H2O2 mediates transcriptome reprogramming during Soybean mosaic virus-induced callose deposition in soybean
2022-02-19TianjieSunXizheSunFukuanLiNanMaMengxuanWangYanChenNaLiuYuanJinJieZhangChunyanHouChunyanYangDongmeiWang
Tianjie Sun, Xizhe Sun, Fukuan Li, Nan Ma, Mengxuan Wang, Yan Chen,Na Liu, Yuan Jin, Jie Zhang, Chunyan Hou,*, Chunyan Yang, Dongmei Wang,*
a State Key Laboratory of North China Crop Improvement and Regulation, Baoding 071000, Hebei, China
b Key Laboratory of Hebei Province for Plant Physiology and Molecular Pathology, Baoding 071000, Hebei, China
c College of Life Sciences, Hebei Agricultural University, Baoding 071000, Hebei, China
d Market Supervision Administration of Chongli District, Zhangjiakou 076350, Hebei, China
e Institute of Cereal and Oil Crops, Hebei Academy of Agriculture and Forestry Sciences, Shijiazhuang 050035, Hebei, China
Keywords:Callose Soybean mosaic virus H2O2 Transcriptome Soybean
ABSTRACT The main defense response to Soybean mosaic virus(SMV)infection in soybean[Glycine max(L.)Merr.]is thought to be blockage of intercellular virus transport by callose deposition on plasmodesmata.But the specific regulatory mechanism remains largely unknown.In this study,we found that hydrogen peroxide(H2O2) signal downstream of NO was associated with the regulation of callose accumulation.Abundant H2O2 was produced on the cell membrane and cell wall in the incompatible combination of soybean cultivar Jidou 7 and SMV strain N3,whereas no obvious H2O2 was observed in the compatible combination of Jidou 7 and strain SC-8.When H2O2 production was inhibited, callose accumulation induced by SMV infection decreased to a level insufficient to restrict virus transport in the incompatible combination.The H2O2-associated transcriptome dynamics of soybean during SMV infection was investigated.Transcriptome and functional analysis using virus-induced gene silencing showed that GmSEOB and GmPAP27, two genes regulated by H2O2, functioned in resistance by positively regulating the accumulation of callose in response to SMV infection.These results lay a foundation for further research on the signal transduction and molecular regulation of callose deposition during soybean resistance to SMV infection.
1.Introduction
Hypersensitive response-programmed cell death(HR-PCD),callose selectively accumulated on plasmodesmata,and RNA interference (RNAi), are the major defense responses restricting intercellular virus spread during plant-virus interaction [1-3].In our previous study[4],HR-PCD failed to interdict the spread ofSoybean mosaic virus(SMV) when callose deposition was inhibited,supporting the decisive role of callose in virus resistance.
As a second intracellular messenger, hydrogen peroxide (H2O2)is involved in many biological responses of plants to environmental stresses such as drought[5],cold damage[6],salt stress[7],and microbial infection[7].Upon TMV infection,NADPH oxidase in theNicotiana tabacumplasma membrane was activated, the level of H2O2increased, and callose was accumulated on the cell wall at the edge of necrotic spots[8].Treatment ofN.tabacumleaves with salicylic acid activated the reactive oxygen species(ROS)signaling pathway and induced callose deposition on plasmodesmata,thereby inhibiting the spread of TMV [9].Microarray analysis in catalase deletion mutantArabidopsisplants showed that H2O2induced the expression of callose synthase(CalS) [10].Mechanical stimulation can induce the production of H2O2, which further regulatesCalS8inArabidopsisand promotes the synthesis of callose in mesophyll cells[11].Both of these findings indicate that H2O2regulates the selective deposition of callose,but the molecular mechanism of this regulation remains unknown.Another intracellular second messenger, nitric oxide (NO), was shown to participate in various physiological and stress response processes in plants[12,13].As a small lipophilic molecule, NO can cross membranes and transduce signal via reversibleS-nitrosylation modification of the target protein [14].The level of NO increased rapidly in the incompatible combination of soybean [Glycine max(L.) Merr.]and SMV, but did not change in the compatible combination, and the accumulation of callose, as well as the restriction to virus transportation, was abolished when nitrate-reductase (NR) or nitric oxide-synthase (NOS) inhibitors were applied prior to SMV inoculation [15].However, the relationship between H2O2and NO signaling pathways remains unknown.
Transcriptional regulation is a key part of cell signal transduction.Mata-Perez et al.[16] discovered by transcriptome sequencing that nitro-fatty acids (NO2-FAs), metabolites of NO, are involved in plant defense responses.Next-generation sequencing(NGS) technology offers unique advantages in mapping traitrelated gene clues, large-scale gene function screening, and signal pathway characterizing.
We have shown that a large amount of callose was produced and deposited mainly on plasmodesmata after virus infection in the incompatible combination of soybean cultivar Jidou 7 and SMV strain N3 [4].Application of 2-deoxy-D-glucose (2-DDG), a callose-synthesis inhibitor, resulted in disease symptoms as well as detection of the SMVcapsid protein(CP) gene in noninoculated upper leaves.In the present study, it was found that H2O2was localized in the cell wall and plasma membrane in the incompatible combination.Hydrogen peroxide regulated the resistance of soybean to SMV by inducing the deposition of callose in plasmodesmata.Plasma membrane H2O2was inhibited by imidazole(an NADPH oxidase-specific inhibitor),and the host transcriptome dynamics was analyzed to identify differentially expressed genes (DEGs) regulated by H2O2.Reverse transcriptionquantitative PCR (RT-qPCR) was used to confirm the expression of several SMV-responsive genes regulated by H2O2.Virusinduced gene silencing (VIGS) was further used to characterize the function of the SMV response genessieve element occlusion Blike(GmSEOB) andpurple acid phosphatase 27(GmPAP27), and silencing these genes reduced callose accumulation upon SMV infection and weakened the resistance of soybean to SMV.
The present study aimed to investigate the relationship between H2O2and SMV-induced callose accumulation in plasmodesmata, to identify by transcriptome analysis the molecular network that regulates virus-induced callose accumulation, and to identify disease-resistance genes involved in this process.
2.Materials and methods
2.1.Plant cultivation and virus inoculation
The soybean cultivar Jidou 7 was used for pharmacological and transcriptomic analyses.The SMV strains N3 and SC-8 were obtained from the National Center for Soybean Improvement,Nanjing Agricultural University.Jidou 7 is resistant to N3 (forming an incompatible combination) and susceptible to SC-8 (forming a compatible combination).SMV was propagated in the soybean cultivar Nannong 1138-2, and sap from diseased leaves was used as virus inoculum as described previously [4].Soybean seedlings were planted in an insect-proof greenhouse with a 14 h light/10 h dark cycle,light intensity of 700 μmol photons m-2s-1,and constant temperature of 25 °C.
Two-week-old soybean plants were used for virus inoculation.To detect NO, H2O2, and callose distribution, SMV-infected soybean leaf sap was mixed with emery powder and gently applied to soybean leaves using a brush as described previously [4].To detect H2O2subcellular localization and the spread of SMV on plants in VIGS experiments, SMV was inoculated in small areas on the leaves using toothpick as described previously [4,15].The leaves were rinsed with ddH2O immediately after inoculation.
2.2.H2O2 in situ detection by the DAB-uptake method
Hydrogen peroxide detection employed the 3,3-diaminobenzidine (DAB)-uptake method as previously described[17,18].Inoculated soybean leaves were cut into pieces of 5 mm2, placed in staining solution (4.67 mmol L-1DAB, pH 3.8),and incubated at 25°C for 8 h.Leaves were cleared in boiling clearing solution (50% ethanol, 16.67% glycerol, 16.67% phenol, and 8.33% lactic acid, v/v) for 2 min.Samples were then stored in 25%glycerol at room temperature.Hydrogen peroxide was visualized as reddish-brown coloration under a fluorescence microscope(BX53, Olympus, Tokyo, Japan).
2.3.Subcellular localization of H2O2
The subcellular localization of H2O2was performed as previously described [4].Leaves inoculated with SMV were cut into pieces of 1 mm2and treated in fixative solution(5 mmol L-1CeCl3and 3% glutaraldehyde) at 4 °C for 24 h.The treated samples were washed three times with washing buffer(5 mmol L-1CeCl3and 0.1 mol L-1Tris-HCl,pH 7.5)and then washed with ddH2O for 30 min(two times).Washed samples were dehydrated in ethanol and acetone and then embedded in Epon812 epoxy resin for 12 h.Samples were incubated at 35°C for 12 h,45°C for 24 h,and 60°C for 48 h.Ultrathin sections were prepared with an ultrathin slicer (LKB-8800 ultramicrotome, LKB-Produkter AB, Bromma, Sweden).The sections were stained with 1.5% uranyl acetate and lead citrate and imaged under a transmission electron microscope (JEM-100CX, JEOL, Mitaka, Japan) at 80 kV.
2.4.Pharmacological treatment
Simple leaves of soybean plants were injected with 50 mmol L-1imidazole or 100 μmol L-1carboxy-PTIO (c-PTIO, Sigma-Aldrich, St.Louis, MO, USA) dissolved in ddH2O to inhibit plasma membrane H2O2production or to scavenge intracellular NO, as described previously [15].The injected soybean plants were cultivated for additional 24 h before SMV inoculation.
2.5.Labeling of NO in soybean leaves
Fluorescent labeling of NO in soybean leaves was performed using diaminofluorescein-FM diacetate (DAF-FM DA, Sigma-Aldrich).Inoculated soybean leaves were cut into pieces of 5 mm2and immersed in phosphate-buffered saline (PBS, 0.1 mol L-1, pH 7.2) containing 100 μmol L-1DAF-FM DA.Soybean leaves were placed under vacuum to facilitate penetration of the dye and rinsed three times with dH2O.Nitric oxide was visualized as yellow-green fluorescence under the fluorescence microscope with Ex/Em = 488 nm/526 nm, as described previously [15].
2.6.Labeling and observation of SMV-induced callose
Soybean leaves after inoculation with SMV were placed in a clearing solution (50% ethanol, 16.67% glycerol, 16.67% phenol,and 8.33% lactic acid, v/v) and boiled for 2 min, washed for 5 min with ddH2O(three times),and treated for 15 min with dyeing buffer(0.01%aniline blue dissolved in 0.1 mol L-1PBS,pH 8.0).Leaves were washed again with ddH2O and observed by fluorescence microscope with Ex/Em = 385 nm/495 nm [4,19].
2.7.Image analysis
ImageJ (https://fiji.sc/) was used to measure band intensity of SMVCPproduct in reverse transcription PCR (RT-PCR), and the fluorescence of callose, H2O2, and NO in microscopic images, as previously described [20].The integrated intensity was calculated as the integrated intensity of the total labeled cells backgroundsubtracted using the integrated intensity of a region outside the cell.
2.8.Isolation of plant RNA and detection of gene expression
Total RNA was isolated with an RNA extraction kit (UNlQ-10 Column Trizol Total RNA Isolation Kit, Sangon Biotech, Shanghai,China)and reverse-transcribed into cDNA with a reverse transcription kit (PrimeScript RT reagent Kit with gDNA Eraser, TaKaRa,Dalian, China).RT-PCR and RT-qPCR experiments were performed using primers listed in Table S1.The SMVCPgene was detected as previously described[4].PCR amplification was performed usingTaqDNA polymerase(EasyTaq,Transgen,Beijing,China).The thermocycling conditions were 94 °C for 5 min (initial denaturation)followed by 40 cycles of 94°C,30 s(denaturation);55°C,30 s(annealing); and 72 °C, 1 min (extension).RT-qPCR was performed using the SYBR Green method (TransStart Tip Green qPCR Super-Mix, Transgen, Beijing, China).GmEF1bwas used as a reference gene in the RT-PCR and RT-qPCR experiments[21].Three biological replicates with their respective three technical replicates were conducted for each sample in the RT-qPCR experiments and calculated using the 2-ΔΔCT method as previously described [21].
2.9.Establishment of transcriptome database
Soybean mosaic viruswas inoculated on soybean leaves preinjected with water or imidazole, and the host transcriptome was determined at 0, 4, 12, 24, and 48 hours post-inoculation (hpi).Because in our previous study the systematic spread of SMV was detected at 42 hpi in the compatible combination[22],the specific time point for the transcriptomic study was determined based on these results.Soybean leaves pre-injected with dH2O were used as control.RNA samples of the three independent biological replicates were mixed in equal amounts and used for the construction of libraries.Library construction and sequencing were performed at the Beijing Genomics Institute (BGI, Shenzhen, China), as described previously [23].Total RNA samples were treated with DNase I, enriched for mRNA with oligo (dT)-conjugated magnetic beads, reverse-transcribed, filled into double-stranded cDNA,ligated with 3’-adapters, and amplified by PCR into fragmented mRNA libraries.Transcriptome libraries were sequenced on a sequencer (HiSeq 2000, Illumina Inc., San Diego, CA, USA) using 90-base pair-ended mode.Raw read data generated by the sequencer were processed to remove adapters, reads with excessive ‘‘N”,and low-quality reads (reads with aQ-value < 10 accounting for> 50% of the entire sequence), resulting in > 4 Gb of clean reads for each library.The Burrows-Wheeler alignment tool (http://biobwa.sourceforge.net/) was used to align the clean reads to the reference soybean genome, Wm82.a2.v1 from SoyBase (https://www.soybase.org/), and to perform secondary quality control.The RNAseq by Expectation-Maximization (RSEM) values were used to quantify the mRNA expression levels.The gene expression levels were calculated as FPKM (fragments per kilobase of transcript per million fragments mapped).Differentially expressed genes were defined as genes with false discovery rate (FDR) ≤0.001 and twofold expression change between corresponding samples based on the Poisson distribution.
2.10.GO functional classification and pathway enrichment analysis
Gene Ontology(GO)and Kyoto Encyclopedia of Genes and Genomes(KEGG)pathway analyses were performed to investigate DEG functions.GO terms that were significantly enriched in DEGs compared to that in the entire genome were identified with Gene Ontology Term Finder (https://www.yeastgenome.org/goTermFinder) and visualized with WEGO (http://wego.genomics.org.cn)[24].
Pathway enrichment analysis was performed by mapping DEGs to the KEGG (https://www.genome.jp/kegg/) pathways as previously described [25].
2.11.Generation of GmPAP27- and GmSEOB-silenced plants
Tobacco rattle virus(TRV)-mediated VIGS was used to generateGmSEOBandGmPAP27-silenced plants.Specific fragments ofGmSEOBandGmPAP27were amplified using the primers listed in Table S1 and inserted into the pTRV2 plasmid betweenEcoR I andXhoI sites.VIGS was performed as previously described [26]using one-week-old soybean seedlings.The empty plasmid pTRV2 was used as control.Agrobacterium tumefacienscarrying pTRV1 and recombinant pTRV2 plasmids were resuspended in the infection buffer (50 mmol L-1MES, 2 mmol L-1Na3PO4, 28 mmol L-1Dglucose, 0.1 mmol L-1acetosyringone, 4.1 mmol L-1L-Cys, and 0.02% (w/v) Silwet L-77) to OD600= 0.5, mixed at a 1:1 ratio, and poured over the roots of soybean seedlings with 5 mL per plant.Before SMV inoculation, silencing effect in the first compound leaves of soybean seedlings was quantified by RT-qPCR three weeks afterA.tumefacienstreatment.
2.12.Detection of SMV CP protein product
Western blotting was used to detect the expression of SMV CP at the protein level.Protein extracts from the SMV inoculation site(IS) and tissue around it at various distances were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis(SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF)membrane.After washing with tris-buffered saline-Tween 20(TBST) (10 mmol L-1Tris-HCl, pH 7.0, 0.25 mol L-1NaCl, and 0.1%Tween 20)containing 5%skim milk,the membrane was incubated in an antibody solution containing 1:2000(v:v)diluted polyclonal antibody prepared with recombinant SMV CP expressed inEscherichia colias the antigen for 1 h at 25°C.The PVDF membrane was incubated in HRP-conjugated antibody and washed with TBST,and protein signal was detected using TMB.Rubisco large subunit(RbcL) was detected as a loading control by Coomassie brilliant blue gel staining.
2.13.Sequence availability
Sequence information of genes involved in this study can be found in SoyBase (https://soybase.org) under the following accession IDs:Glyma.04G073900.1 (GmDCAF1), Glyma.20G204500.2(GmSEOB), Glyma.12G217400.2 (GmSALI3-2), Glyma.06G170300.1(GmPAP27), Glyma.07G014500.1 (GmVSPA), Glyma.14G089800.1(GmHRGP), and Glyma.02G276600.1 (GmEF1b).
2.14.Statistical analysis
Statistical analysis was performed using Excel software(version 16.0.12624.20278,Microsoft,Redmond,WA,USA).The significance of differences was assessed with one-way ANOVA/Duncan or Student’st-test atP< 0.05.
3.Results
3.1.H2O2 participated in soybean defense response to SMV
In the incompatible combination, a small amount of H2O2was observed at 2 hpi,and the distribution area of H2O2increased with time (Fig.1).Necrotic plaques appeared at 72 hpi, and a large amount of H2O2was observed around them.In the compatible combination, H2O2was produced after 2 hpi, but the amount of H2O2was lower than that in the incompatible combination.Only a small amount of H2O2was observed at 2-12 hpi, whereas no H2O2was observed after 24 hpi in the mock treatment.The difference in H2O2production in the incompatible and compatible combinations suggested the involvement of H2O2in the signal transduction process of defense response against SMV infection.
3.2.H2O2 induced by SMV was distributed in the cell wall and plasma membrane
H2O2was observed by TEM as a high-electron-density substance on the cell wall at 2-48 hpi (Fig.2b-f, i, j, arrows), and on the cell membrane(arrowheads)at 48 and 72 hpi(Fig.2f,g).However, H2O2was not observed at 0 h post-SMV inoculation (Fig.2a)and 24 hpi in the mock (Fig.2h).
3.3.Pre-injection of imidazole impeded SMV-induced callose and resistance
Since H2O2induced by SMV was observed on the cell wall and plasma membrane, we speculated that this process relies on the activity of NADPH oxidase.The production of SMV-induced H2O2was reduced in plants pre-injected with NADPH oxidase inhibitor imidazole (Fig.1), confirming our speculation.To further reveal the relationship between H2O2and callose, imidazole-pretreated leaves were inoculated with SMV, and callose was observed in these leaves (Fig.3a).In leaves pretreated with water, callose induced by SMV could be observed at 6 hpi.Callose accumulation gradually increased with the infection process.At 72 hpi, a large amount of callose was observed around necrotic plaque.However,only a small amount of callose was observed at 6-72 hpi in imidazole pre-injected leaves, less than that observed in plants preinjected with water (Fig.3a, b).Thus, inhibition of H2O2reduced callose induced by SMV.
We also observed disease symptoms(Fig.3c)and tested for the presence of viralCPin non-inoculated upper leaves at 10 days after inoculation(Fig.3d,e).All tested leaves showed no signs of disease when pre-injected with water before SMV N3 inoculation, the same outcome as for plants inoculated with healthy leaf sap(Mock).However, a clear curling phenomenon with necrotic spots was observed in the non-inoculated upper leaves of imidazole preinjected and compatible plants (Fig.3c).ViralCPwas detected using RT-PCR in the upper leaves of imidazole-treated plants(Fig.3d).The SMVCPwas induced (P< 0.05) in non-inoculated upper leaves pre-injected with imidazole in comparison with leaves pre-injected with water (Fig.3e).
At the end of this time he had fallen in love with a charming Princess, but that when he had shut himself up into a room with her, and had thrown off his snake s skin, her parents had forced their way into the room and had burnt the skin, whereupon the Prince, changed into the likeness29 of a dove, had broken a pane of glass in trying to fly out of the window, and had wounded himself so badly that the doctors despaired of his life
3.4.Nitrogen oxide functioned upstream of H2O2
To characterize the relationship between NO and H2O2signals,we used the NO-specific fluorescent probe DAF-FM DA to label NO in the incompatible combination pretreated with imidazole.NO signal induced by SMV at 24 hpi in the imidazole preinjected leaves was not significantly different from that in the control group (Fig.4a, c), whereas H2O2production was reduced (P<0.05) in leaves pretreated with c-PTIO (Fig.4b, d), a specific scavenger of NO, indicating that NO acts upstream of H2O2signaling pathways in the incompatible interaction.
3.5.H2O2 regulated transcriptome dynamics in soybean-SMV interaction
We analyzed transcriptomes at 0, 4, 12, 24, and 48 hpi in the incompatible combination and investigated the regulatory effect of H2O2on the transcriptome.The quality control of the sequencing results of the samples showed that Clean reads accounted for 97% of the total sequence in the 10 samples tested in this experiment.The proportion accounted for by adapters and low-quality reads did not exceed 3%,indicating that the quality of the NGS data was acceptable (Fig.S1).
Based on the clean reads filtered from each sample, the numbers of DEGs affected by imidazole at 4, 12, 24, and 48 hpi were determined.The large number of genes regulated by H2O2(Fig.S2a) suggested the scale of H2O2signaling in transcriptionlevel regulation.The DEGs in the transcriptome after imidazole treatment showed the lowest correlation with DEGs at 24 hpi(Fig.S2b), and the scale of DEGs affected by H2O2also peaked at 24 hpi(Fig.S2a).Thus,in the soybean-SMV interaction,H2O2signal showed the highest impact on DEGs at 24 hpi in the transcriptome.
3.6.Characteristics of DEGs and their involvement in signal transduction pathways

Fig.1.Detection of SMV-induced H2O2 in soybean.DAB stained H2O2 in leaves induced by Soybean mosaic virus (SMV) strain N3 or SC-8 was observed by fluorescence microscopy.Pre-injection of imidazole was used to inhibit H2O2.Time points post-SMV inoculation are indicated.Mock, material inoculated with healthy leaf sap.Representative images of three biological replicates are shown.Scale bars, 50 μm.hpi, hours post-inoculation.
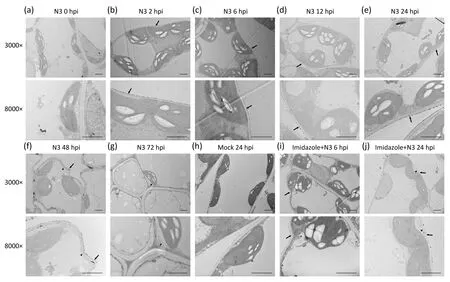
Fig.2.Intracellular distribution of H2O2 induced by SMV.(a-g)Distribution of H2O2 induced by Soybean mosaic virus(SMV).(h)Cells 24 hours post-inoculation with healthy leaf sap.(i, j) Distribution of H2O2 induced by SMV in tissues pre-injected with imidazole.Cells were observed by transmission electron microscopy.The 3000× and 8000×labels indicate magnification.Arrows indicate cell wall and arrowheads indicate cell membrane.Imidazole:materials pre-injected with imidazole.Representative images of three biological replicates are shown.Scale bars, 2 μm.hpi, hours post-inoculation.
The enrichment pattern of GO in the transcriptome is shown in Fig.S3.The GO components of genes affected by H2O2at 4,12,24,and 48 hpi were enriched mainly in cellular process, metabolic process, response to stimulus process, and single-organism process.Molecular functions of the affected genes were enriched mainly in binding, catalytic activity, and transporter activity.Although DEGs in host varied greatly with SMV infection, the GO components of genes regulated by H2O2were conserved at all time points.
We further classified the NGS gene data according to the annotations on the KEGG database and determined the enrichment degree in the top 20 enriched pathways.At 4 hpi, DEGs were enriched mainly in the ‘‘spliceosome” pathway and the ‘‘starch and sucrose metabolism” pathway (Fig.5a).At 12, 24, and 48 hpi, the DEGs were enriched mainly in the ‘‘plant-pathogen interaction” pathway and the ‘‘biosynthesis of secondary metabolites”pathway (Fig.5b-d).The changes in the DEGs regulated by H2O2upon infection in the ‘‘plant-pathogen interaction” pathway revealed several regulatory patterns (Fig.5e).Following preinjection with imidazole, genes whose expression was affected at 4 and 12 hpi included the genes encoding mitogen-activated protein kinase 20 (Glyma.02G286800) and disease resistance protein RPM1 (Glyma.17G179200andGlyma.06G267400).Genes whose expression was affected by imidazole at 24 and 48 hpi included those encoding L-type lectin-domain containing receptor kinase VII.2 (Glyma.12G205100), ZIM domain-containing protein TIFY8(Glyma.06G072700), and transcription factor PIF4 (Glyma.10G042800).In the plant-pathogen interaction pathway, genes affected by imidazole at 4,12,24,and 48 hpi included those encoding serine/threonine-protein kinase PBS1 (Glyma.08G360600andGlyma.08G360600), tobacco mosaic virus (TMV) resistance protein N (Glyma.13G076200), interleukin-1 receptor-associated kinase 4(Glyma.18G233500), and cyclic nucleotide-gated channel(Glyma.08G241600).
3.7.RT-qPCR verification of gene expression in the transcriptome
We selected six genes with transcription-level changes in response to SMV infection and imidazole pre-injection and measured their expression by RT-qPCR.The relative transcription levels of DDB1- and CUL4-associated factor homolog 1 (GmDCAF1,Fig.6a),GmSEOB(Fig.6b), BURP domain-containing protein SALI3-2(GmSALI3-2,Fig.6c),GmPAP27(Fig.6d),vegetative storage protein A (GmVSPA, Fig.6e), and hydroxyproline-rich glycoprotein(GmHRGP, Fig.6f) were suppressed(P<0.05)by the application of imidazole, and were very close to the expression levels in the transcriptome.
3.8.GmSEOB and GmPAP27 positively regulated soybean resistance to SMV
We next tested the function of genes that were induced by SMV infection and affected by H2O2for SMV resistance.By introducing TRV-mediated VIGS,we silencedGmSEOBandGmPAP27in soybean seedlings to study the function of the two H2O2-regulated genes on SMV-induced callose and resistance.Silencing was confirmed by RT-qPCR analysis (Fig.S4).The area of cells with callose fluorescence induced by SMV increased at 24 and 72 hpi onGmSEOB- orGmPAP27-silenced plants (Fig.7a, b), whereas the average relative integrated intensity was lower (P<0.05) than that of the control(Fig.7a, c).To further characterize the resistance ofGmSEOB- andGmPAP27-silenced plants to SMV, we tested the expansion of the virus at the transcription and protein levels.SMV was inoculated in a small area of soybean leaves, and was tested in the IS and tissues extending from IS at several distances(Fig.S5).At 72 hpi,SMVCPdetected by RT-PCR extended only 1 mm outside the IS of the control plants, whereas inGmSEOB-silenced plants, a weak signal ofCPwas detected 3 mm beyond IS, and similar results were obtained inGmPAP27-silenced plants(Fig.7d).The protein product of SMV CP was detected only in the IS of the control plants,whereas inGmSEOB- andGmPAP27-silenced plants, a weak signal could be detected at 2 mm outside the IS (Fig.7e).
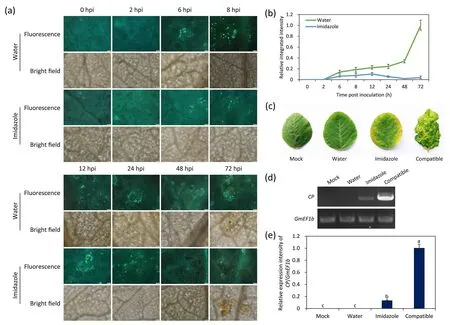
Fig.3.H2O2 regulated callose accumulation and resistance.(a)Effect of H2O2 on Soybean mosaic virus(SMV)-induced callose accumulation.SMV strain N3 was inoculated on soybean leaves pretreated with water or imidazole and observed at indicated time points.Scale bars,50 μm.Representative images of three biological replicates are shown.(b)The integrated intensity of callose fluorescence relative to that measured in water at 72 hpi in(a).Values are mean±SE of three biological replicates.(c)Disease symptoms of non-inoculated upper leaves pre-injected with imidazole.Viral symptoms were observed 10 days post-SMV inoculation.Mock, upper leaves in plants inoculated with healthy leaf sap.Plants inoculated with SC-8 (compatible) were used to show viral symptoms.(d) RT-PCR detection of SMV CP gene in the non-inoculated upper leaves.GmEF1b was used as a reference gene.Representative images of three biological replicates are shown in(a),(c),and(d).(e)Expression of SMV CP relative to that of GmEF1b in(d).Different letters indicate significant difference tested by one-way ANOVA/Duncan(P<0.05).Values are mean±SE of three independent measurements and calculated as the band intensity relative to that of compatible samples.Error bars indicate standard error of the mean in (b) and (e).hpi, hours post-inoculation.
4.Discussion
Hydrogen peroxide plays an important role in regulating the permeability of plasmodesmata.InArabidopsis, the mechanical stimulation-induced plasmodesmata closure is associated with the promotion of callose deposition by H2O2throughCalS8[11].We detected systemic infection of SMV in an incompatible combination when callose synthesis was inhibited [4], suggesting that the deposition of callose on the plasmodesmata is the main defense to restrict SMV transportation.In this study,H2O2was produced around necrotic spots in the incompatible combination,and H2O2was distributed mainly in the cell wall and plasma membrane.The application of imidazole, a plasma membrane NADPH oxidase inhibitor [27], resulted in a decreased callose deposition in the incompatible combination.As a result, disease symptoms and systematic transportation of SMV were detected in the noninoculated upper leaves, indicating that callose deposition and resistance rely on the presence of NADPH oxidase-produced H2O2.The observation of H2O2in samples pre-injected with imidazole hints at the presence of H2O2synthesis pathways other than NADPH oxidase in response to SMV infection, although they are certainly not the main source of H2O2.
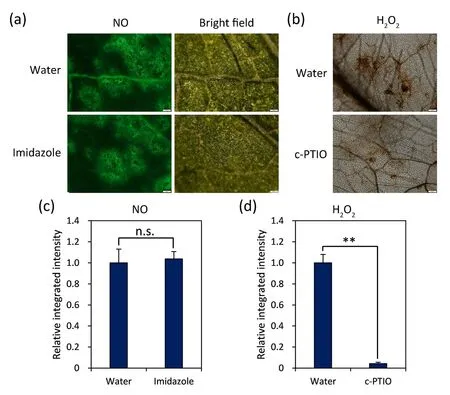
Fig.4.The relationship between NO and H2O2.(a)Visualization of Soybean mosaic virus(SMV)-induced NO at 24 hpi in plants pre-injected with imidazole.NO,fluorescence of NO labeled with DAF-FM DA; BF, bright field.(b) Detection of SMV-induced H2O2 at 24 hpi in c-PTIO-pretreated plants.Tissues were observed by fluorescence microscopy.Water:plants pre-inoculated with water.Representative images of three biological replicates are shown in (a) and (b).Scale bars, 100 μm.(c) Statistics of the integrated density of the DAF-FM DA-labeled area in the imidazole-pretreated leaves relative to that measured in water in(a).(d)Statistics of the integrated density of the DAB-stained area in the c-PTIO-pretreated leaves relative to that measured in water in (b).Values are mean±SE of 30 independent measurements.Standard errors are shown as bars above columns; n.s., not significant;**, significant difference as determined by Student’s t-test at P < 0.05.
Another intracellular signaling molecule,NO,was found to facilitateS-nitrosylation modification and to trigger other intracellular signaling pathways to fine-tune plant responses to adversity.In red kidney bean (Phaseolus vulgarisL.), the increased level of NO in response toPseudomonas fluorescensinfection was able to induce resistance againstP.fluorescensandRhizoctonia solaniby augmenting the level of H2O2[30].Hydrogen peroxide acts downstream of NO to mediate induction of RNA-dependent RNA polymerase 1(RDR1), an element of the RNA silencing pathway that restricts TMV systemic infection in tobacco andArabidopsis[31].Previously,we confirmed that NO was involved in the regulation of callose deposition in the incompatible combination of soybean and SMV[13].In the present study, the inhibition of H2O2did not affect the production of NO, while the scavenging of NO inhibited H2O2,suggesting that nitric oxide may act upstream of H2O2to regulate defense response to SMV.
Numerous studies have shown that genes that confer resistance in plants were usually enriched in the plant-pathogen interaction pathway in KEGG.Pathways in the categories of photosynthesis,pigment metabolism, and plant-pathogen interaction in tobacco were significantly regulated at the transcription level following inoculation withCucumber mosaic virus[32].DEGs in response toStrawberry vein banding viruswere abundantly distributed in the pathways of pigment metabolism, photosynthesis, and plantpathogen interactions in woodland strawberry (Fragaria vescaL.)[33].We accordingly speculate that these pathways are universal transcriptional regulatory pathways in plants that counteract virus infection.Considering that there are numerous genes without clear annotation in the soybean genome, and the plant-pathogen interaction pathway in KEGG contains many genes associated with plant resistance to fungal and bacterial pathogens rather than viruses, we combined gene number and rich factor to determine the level of gene enrichment in the pathway.In the plantpathogen interaction pathway, genes whose expression was regulated by H2O2at 4-and 12 h post-SMV inoculation may be involved in identification of the pathogen and initiation of defense responses.At 24- and 48 hpi, H2O2may manipulate the cellular response to phytohormone (especially JA) signals by regulating the expression of hormone-responsive genes, such asGlyma.06G072700, to implement the deployment of defense reaction.Genes continuously manipulated by H2O2may be modulators of defense response, thereby precisely regulating the process of defense responses.As the second intracellular messenger, hydrogen peroxide may directly regulate the expression of only a small number of genes, implying a decisive role for these genes.
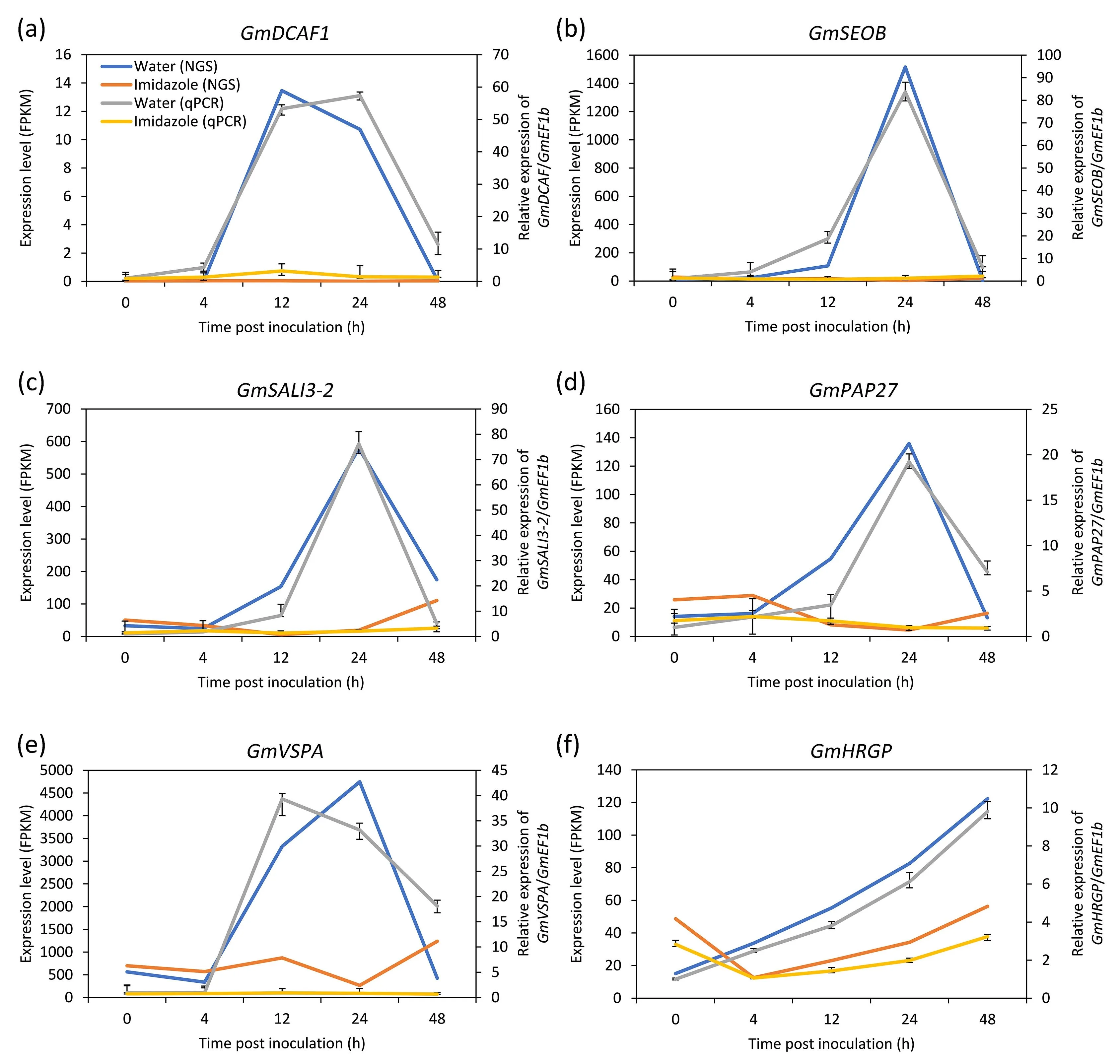
Fig.6.RT-qPCR verification of relative expression of selected genes.The relative expressions of GmDCAF1 (a), GmSEOB (b), GmSALI3-2 (c), GmPAP27 (d), GmVSPA (e), and GmHRGP(f)were detected using RT-qPCR and normalized to the values measured at 0 hpi.The primary axis shows the expression level of genes in soybean-Soybean mosaic virus(SMV)interaction at 0,4,12,24,and 48 hpi without(Water)and with(Imidazole)the application of imidazole in the transcriptome(NGS),and the secondary axis shows the relative expression level of genes detected by RT-qPCR(qPCR).Values in the transcriptome are reads observed for each gene(fragments per kilobase of exon per million fragments mapped, FPKM analysis). GmEF1b was used as a reference gene.Each value represents the mean±SE of three biological replicates.
The DDB1- and CUL4-associated factor is a substraterecognition receptor for E3 ubiquitin ligases in human, and a homolog of this protein was essential for embryonic development inArabidopsis[34].Sieve element occlusion B belongs to the structural phloem proteins, homologs of which inArabidopsis[35] andN.tabacum[36] participate in rapid sealing of sieve tubes.SALI3-2 is a plant-specific protein,involved in response to drought stress inBrassica napus[37] andMedicago truncatula[38].Purple acid phosphatase was localized in the cell wall of tobacco cultured cells[39],and mediated activation of beta-glucan synthases.Vegetative storage protein A participated in late blight resistance in potato(Solanum tuberosumL.) [40].Hydroxyproline-rich glycoprotein has been found to accumulate in tobacco inoculated with a strain ofPotato virus Y(PVYN), thereby inducing systemic acquired resistance (SAR) to combatErysiphe cichoracearuminfection [41].The induction of these genes under SMV infection was suppressed bypretreatment with imidazole, suggesting that these genes may be directly or indirectly involved in the defense response under the regulation of H2O2.
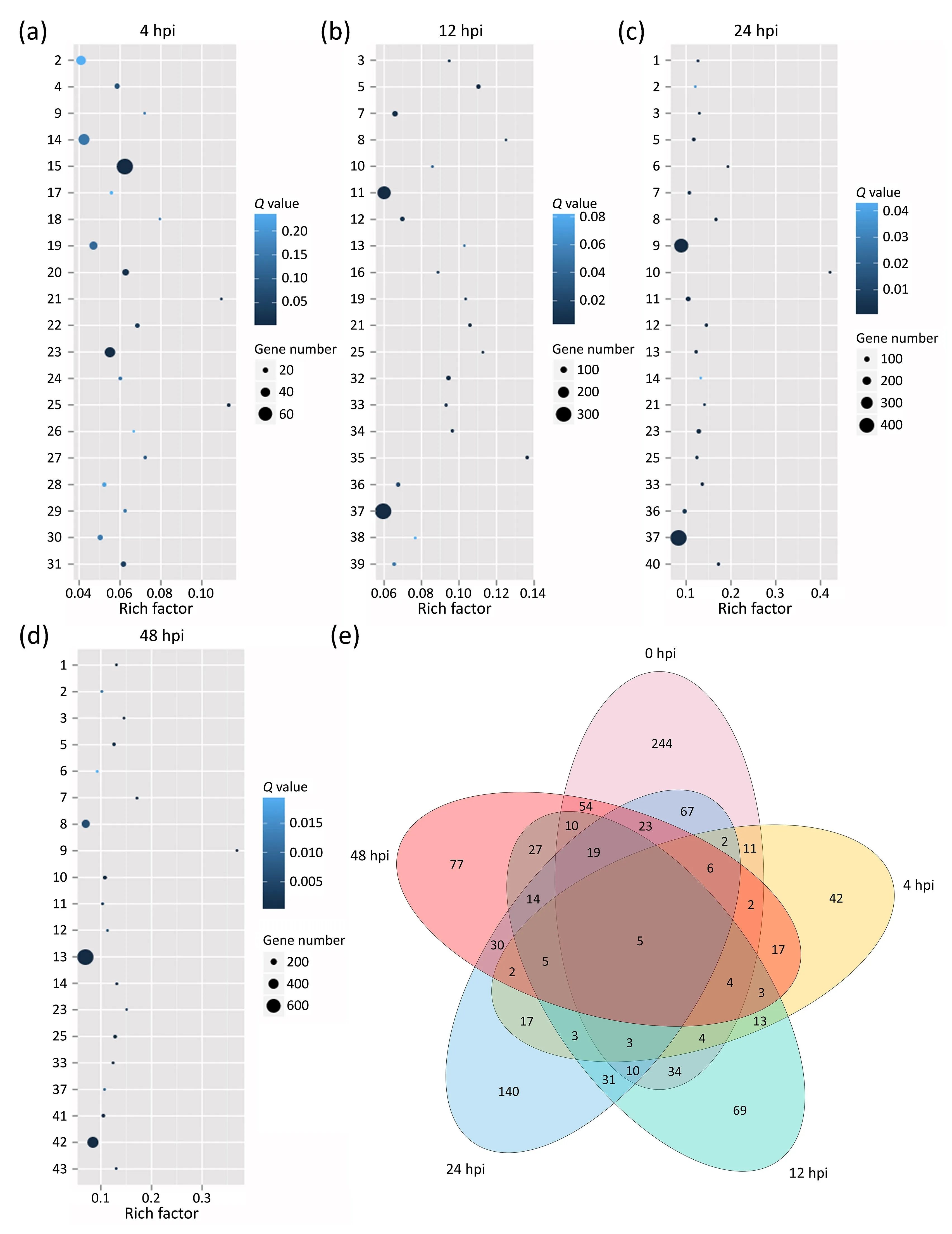
Fig.5.Pathway enrichment analysis of DEGs affected by H2O2 at 4(a),12 (b),24(c),and 48(d) hours post-SMV inoculation.1,valine,leucine and isoleucine degradation;2,tryptophan metabolism; 3, stilbenoid, diarylheptanoid, and gingeroid biosynthesis; 4, starch and sucrose metabolism; 5, plant-pathogen interaction; 6, photosynthesisantenna proteins; 7, phenylpropanoid biosynthesis; 8, limonene and pinene degradation; 9, isoquinoline alkaloid biosynthesis; 10, flavonoid biosynthesis; 11, flavone and flavonol biosynthesis; 12, circadian rhythm-plant; 13, biosynthesis of secondary metabolites; 14, beta-alanine metabolism; 15, arginine and proline metabolism; 16, ABC transporter; 17, ubiquitin mediated proteolysis; 18, tropane, piperidine and pyridine alkaloid biosynthesis; 19, spliceosome; 20, sphingolipid metabolism; 21,sesquiterpenoid and triterpenoid biosynthesis; 22, RNA degradation; 23, pyruvate metabolism; 24, non-homologous end- joining; 25, nitrogen metabolism; 26, mRNA surveillance pathway; 27, lysine degradation; 28, histidine metabolism; 29, ether lipid metabolism; 30, carbon fixation in photosynthesis organisms; 31, alanine, aspartate and glutamate metabolism;32,ubiquinone and other terpenoid-quinone biosynthesis;33,propanoate metabolism;34,other types of O-glycan biosynthesis;35,linoleic acid metabolism; 36, isoflavonoid biosynthesis; 37, cyanoamino acid metabolism; 38, cutin, suberine and wax biosynthesis; 39, alpha-linolenic acid metabolism; 40, terpenoid backbone biosynthesis; 41, tyrosine metabolism; 42, phenylalanine metabolism; and 43, metabolic pathways.Rich factor, the ratio of the number of genes in the pathway entry of the DEGs to the total number of genes in the pathway entry of all annotated genes.(e) The relationship between the distribution of DEGs in the plant-pathogen interaction pathway and the process of Soybean mosaic virus(SMV)infection,numbers in the Venn diagram indicate DEGs that meet the corresponding conditions.hpi,hours post-inoculation.
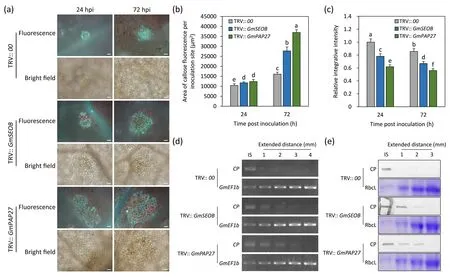
Fig.7. Roles of GmSEOB and GmPAP27 in SMV-induced callose accumulation and disease resistance.(a)The effect of GmSEOB and GmPAP27 silencing on Soybean mosaic virus(SMV)-induced callose.Callose was observed in the leaves of GmSEOB-and GmPAP27-silenced plants generated by TRV-mediated VIGS inoculated with SMV at 24 and 72 hpi.Plants inoculated with TRV empty vector(TRV::00)were used as control.Scale bars,50 μm.Area of callose fluorescence in single SMV infection sites(b)and the integrated intensity of callose fluorescence relative to TRV:: 00 at 24 hpi (c) in GmSEOB- and GmPAP27-silenced plants was calculated.Different letters indicate significant difference tested by one-way ANOVA/Duncan(P<0.05).Values are mean±SE of 30 independent measurements.Standard errors are shown as bars above the columns.Extended range of SMV in GmSEOB-and GmPAP27-silenced plants was detected by RT-PCR(d)and Western blotting(e).Mixed samples from 10 independent inoculation experiments at 72 hpi were applied to detect the CP transcript and its protein product. GmEF1b and Rubisco large subunit (RbcL) was used as a loading control in (d) and (e), respectively.Representative images of three biological replicates are shown in (d) and (e).
To investigate whether genes regulated by H2O2mediate regulation of callose deposition as well as resistance,we further characterized the function of some genes of interest in the SMV-induced callose accumulation.Using TRV-VIGS, we found that silencing ofGmSEOBandGmPAP27reduced the resistance of soybean plants to SMV.AlthoughGmSEOB- andGmPAP27-silenced plants showed reduced resistance to SMV,the area of callose fluorescence induced by the virus increased.Silencing ofGmSEOBandGmPAP27may have reduced the level of callose accumulation in the SMVinfected cells, and the reduced callose may have failed to sufficiently restrict the intercellular transport of the virus, resulting in a greater area of cells with callose.Given that purple acid phosphatase in tobacco can activate the synthesis of callose and cellulose [39], we speculate that GmPAP27 increases the accumulation of callose on the plasmodesmata in a similar way to combat SMV.The specific molecular mechanism by whichGmSEOBandGmPAP27regulate callose accumulation and resistance, the relationship between the two callose regulators downstream of H2O2signaling, and the determination of whether other regulatory factors control the SMV-induced callose deposition,await further research.These findings lay a foundation for further study of the H2O2-regulated signal transduction pathway that controls the selective deposition of callose on plasmodesmata.
Data availability statement
The data that support the findings of this study are available on request from the corresponding authors.
CRediT authorship contribution statement
Tianjie Sun, Xizhe Sun, Fukuan Li, Chunyan Hou, Chunyan Yang,and Dongmei Wangconceived and designed the experiments.Tianjie Sun, Xizhe Sun, Fukuan Li, and Nan Maperformed the experiments.Tianjie Sun,Xizhe Sun,Fukuan Li,Yan Chen,Na Liu,Jie Zhang,and Dongmei Wanganalyzed the data.Nan Ma,Mengxuan Wang,Na Liu,Yuan Jin,Jie Zhang,Chunyan Yang,and Dongmei Wangprovided reagents, materials, or analysis tools.Tianjie Sun,Fukuan Li,Nan Ma,and Dongmei Wangwrote the paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr.Haijian Zhi (Nanjing Agricultural University,China)for providing the virus strains and advice on their propagation and Professor Mengchen Zhang (Institute of Cereal and Oil Crops,Hebei Academy of Agriculture and Forestry Sciences,China)for providing the soybean seeds.This work was supported by the National Natural Science Foundation of China (30971706 and 31471421), ‘‘973” Preliminary Program (2014CB160318), and Hebei Natural Science Foundation (C2020204132).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.04.005.
杂志排行
The Crop Journal的其它文章
- Brief Guide for Authors
- Heavy soil drying during mid-to-late grain filling stage of the main crop to reduce yield loss of the ratoon crop in a mechanized rice ratooning system
- GmNMHC5 may promote nodulation via interaction with GmGAI in soybean
- Contrasting patterns of accumulation,partitioning,and remobilization of biomass and phosphorus in a maize cultivar
- TaMADS2-3D, a MADS transcription factor gene, regulates phosphate starvation responses in plants
- Calcineurin B-like protein 5 (SiCBL5) in Setaria italica enhances salt tolerance by regulating Na+ homeostasis
