corporation,Redmond, WA, USA).A one-way analysis of variance (ANOVA) least significant difference test (LSD) was used to compare the significance of differences between WT and mutants at the 0.01 probability level.Origin 2017 (Origin Lab, Northampton, MA, USA) was used for drawing boxplots and histograms.
2.3.Protein interaction and subcellular localization
To further investigate the mechanism by which GmNMHC5 promotes soybean nodule formation and to identify metabolic pathways in which GmNMHC5 might be involved, we used BD-GmNMHC5 as bait in a yeast-two-hybrid (Y2H) experiment.BD-GmNMHC5 and AD were transformed into AH109 strain to test whether BD-GmNMHC5 as the bait protein showed self-activation(pGADT7-largeT + pGBKT7-p53 as positive control; pGADT7-lar geT + pGBKT7-laminC as negative control).After the absence of autoactivation was confirmed, the Yeastmaker Yeast Transformation System 2 (Clontech, Mountain View, CA, USA) was used to screen the cDNA library with pGBKT7-BD-GmNMHC5 as bait.The transformants were seeded on Trp- and Leu-deficient plates, and Trp-, Leu-, Ade-, and His-deficient plates.Finally, suspected positive clones were verified by Y2H backcross experiment.Bimolecular fluorescence complementation(BiFC)was used to further verify the protein interaction.The constructed vectors pCAMBIA1300-35S-nYFP173-GmNMHC5 and pCAMBIA1300-35SYFPc155-GmGAI were separately transferred toAgrobacteriumstrain GV3101.About four-week-old tobacco seedlings were used as the material for injection, and injected leaves were placed on a glass slide after 48 h of incubation.Confocal microscopy (Leica SP8, Solms, Germany) was used to observe the expression of fluorescent protein in tobacco leaves,which was photographed with a digital camera.For GST pulldown assay, GmGAI and GmNMHC5 were bacterially expressed and purified as His-tag and GST fusion protein,respectively.His-GmGAI was mixed with GST-GmNMHC5 or GST and incubated with GST resin.The GST resin was washed with phosphate buffer saline(PBS)three times.The bound proteins were loaded onto a 15% SDS-PAGE gel and visualized by Western blotting with anti-GST and anti-His tag.
The constructed pCAMBIA1302-35S-eGFP-GmGAI vectors were transferred toAgrobacteriumstrain GV3101 for subcellular localization of GmGAI.TheAgrobacteriumGV3101 was inoculated into 10 mL YEB liquid medium,cultured at 170 r min-1for 1 h,and centrifuged to remove the supernatant.GV3101 was resuspended with 10 mmol L-1MgCl2(containing 120 mol L-1Acetosyringone) and adjusted toA600of 0.6.Tobacco plants cultured (12 h light) for one month were selected and injected in the lower epidermis of leaves with 1 mL GV3101.After injection, the leaves were incubated for 48 h in the dark.GFP signals were observed using a confocal microscope Nikon C2-ER (Nikon, Kyoto, Japan).
2.4.Quantitative determination of GA3 concentrations
Plant roots were washed with distilled water and weighed to obtain about 100 mg (not less than 50 mg).They were homogenized at a proportion (saline or PBS buffer) of 100 mg/mL and centrifuged to extract the supernatant.The samples were held at -20 °C overnight.After these treatments, GA3content was detected using a Plant GA3ELISA Kit (TSZ, Lexington, M A, USA).The OD value of GA3was detected using a standard microplate reader capable of measuring absorbance at 450 nm.A linear regression was fitted expressing the concentration of the GA3standard(Sigma-Aldrich, Darmstadt, Germany) as a function ofA450value.Substituting theA450of the sample into the regression equation yielded the concentration of the sample.
3.Results
3.1.Overexpression of GmNMHC5 promoted nodulation in soybean,whereas Gmnmhc5 reduced nodulation
Nodule numbers were recorded on day 28(14 days after inoculation)(Fig.1a,b).As shown in Fig.1,the mean nodule numbers of theGmNMHC5-#5,GmNMHC5-#25,andGmNMHC5-#32lines were much higher than that in the WT, whereas CRISPR/Cas9 edited strains ofGmnmhc5showed significantly fewer nodules.The expression levels of genes which involved in nodule development(GmENOD40-1,GmENOD40-2,GmNFR1α,GmNFR5α, andGmNIN)andGmNMHC5in the transgenic mutants and WT were measured.A significant increase was observed in the35S:GmNMHC5mutants but a decrease inGmnmhc5mutants (Fig.1c).
3.2.GmNMHC5 interacts directly with GmGAI
Self-activation experiments confirmed that GmNMHC5 showed no autonomous activation (Fig.2a), and Y2H was then used for screening the proteins interacting with GmNMHC5.GmGAI, a key regulatory factor in the GA pathway showing a positive interaction with GmNMHC5,was identified.To confirm the virtual interaction between GmNMHC5 and GmGAI,a Y2H backcross experiment was performed, revealing that the transformed yeast of fusion constructs (pGADT7-AD-GmGAI + pGBKT7-BD-GmNMHC5) grew well on selective SD/-Leu/-Trp medium and also on selective SD/-Leu/-Trp/-His/-Ade medium (Fig.2b).BiFC confirmed the interaction between GmNMHC5 and GmGAI.In contrast to the total absence of fluorescence in the negative control (GmNMHC5-YFPn + YFPc,YFPn + GmGAI-YFPc, YFPn + YFPc), interaction of GmNMHC5-YFPn (N-terminal fragment of yellow fluorescent protein) and GmGAI-YFPc (C-terminal fragment of yellow fluorescent protein)produced fluorescent signals (Fig.2c).In a GST pull-down assay to determine whether there was a direct interaction between GmGAI and GmNMHC5in vitro, as shown in Fig.2d, GmGAI-His was pulled down by GST-GmNMHC5 but not by GST alone,indicating direct binding between GmGAI and GmNMHC5in vitro.
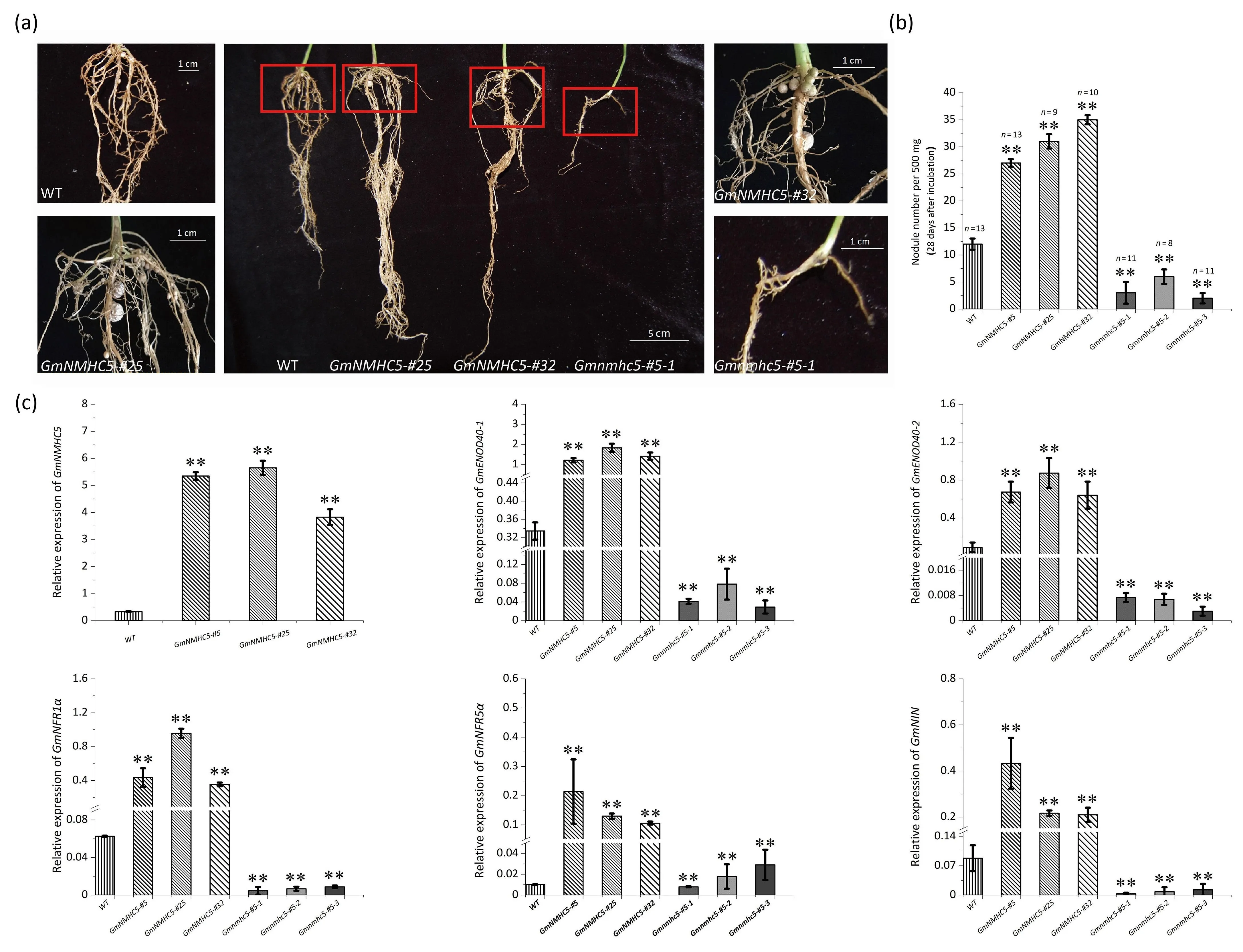
Fig.1.Nodule phenotype and nodule number statistics on day 28 and the relative expression of genes detected by qPCR.(a) Nodule phenotype of WT, GmNMHC5-#25,GmNMHC5-#32, and Gmnmhc5#5-1 (with overviews in the middle and close-up views on either side).(b) Mean nodule number per 500 mg of root on day 28.(c) Relative expression of GmNMHC5 and expression of the nodule-associated genes GmENOD40-1, GmENOD40-2, GmNFR1α, GmNFR5α, and GmNIN.**, P < 0.01 by t-test.
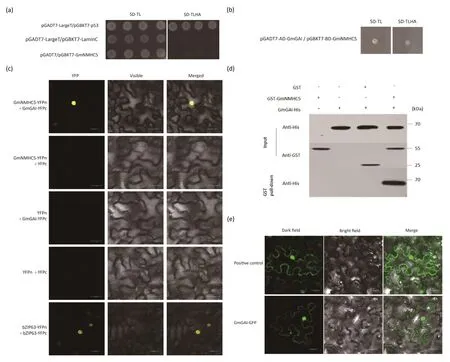
Fig.2.Detection and results of protein interaction.(a)The left panel shows the SD/-Leu/-Trp plate,the right panel shows the SD/-Leu/-Trp/-His/-Ade plate,and the plaques are pGADT7-LargeT/pGBKT7-p53(as positive control), pGADT7-LargeT/pGBKT7-LaminC(as negative control), and pGADT7/pGBKT7-GmNMHC5 from top to bottom.(b)The results of a Y2H backcross experiment using pGADT7-AD-GmGAI and pGBKT7-BD-GmNMHC5.(c) Bimolecular fluorescence complementation assay.GmNMHC5-YFPn+GmGAI-YFPc was the experimental group;GmNMHC5-YFPn+YFPc,YFPn+GmGAI-YFPc,and YFPn+YFPc were used as negative control;bZIP63-YFPn+bZIP63-YFPc was used as positive control.Scale bars, 20 μm.(d) His-GmGAI interacts directly with GST-GmNMHC5 but not GST by in vitro GST pulldown.(e) Subcellular localization of eGFP and GmGAI-eGFP fusion proteins.Photographs were taken in a dark field for green fluorescence (left column) and a bright field for cell morphology (middle column).The right column shows the merged view of the dark and bright fields for comparative clarity.Scale bars, 20 μm.
Following the identification of the protein GmGAI interacting with GmNMHC5,based on the previous[7]subcellular localization of GmNMHC5 in onion epidermis cells (under the control of the CAMV 35S promoter) in the nucleus, we performed a subcellular localization experiment for GmGAI in tobacco leaves.Confocal microscopy showed that the GmGAI-GFP fusion protein displayed strong fluorescence in the nucleus.Although there was a small amount of expression on the cell membrane, the GFP fluorescence was weaker than in the positive control.Thus,GmGAI is expressed mainly in the nucleus.In contrast,the free GFP protein in the control group was dispersed throughout the entire cell (Fig.2e).
3.3.GmNMHC5 adjusted the concentration of GA3
The expression levels of key genes in the GA synthesis pathway(GmGA3ox1,GmGA3ox2,andGmGA3ox5)andGmGAIwere measured via qPCR (Fig.3).Their expression levels decreased in the35S:GmNMHC5mutants and increased in theGmnmhc5mutants.GmNMHC5also regulated the gene expression ofGmGAI.Overexpression ofGmNMHC5reduced the content of GA3,but the opposite trend was observed forGmnmhc5(Fig.3).
4.Discussion
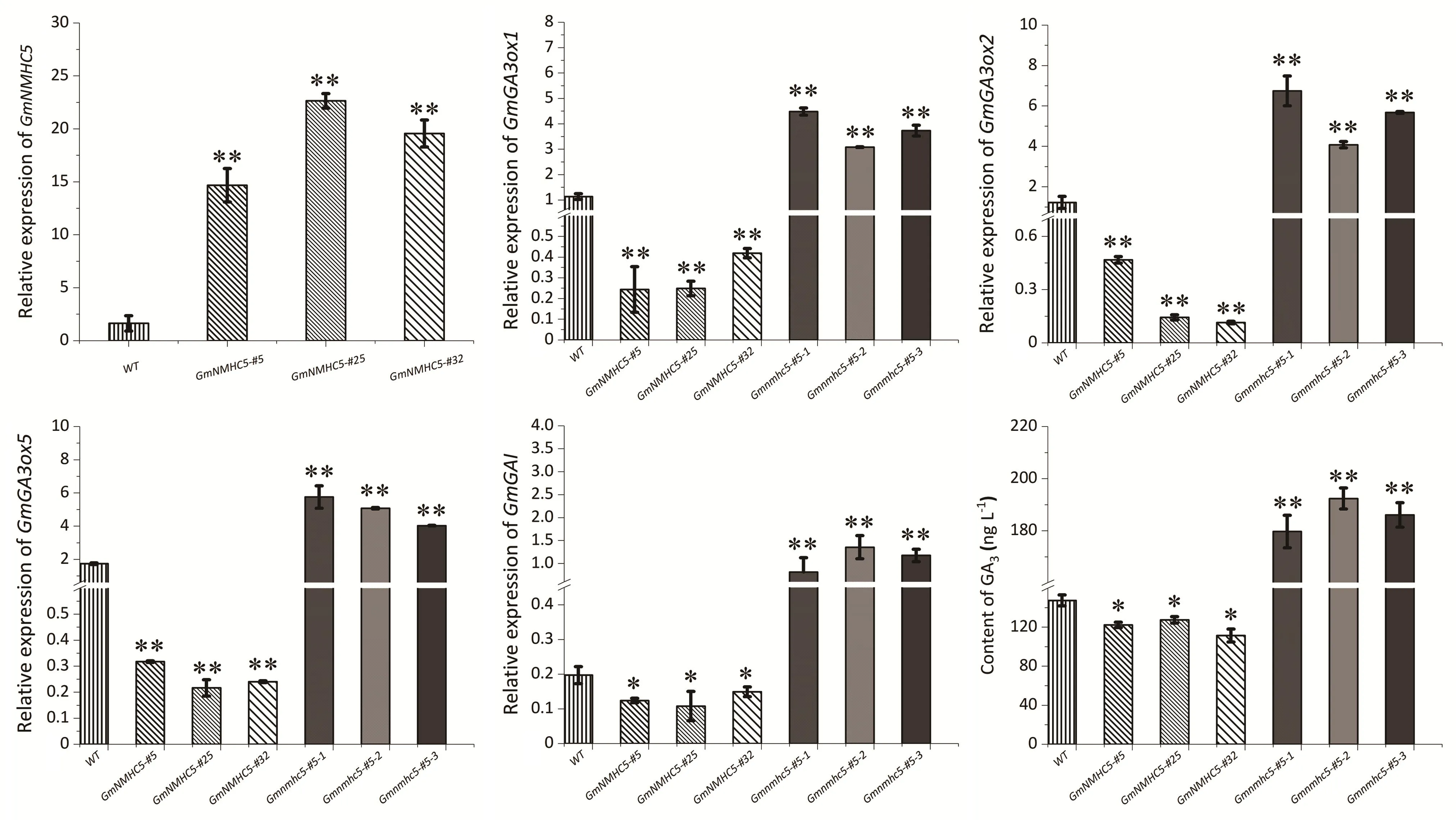
Fig.3.Relative expression of GmNMHC5,GmGAI and genes involved in GA3 synthesis in roots and content of GA3 in roots.Root materials were selected at 48 h after injection of rhizobia.Relative expression levels are normalized to GmActin.*, P < 0.05; **, P < 0.01 by t-test.
Nitrogen is essential for soybean production, and nitrogen use by soybean depends mainly on nodulation.To avoid the lack of regulation and signal transduction in the hairy root system compared with the whole plant, we used the 35S:GmNMHC5mutants andGmnmhc5mutants to further research the effect ofGmNMHC5on nodulation in this study.The observation thatGmNMHC5-#5,GmNMHC5-#25,andGmNMHC5-#32lines showed increased nodulation suggested that theGmNMHC5 gene increased nodulation in overexpressing mutants.In contrast, the CRISPR/Cas9-edited plant showed significantly reduced nodule numbers, further supporting the positive effect of theGmNMHC5gene on soybean nodule formation.The results of the detection of genes related to the nodulation process showed that the overexpression ofGmNMHC5promoted the expression of these genes which belong to the autoregulation of nodulation(AON)pathway in legumes,providing a theoretical basis for functional localization ofGmNMHC5.In view of the promoting role ofGmNMHC5in nodulation process,it will be associated with breeding practice in subsequent studies, which is believed to provide a new idea for improving the nitrogen fixation efficiency of soybean nodule in breeding cultivars for low-nitrogen soils.
Given the fierce energy competition between the vegetative growth process represented by the formation of nodules and the reproductive growth represented by flowering, the balance between the two processes is worth considering in breeding and production.GmNMHC5was shown in our previous study[8]to promote soybean flowering and maturity.GmNMHC5has the dual effect of synergistically regulating soybean flowering and nodulation, opening a new research direction to investigate the coordinated regulation of the two important physiological processes.
With respect to the influence of GA concentration on nodule formation, previous studies [4,35-37] have shown that GA3inhibits the formation of root nodules above a certain threshold.The nodule number of GA synthetic mutants in pea was reduced [27].The GA pathway is regulated by a complex network, in which the key protein DELLA dynamically regulates the concentration of GA3[38].Because DELLA proteins lack a canonical DNA binding domain,it is likely that they regulate the expression of their target genes by interacting with other transcription factors.In this study,we confirmed the interaction between GmNMHC5 (a MADS-box transcription factor) and GmGAI (a member of DELLA proteins).Previous studies have confirmed that DELLA proteins function by feedback regulation of GA signaling, including transcriptional regulation of the GA synthesis geneGA3ox.This study also revealed that the expression levels ofGmGA3ox1,GmGA3ox2,andGmGA3ox5in the mutants were changed relative to those in the wild type,and the contents of GA3in the root were also changed.We accordingly hypothesize that GmNMHC5 is an indispensable interaction protein in DELLA function and a new member in the classical GA pathway participating in GA3homeostasis in plants.
Fig.4 shows a hypothetical model of the mechanism by which this interaction leads to a change in GA3concentration in soybean roots and thus affects the number of nodules.In the GA pathway,GA-GID1 and SCFSLY1/GID2promote DELLA degradation, so that the GA response switch is turned on.Active GA then begins to accumulate.InGmnmhc5mutants,owing to the lack of GmNMHC5 protein,DELLA loses its feedback regulation function of the GA pathway,causing the concentration of GA to be higher than the threshold required for nodulation and finally to inhibit nodulation.In the 35S:GmNMHC5mutants, when the concentration of GA is high,DELLA combines with GmNMHC5 to exert a feedback-regulation function to inhibit the GA synthesis geneGA3ox, leading to a decrease of GA-GID1 and thereby reducing the degradation of DELLA.The GA response switch is then turned off, and when the GA concentration is lower than the required threshold for nodulation,the feedback regulation is activated.At this time,GA is resynthesized while DELLA is degraded.The GA response switch is again turned on.The process is repeated to adjust the GA concentration dynamically to its optimal level, ultimately promoting soybean nodulation.The specific regulatory mechanism in the process awaits further verification and refinement and the specific structural domain of GmNMHC5 interaction with GmGAI awaits identification.
CRediT authorship contribution statement

Fig.4. A hypothetical model of the interaction between GmNMHC5 and GmGAI in regulating soybean nodulation.Brown lines and arrows represent common processes.The blue arrows and dotted circle on the left represent the regulatory process of Gmnmhc5 mutants created by CRISPR/Cas9, and the pink arrows and dotted circle on the right represent the regulation of GA signaling by 35S:GmNMHC5 mutants.The X indicates that the regulatory pathway is not functional.The number of GA modules represents the concentration of active GA3.Owing to the lack of feedback regulation function in Gmnmhc5 mutants, GA3 concentration is too high to inhibit nodulation.In 35S:GmNMHC5 mutants, the optimal concentration of GA3 is maintained to promote nodule formation by feedback regulation via interaction between GmNMHC5 and GmGAI.
Wenting Wang:Data curation, Formal analysis, Investigation,Writing - original draft.Zhili Wang:Data curation, Formal analysis, Investigation.Wensheng Hou:Methodology.Li Chen:Data curation, Methodology.Bingjun Jiang:Validation.Wenya Ma:Investigation, Methodology.Lijuan Bai:Investigation.Wenwen Song:Validation.Cailong Xu:Validation.Tianfu Han:Supervision.Yongjun Feng:Conceptualization, Formal analysis,Supervision, Writing - review & editing.Cunxiang Wu:Conceptualization,Formal analysis,Validation,Supervision,Writing-review& editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31271636) and the China Agriculture Research System (CARS-04).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.03.019.
杂志排行
The Crop Journal的其它文章
- Brief Guide for Authors
- Heavy soil drying during mid-to-late grain filling stage of the main crop to reduce yield loss of the ratoon crop in a mechanized rice ratooning system
- H2O2 mediates transcriptome reprogramming during Soybean mosaic virus-induced callose deposition in soybean
- Contrasting patterns of accumulation,partitioning,and remobilization of biomass and phosphorus in a maize cultivar
- TaMADS2-3D, a MADS transcription factor gene, regulates phosphate starvation responses in plants
- Calcineurin B-like protein 5 (SiCBL5) in Setaria italica enhances salt tolerance by regulating Na+ homeostasis
