TaMADS2-3D, a MADS transcription factor gene, regulates phosphate starvation responses in plants
2022-02-19YingchunHnLiuChungLiShuiwuWngLihuJiRuiZhngHuiLiJinfngTnHongweiXueWenmingZheng
Yingchun Hn, N Liu, Chung Li, Shuiwu Wng, Lihu Ji, Rui Zhng, Hui Li, Jinfng Tn,c,*,Hongwei Xue, Wenming Zheng,*
a Collaborative Innovation Center of Henan Grain Crops/State Key Laboratory of Wheat and Maize Crop Science, College of Life Sciences, Henan Agricultural University,Zhengzhou 450002, Henan, China
b College of Resource and Environment, Henan Agricultural University, Zhengzhou 450002, Henan, China
c School of Agriculture, Sun Yat-sen University, Guangzhou 510275, Guangdong, China
d School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China
Keywords:MADS-box transcription factor Phosphate starvation response Wheat Arabidopsis
ABSTRACT Soil inorganic phosphate (Pi) levels are frequently suboptimal for the growth and development of crop plants.Although MADS-box genes participate in diverse plant developmental processes, their involvement in phosphate starvation responses (PSRs) remains unclear.We identified a type I MADS-box transcription factor gene, TaMADS2-3D, which was rapidly induced under low-Pi stress in roots of wheat(Triticum aestivum).A TaMADS2-3D-GFP fusion protein was found located in the nucleus.Transgenic Arabidopsis plants overexpressing TaMADS2-3D (TaMADS2-3DOE) showed shortened primary roots,increased lateral root density,and retarded seedling growth under high-Pi(HP)conditions,accompanied by increased Pi contents in their shoots and roots.The Arabidopsis TaMADS2-3DOE plants showed similar PSR phenotypes under low Pi (LP) conditions.These results indicate constitutive activation of PSRs by overexpression of TaMADS2-3D in Arabidopsis.Reactive oxygen species (ROS), H2O2 and O2-, levels were increased in root tips of Arabidopsis TaMADS2-3DOE plants under HP conditions.Transcriptome analysis of Arabidopsis TaMADS2-3DOE plants under different Pi regimes revealed expression changes for a variety of PSR genes including AtZAT6.Overexpression of TaMADS2-3D in wheat also led to constitutive activation of PSRs.We propose that TaMADS2-3D regulates plant PSRs probably by modulating ROS homeostasis,root development, PSR gene expression, and Pi uptake.This study increases our understanding of plant PSR regulation and provides a valuable gene for improving phosphorus-use efficiency in wheat and other crops.
1.Introduction
Phosphorus is an essential nutrient for plant growth and development.It participates in the regulation of enzymatic reactions,signal transduction processes, and metabolic pathways [1].Owing to its uneven distribution, high rate of fixation, and slow diffusion in most soils,inorganic phosphate(Pi)is a key factor restricting the growth and development of crop plants [2].To cope with the low availability of Pi, plants have evolved a set of complex adaptive responses, known as phosphate starvation responses (PSRs), to maintain Pi homeostasis[3].Phosphate starvation results in adaptive morphological modifications to the root system architecture(RSA), including a primary root (PR), lateral roots (LRs), and root hairs in most plants [4].Plants can also mobilize multiple metabolic responses to alleviate Pi starvation and ensure the metabolic balance of phosphorus at the whole-plant level.All of these responses are finely tuned by a complex gene-regulatory network.
Transcription factors (TFs) are crucial components integrating environmental cues and responses of plants at the cellular level,and function in the regulation of PSRs.Among the well-studied TFs that affect PSRs, MYB TF, PHOSPHATE STARVATION RESPONSE 1(AtPHR1),functions as a central regulator of phosphate signaling pathways [5].AtWRKY6 [6], AtWRKY42 [7], and AtWRKY45 [8]modulate phosphate uptake and homeostasis by regulating the expression ofAtPHO1andAtPHT1;1inArabidopsis thaliana(L.)Heynh.OsWRKY74 [9], OsWRKY28 [10], OsPTF1 [11], and AtZAT6[12] regulate Pi homeostasis by affecting RSA.AtZAT6 has also been shown [13] to trigger ROS-mediated increases of flavonoids and anthocyanins inArabidopsis via direct regulation of theAtTT4,AtTT5, andAtTT7genes.
The function of the MADS-box TF family in PSR regulation has been little studied.MADS-box TFs,which function in diverse regulatory networks underlying multiple developmental processes in plants [14], are characterized by the presence of a MADS-box domain in their N-terminal region.The MADS-box TF family is divided into two lineages, type I and type II, based on protein domain structures.Type I MADS-box TFs are further classified into three subclasses:Mα,Mβ,and Mγ,whereas type II MADS-box TFs,also called MIKC-type TFs, contain four protein domains [15].MADS-box TFs regulate expression of target genes by binding to the CArGciselement in the form of heterodimers or heterotetramers[16].Previous studies[17-19]have revealed the functions of MADS-box TFs in controlling floral organ formation, flowering time,root growth,and fruit development and maturation in plants.
Some MADS-box family TFs have been shown to regulate plant responses to different abiotic stresses, including drought [20,21],cold [22], and salinity [23].However, the function of MADS genes in plant nutrient response regulation has received little attention.AtANR1/AtAGL44, a type II MADS gene, is a positive regulator of LR development inArabidopsisthat responds to nitrate availability[24].Among the fiveANR1orthologs of rice (Oryza sativaL.),OsMADS25[25] andOsMADS27[26] modulate root development in a nitrate-dependent manner.Ma et al.[27] suggested that type I MADS genes are involved in regulation of stress responses,including the PSRs in wheat (Triticum aestivumL.).
Wheat is a globally important staple food crop providing protein, starch, and energy for human diets.Low availability of Pi has become one of the main factors limiting wheat production worldwide [28].It is therefore desirable to uncover the molecular regulatory mechanism, and to identify the key TFs involved, in the PSRs of wheat.Using cDNA microarray analysis, Shi et al.[29]found that several MADS-box genes might be involved in Pi deprivation stress in wheat; however, the roles of these genes remain unknown.In our previous study,we found that a wheat expressed sequence tag(GenBank accession number CA658343),which came from the nucleotide sequence of TaMADS2-3D though Nucleotide BLAST analysis, was differentially affected by Pi availability in the roots of wheat cultivar ZM9023 in a microarray assay experiment.The expression ofTaMADS2-3Dwas down-regulated in the compatible interaction between wheat and stripe rust (caused byPuccinia striiformisf.sp.tritici) [30] and decreased by cold stress in many wheat cultivars[31],but there is no information on its function in the regulation of plant PSRs.
To investigate the role ofTaMADS2-3Din the regulation of PSRs,the expression and location ofTaMADS2-3Dwere detected,and the transgenicArabidopsisand wheat overexpressingTaMADS2-3Dwere developed to explore the changes of physiological and morphological PSR phenotypes.The comparative transcriptomes ofArabidopsistransgenic lines and WT control were used to identify the changes in gene expression levels.The findings shed light on the role of MADS-box TFs in plant response to low-Pi stress.
2.Materials and methods
2.1.Plant materials and growth conditions
Zhengmai 9023 (ZM9023), an elite winter wheat cultivar, was used for amplifying the coding sequence (CDS) and measuring the expression ofTaMADS2-3D.Fielder, a spring wheat cultivar,and theArabidopsisecotype Columbia-0 (Col-0) were used for developing transgenic lines overexpressingTaMADS2-3D.
The growth conditions were adapted to the plant materials used.Petri dish culture was used for studying the PSRs ofArabidopsis TaMADS2-3DOElines.Arabidopsisseeds were surface-sterilized and cold-treated at 4 °C for 48 h.Seeds were germinated initially on 0.5× Murashige and Skoog (MS) medium supplemented with 1% (w/v) sucrose and 1% (w/v) agar and grown at 22 °C under a 16-h light/8-h dark cycle.The HP and LP MS media contained 625 μmol L-1and 10 μmol L-1KH2PO4,respectively.For measuring fresh weight, the growth media were supplemented with 0.01,0.05,0.3125,0.625,2.5,or 5.0 mmol L-1KH2PO4.Agar plates were inclined at 65° to allow growth of roots along the agar surface.
Hydroponic culture was performed to detect expression and investigate the phenotypes of wheat materials.Wheat seeds were germinated and grown in Petri dishes containing distilled water.On the 7th day,seedlings were transferred to modified hydroponic solution containing 1.25 mmol L-1KNO3,1.50 mmol L-1Ca(NO3)2,0.75 mmol L-1MgSO4·7H2O, 10 μmol L-1MnSO4·H2O, 2 μmol L-1ZnSO4,1.5 μmol L-1CuSO4,50 μmol L-1KCl,72 μmol L-1Fe-EDTA,50 μmol L-1H3BO3, 0.075 μmol L-1Na2MoO4·2H2O, and 10 μmol L-1Na2SiO3·9H2O.The culture solution was supplemented with 200 μmol L-1or 10 μmol L-1KH2PO4for HP or LP treatments.After 7 days, seedling roots were sampled for RNA extraction to detect expression ofTaMADS2-3D.Roots grown under HP conditions were used as a control.For the short-term low-Pi treatment, seedling roots were sampled at 0, 3, 6, 9, 12, and 24 h for RNA extraction to detect expression ofTaMADS2.For observation of phenotype,transgenic wheat lines overexpressingTaMADS2-3Dand their wild-type (WT) control (Fielder) were grown under HP or LP conditions for an additional 21 d.Plants were grown under greenhouse conditions with a 16-h light/8-h dark photoperiod at 25 °C.
2.2.Plasmid construction and plant transformation
To generate theTaMADS2-3Dtransgenic expression vector,TaMADS2-3DCDS was amplified from the cDNAs of ZM9023 roots.ForArabidopsistransformation, theTaMADS2-3Dsequence was cloned into theSalI andKpnI sites of a modified pCAMBIA1300 plasmid that carriesGFPCDS under a superpromoter [32], and the resulting construct contained the TaMADS2-3D-GFP expression cassette, which was transferred intoAgrobacterium tumefaciensstrain GV3101 for transformingArabidopsisCol-0 plants by the floral dip method [33].Stable transgenic lines were selected on 0.5×MS medium containing hygromycin(50 mg mL-1).For wheat transformation, theTaMADS2-3Dsequence was cloned into theBamHI andSmaI sites of the pCUB vector [34], resulting in an expression cassette ofTaMADS2-3Dunder the ubiquitin gene promoter (pUbi).This construct was transformed into wheat cv.Fielder byAgrobacterium-mediated transformation with the assistance of the State Key Laboratory of Crop Stress Biology for Arid Areas in Northwest Agriculture and Forest University, Yangling,Shaanxi province.The PCR primers used in these experiments are listed in Table S1.
2.3.Subcellular localization
For the subcellular localization assay, theTaMADS2-3Dsequence was cloned into a modified pCAMBIA1300:GFP vector usingXholI andSalI, resulting in a construct expressing TaMADS2-3D-GFP under the 35S promoter.Plasmids were transformed intoA.tumefaciensstrain GV1301.Agrobacteriumcells were harvested by centrifugation, suspended in solution containing 10 mmol L-1MES (pH 5.7), 10 mmol L-1MgCl2, and 200 μmol L-1acetosyringone, and adjusted to OD6001.0.After incubation for 3 h at room temperature, theAgrobacteriumsuspension was infiltrated into tobacco (Nicotiana benthamiana) leaves with a needleless syringe.Two days after infiltration, GFP fluorescence in the transiently transformedN.benthamianacells was imaged using a laser-scanning confocal microscope (FLUOVIEW-FV10i,Olympus, Tokyo, Japan).The primers used are listed in Table S1.
2.4.RNA-seq analysis
For RNA-seq experiments,Arabidopsis TaMADS2-3DOEand WT plants were grown under the HP and LP conditions for 7 d prior to root harvest.Total RNA was isolated from collected root materials using TRIzol Reagent (Cat.15596026, Invitrogen, Carlsbad, CA,USA).RNA integrity and concentration were assessed using an RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies,Santa Clara,CA,USA).RNA-seq was performed commercially(Annoroad Gene Technology Corporation,Beijing,China),with three biological replicates independently sequenced for each sample.RNA sequencing libraries were generated using a NEB Next Ultra RNA Library Prep Kit for Illumina (#E7530L, NEB, Ronkonkoma, NY, USA) following the manufacturer’s recommendations,which were sequenced on Illumina Hiseq (Illumina, San Diego,CA, USA) to generate 150-base paired-end reads.A Perl (Perl 5.16.2,https://www.metacpan.org/release/RJBS/perl-5.16.2/)script was used to filter the original data to ensure quality data for further analysis.All subsequent analyses were performed with highquality clean-read datasets that were aligned against theA.thalianareference genome sequence (TAIR 10 Genome Release 27,http://plants.ensembl.org/index.html).Fragments per Kilobase per Million Mapped Reads (FPKM) were calculated to estimate gene expression levels in each sample.Thresholds ofq≤0.05 and|log2-Ratio| ≥1 were used to identify differentially expressed genes(DEGs)among groups.
Gene ontology (GO) enrichment tests of DEGs were performed using a hypergeometric test (http://geneontology.org/).GO terms with correctedP≤0.05 were assigned as significantly enriched by DEGs compared to the genome background.Venn diagrams were constructed with Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/).Expression heat maps of genes were created with Morpheus (https://software.broadinstitute.org/morpheus/).
2.5.Reverse transcription quantitative PCR (qRT-PCR) analysis
First-strand cDNA was synthesized using a Reverse Transcription System (Cat.R212-01, Vazyme, Nanjing, China).Reversetranscription quantitative PCR was performed with SYBR Green PCR Master Mix (Cat.No.208154, Qiagen, Hilden, Germany) on a StepOne Real Time PCR System (Applied Biosystems, Foster City,CA, USA).Three independent experiments were performed for qRT-PCR and three technical replicates were used for each sample in each independent experiment.Expression levels of the analyzed genes were calculated using the 2-ΔΔCTmethod for relative expression quantification[35].Ta26Swas used as an internal control.Two primer sets were used for qRT-PCR analysis:one (CA658343_RT)was specific forTaMADS2-3Dtargeting the 3ʹ-UTR of its mRNA(GenBank:JN248615.1), and the other (TaMADS2_RT) recognizedTaMADS2-3Dand its two homoeologs (TaMADS2-3Aand -3B).Primers used for qRT-PCR analysis are listed in Table S1.
2.6.Measurement of plant phenotypes
Arabidopsisseedlings were grown on Petri dishes under HP or LP conditions for 7 days.Wheat seedlings were grown in HP or LP hydroponic culture for 21 days.Plant height and root length(maximum root length in wheat)were measured with a ruler.ArabidopsisLR numbers including lateral root primordia were measured using a dissecting microscope.LR density was calculated as follows:LR density = LR number/PR length.
Biomass (fresh weight) of shoots and roots in wheat was measured in seedlings grown in HP or LP hydroponic culture for 21 days.For measuringArabidopsisbiomass (fresh weight), shoots and roots were excised from plants grown on media containing different Pi concentrations for 10 days.For each genotype,three independent measurements (biological repeats) were conducted, with each measurement using 10 different plants.
2.7.Measurement of Pi content
Fresh samples of shoots and roots were harvested for Pi determination following Zhou et al.[36].Specifically, the samples were homogenized in 100 μL 10%(w/v)perchloric acid with an ice-cold mortar and pestle.The homogenate was diluted 10-fold with 5%(w/v) perchloric acid and incubated on ice for 30 min.After centrifugation at 10,000×gfor 10 min at 4°C,the supernatant,as sample solution, was used for Pi measurement by the molybdenum blue method.Ammonium molybdate (0.4%, w/v) dissolved in 0.5 mol L-1H2SO4(solution A) was mixed with 10% ascorbic acid(solution B) in a ratio of 6:1, and the mixed reagent was used as the working solution.A 200-μL aliquot of this working solution was added to 100 μL of the sample solution and incubated at 42 °C for 20 min.The absorbance at 820 nm was measured using a SpectraMax M2e 96-well plate reader(Molecular Devices,Sunnyvale, CA, USA).Three independent measurements were performed for each genotype and each Pi treatment.
2.8.Anthocyanin measurement
Four-day-oldArabidopsisseedlings were placed in HP or LP medium for 7 days.Shoots were harvested for anthocyanin measurement following Shi et al.[13].Shoots (0.2 g for each sample)were treated with 1 mL of an extraction buffer,prepared by mixing methanol,hydrochloric acid,and water in the ratio 25:5:70(v:v:v),in the dark for 4 h, followed by centrifugation at 12,000×gfor 10 min.The supernatant was retained and its light absorbance was measured at 525 and 657 nm,with anthocyanin content calculated as (A525- 0.25 × A657) g-1fresh weight (FW).Each experiment was performed in three replicates, with shoots of 15 seedlings in each replicate.
2.9.Histochemical staining for ROS detection
To detect O2-in the roots of WT andTaMADS2-3DOEplants grown under HP or LP conditions,Arabidopsisseedlings were placed in tubes, immersed in 0.5 mg mL-1nitroblue tetrazolium(NBT) staining solution in 50 mmol L-1sodium phosphate buffer(pH 7.5),and held overnight at 28°C.After removal of the staining solution, seedlings were immersed in absolute ethanol, followed by heating in a boiling water bath for 5 min to remove chlorophyll pigments.Hydrogen peroxide in the roots was detected by 3,3′-diaminobenzidine (DAB) staining.Seedlings were placed in tubes and immersed in 1 mg mL-1DAB.After overnight incubation at 28 °C, the seedlings were preserved in absolute ethanol.The NBT and DAB staining signals in the primary root tips and the lateral root primordia of the stained seedlings were examined and imaged under a light microscope (Eclipse Ni-U, Nikon, Tokyo, Japan).
2.10.Gene structure and phylogenetic analysis
Genomic and CDS sequences ofTaMADS2-3Dand its homologous inT.aestivumandAegilops tauschiiwere obtained from EnsemblPlants(http://plants.ensembl.org/Triticum_aestivum/Info/Index) and Triticeae Multi-omics Center (http://202.194.139.32/getfasta/index.html) based on International Wheat Genome Sequencing Consortium Reference Sequence v 1.0 (IWGSC RefSeq v 1.0).The deduced amino acid sequences were aligned using ClustalW (https://www.genome.jp/tools-bin/clustalw) with default parameters.A neighbor-joining phylogenetic tree was reconstructed using MEGA 6.0 (https://www.megasoftware.net/).Tree nodes were tested by bootstrap analysis with 1000 replicates.
2.11.Statistical analysis
Statistically significant differences between means were determined by one-way ANOVA.Statistical analyses were performed with Microsoft Excel (Microsoft, Redwood, MS, USA) and PASW Statistics 18 (IBM, Inc., Chicago, IL, USA).
3.Results
3.1.TaMADS2-3D is induced at an early stage by low-Pi stress in wheat
To confirm whetherTaMADS2-3Dwas responsive to low-Pi stress, we used qRT-PCR to verify its expression level in ZM9023 roots with the primer set (CA658343_RT, Table S1) specific forTaMADS2-3D.As shown in Fig.1A, the expression level ofTaMADS2-3Dwas significantly down-regulated in roots under LP conditions in comparison with its expression under HP conditions.A similar expression profile was observed forTaMADS2using the primer set (TaMADS2_RT, Table S1) recognizingTaMADS2-3Das well as its two homoeologs(TaMADS2-3Aand-3B)(Fig.1B).Owing to the high nucleotide sequence similarity betweenTaMADS2-3Aand -3B, we were unable to design primer sets specific forTaMADS2-3Aand -3Bseparately.Consequently, we conducted a more detailed time course qRT-PCR analysis using the primer set TaMADS2_RT.TheTaMADS2transcript level was clearly induced at 3 h post-LP treatment before it was down-regulated at later time points in ZM9023 roots (Fig.1C).These results indicate that the expression ofTaMADS2-3D, as well as its two homoeologs(TaMADS2-3Aand -3B), was dynamically regulated by Pi availability.
3.2.TaMADS2-3D belongs to type I MADS-box subfamily
By searching against the genomic sequence of Chinese Spring in EnsemblPlants (http://plants.ensembl.org),TaMADS2-3Dwas found to be identical toTraesCS3D02G428000.1located on chromosome 3D.Its homoeolog on chromosome 3B(TraesCS3B02G470000.1), previously designatedTaAGL33[37], was renamedTaMADS2-3B, and its homoeolog on chromosome 3A(TraesCS3A02G435000.1)was designatedTaMADS2-3A(Fig.2).Phylogenetic analysis of deduced amino acid sequences placed TaMADS2-3D and homologous proteins, including AetMADS51-like, HvOS2, TuMADS56, OsMADS51, and OsMADS84, in the type I MADS-box subfamily (Fig.2A).TaMADS2-3Dshowed the highest similarity toAetMADS51-likeinAe.tauschiithat carries a wide range of resistance/tolerance factors responding to abiotic stresses,with the two genes having the same exon and intron structure with identical exon sizes (Fig.2B).The deduced protein sequence of TaMADS2-3D contained 159 amino acids and was identical to that ofAetMADS51-like(Fig.2C).Thus, we focused on analyzing the potential role ofTaMADS2-3Din plant response to low-Pi stress.
3.3.TaMADS2-3D located in the nucleus
To test whether TaMADS2-3D functions as a nucleus-localized TF,the TaMADS2-3D-GFP fusion protein was transiently expressed in tobacco leaf cells.In repeated examinations, the TaMADS2-3DGFP fusion protein was observed to reside in the nucleus,whereas GFP alone was distributed throughout the cell(Fig.3),thus demonstrating that TaMADS2-3D localized in cell nucleus.
3.4.Overexpressing TaMADS2-3D in Arabidopsis altered root architecture
To investigate the function ofTaMADS2-3D,we generated transgenicArabidopsislines overexpressing a GFP-tagged TaMADS2-3D fusion protein in the Col-0 ecotype.Alteration of RSA, as reduced PR length and increased LR number and density, is a major adaptive response to low-Pi stress inArabidopsis.To investigate whetherTaMADS2-3Dmodulates RSA inArabidopsis, we measured PR length, LR number, and LR density of twoTaMADS2-3DOElines(SM109 and SM110), with the WT Col-0 as control.Compared to the WT control, theTaMADS2-3DOElines showed reduced PR length but increased LR number and density under HP (625 μmol L-1Pi) conditions (Fig.4A-D).Under LP (10 μmol L-1Pi) conditions, PR length was substantially decreased in bothTaMADS2-3DOElines and the WT control,with the degree of decrease tending to be smaller in the transgenic plants than in the WT (Fig.4A, D).Number of LR was increased by LP treatment in the WT control,but reduced (P≤0.05) in SM109 (Fig.4B, D).LR density was elevated in bothTaMADS2-3DOElines and the WT control by LP treatment, with the degree of elevation tending to be reduced inTaMADS2-3DOEplants (Fig.4C, D).The root phenotypes ofTaMADS2-3DOElines under HP conditions, including decreased PR length and increased LR number and density, resembled the typical RSA changes induced by low-Pi stress,suggesting that overexpressingTaMADS2-3DinArabidopsisled to constitutive activation of PSRs.Under LP conditions, the WT plants showed the anticipated RSA changes:shortening of PR length and increases in LR number and density.TheTaMADS2-3DOEplants behaved like the WT control with respect to PR length and LR density,but their LR number did not increase as expected.Thus,low-Pi induced RSA changes were partly impaired inTaMADS2-3DOEplants, possibly owing to the presence of constitutive PSRs in these plants.Collectively, these findings lend support for the regulation of PSRs byTaMADS2-3Din plants, but artificially overexpressingTaMADS2-3Daltered RSA under either HP or LP conditions.
3.5.Arabidopsis TaMADS2-3DOE plants showed retarded vegetative growth
The effects of overexpressingTaMADS2-3Don plant vegetative growth were further investigated using theTaMADS2-3DOElines.The fresh weight (biomass) of shoots and roots of theTaMADS2-3DOEand WT seedlings grown on solid medium containing 0.01 to 5 mmol L-1Pi for 10 days were measured.As expected, shoot biomass decreased with exogenous Pi concentration in both WT andTaMADS2-3DOEplants (Fig.4E).The biomass ofTaMADS2-3DOEplants (shoots and roots) was generally lower than that of WT plants with exogenous Pi concentrations of 0.05 to 5 mmol L-1(Fig.4E, F).However, there was no difference betweenTaMADS2-3DOEand WT plants under the low-Pi condition(0.01 mmol L-1).Thus, ectopic expression ofTaMADS2-3Dcaused a decrease in both shoot and root biomass ofArabidopsisrelative to the WT control, except at low Pi.Thus, overexpressingTaMADS2-3Dimpaired the growth of both roots and shoots of plants.

Fig.1.Expression changes of TaMADS2-3D and TaMADS2 in wheat under HP and LP conditions.(A,B)Expression changes of TaMADS2-3D and TaMADS2 in roots of ZM9023.Wheat seedlings germinated for 7 days under normal conditions were transferred to modified hydroponic solutions containing 200 μmol L-1 Pi(HP)or 10 μmol L-1 Pi(LP),followed by culture for another 7 days.Their roots were then harvested for RNA extraction and qRT-PCR analysis with the primer sets CA658343_RT(A)or TaMADS2_RT(B).(C)Time course of TaMADS2 expression level changes in roots of ZM9023.Wheat seedlings were subjected to LP treatment as described in(A,B),with root samples collected at 0, 3, 6, 9, 12, and 24 h for RNA extraction and qRT-PCR analysis with the primer set TaMADS2_RT.Error bars indicate SD (three biological replicates).Asterisks indicate means statistically different at P ≤0.01 (one-way ANOVA followed by LSD and Duncan tests).
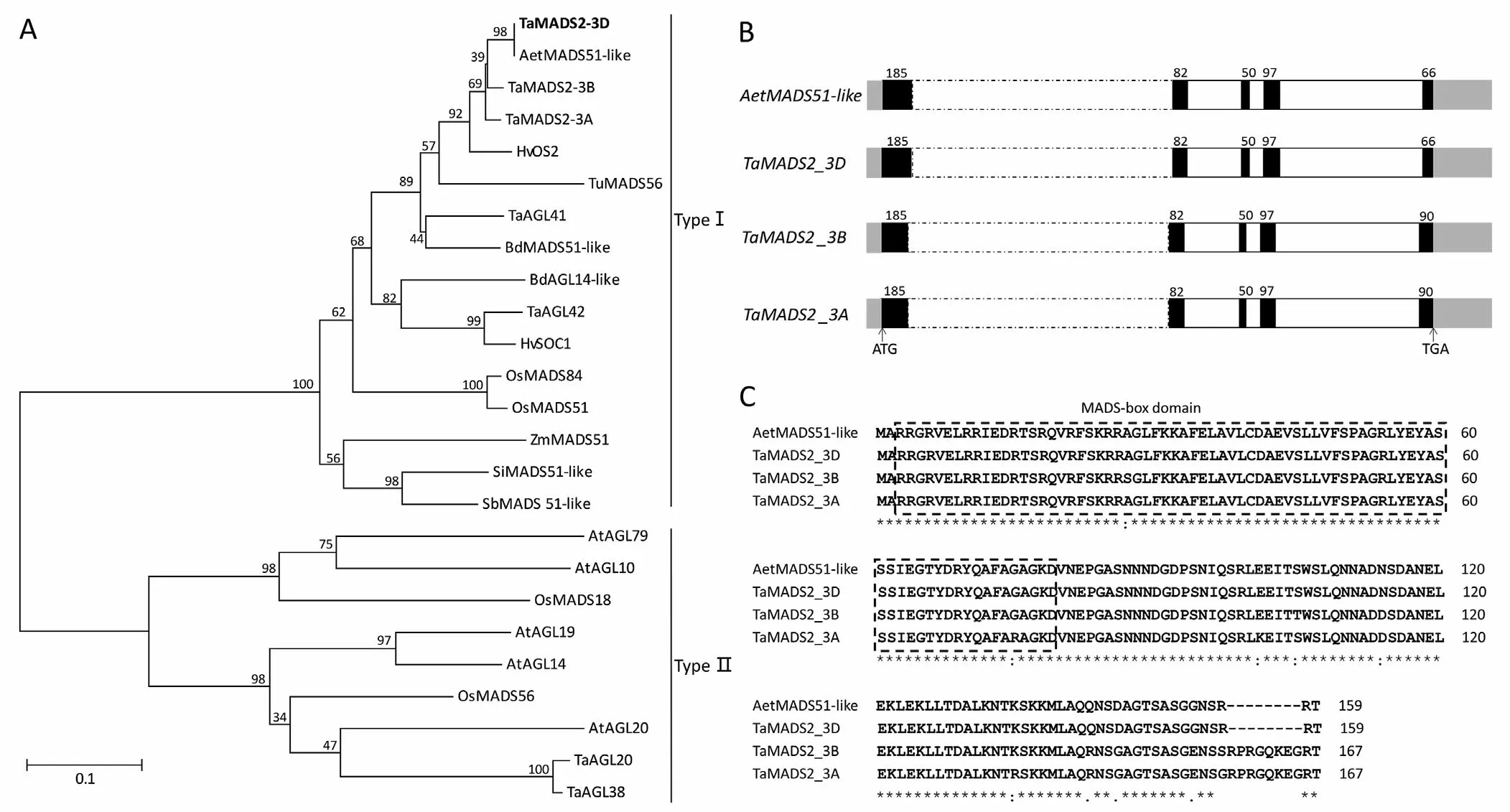
Fig.2.Phylogenetic analysis of TaMADS2-3D,exon-intron structures of TaMADS2-3D and homologous genes,and protein sequence alignment of TaMADS2-3D and homologs.(A) Phylogenetic tree of TaMADS2-3D and related MADS-box proteins.The neighbor-joining method was used to construct a phylogenetic tree of MADS-box proteins from Triticum aestivum(Ta),T.urartu(Tu),Aegilops tauschii(Aet),Hordeum vulgare(Hv),Oryza sativa(Os),Zea mays(Zm),Brachypodium distachyon(Bd),Setaria italica(Si),Sorghum bicolor (Sb), and Arabidopsis thaliana (At).Numbers at branch points represent the bootstrap value supporting each branch.(B) Exon-intron structures of TaMADS2-3D and homologous genes.Exons and introns are represented as black and white boxes,respectively.Numbers are numbers of nucleotides in each exon.Dotted white boxes indicate extremely long introns.ATG and TGA are start and termination codons,respectively.(C)Amino acid sequence alignment of TaMADS2-3D and homologous proteins performed by ClustalW.The sequences in dotted box represent the MADS-box domain predicted at NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
3.6.Pi content was increased in Arabidopsis TaMADS2-3DOE plants
To investigate whetherTaMADS2-3Dregulates the uptake or distribution of Pi, Pi contents in the shoots and roots of theTaMADS2-3DOEand the WT plants were measured.Pi content increased in shoots (by 49%-58%) and roots (by 37%-74%) ofTaMADS2-3DOElines relative to the contents of the WT control under HP conditions (Fig.4G).However, under LP conditions, an increase in Pi content in theTaMADS2-3DOEshoots and roots was not significant, and the amplitude of variation in Pi content betweenTaMADS2-3DOEand WT plants was narrow compared with that under HP conditions(Fig.4H).Nor was there a significant difference betweenTaMADS2-3DOEand WT plants in the root-toshoot Pi content ratio (Piroot/Pishoot) under HP or LP conditions.Thus,overexpressingTaMADS2-3Dincreased Pi uptake inArabidopsisseedlings on HP medium,consistent with the foregoing suggestion of constitutive PSRs inTaMADS2-3DOEplants.OverexpressingTaMADS2-3Dalso promoted Pi uptake by roots ofTaMADS2-3DOEplants under LP conditions,although the elevated root Pi content ofTaMADS2-3DOEplants did not lead to an increase of Pi accumulation in their shoots (Fig.4H).Thus, overexpression ofTaMADS2-3Dincreased Pi uptake by roots but not translocation from root to shoot inArabidopsis.
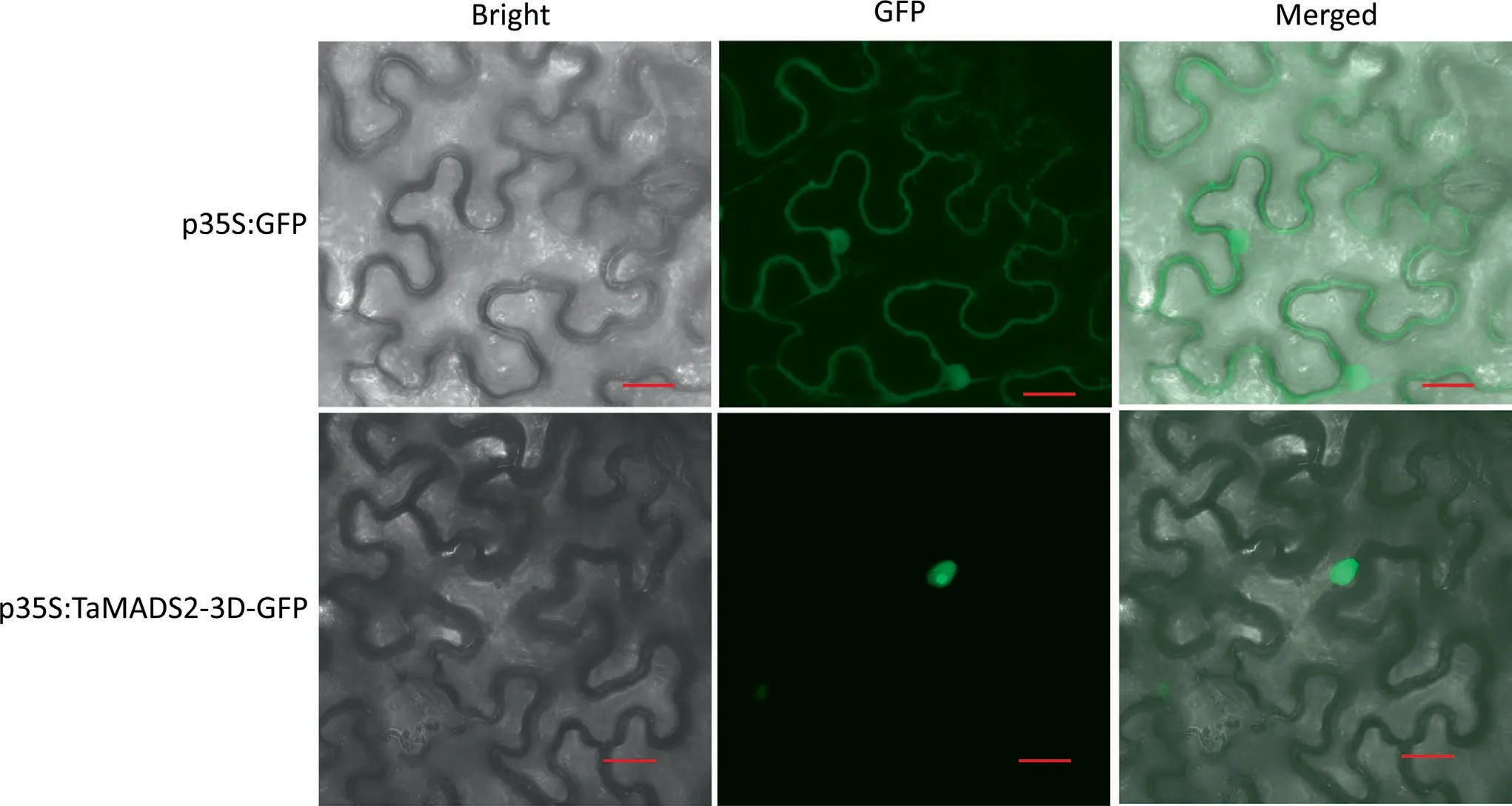
Fig.3.Subcellular localization of TaMADS2-3D.Confocal images of tobacco epidermal cells expressing TaMADS2-3D-GFP fusion protein or free GFP that served as a control.TaMADS2-3D-GFP and GFP were expressed from the constructs p35S:TaMADS2-3D-GFP and p35S:GFP,respectively.The data shown are representative of two independent experiments.Scale bars, 20 μm.
3.7.Transcriptomic comparison of Arabidopsis TaMADS2-3DOE and WT plants
To investigate the mechanism underlying the regulation of PSRs byTaMADS2-3D, the root transcriptomes ofTaMADS2-3DOElines and WT control were subjected to RNA sequencing, with changes in gene expression identified.Three comparisons were made:(1)TaMADS2-3DOE(OE) (HP) vs.WT (HP) for identifying DEGs inTaMADS2-3DOEroots under HP conditions, (2) WT (LP) vs.WT(HP) for identifying DEGs regulated by LP treatment in WT roots,and (3) OE (LP) vs.OE (HP) for identifying DEGs regulated by LP treatment inTaMADS2-3DOEroots(Fig.5A-C).A total of 206 DEGs were found in the group of OE(HP)vs.WT(HP),among which 117 and 89 genes were significantly up- or down-regulated (Fig.5A;Table S2).A set of 1028 DEGs was identified by comparing WT(LP) vs.WT (HP), among which 675 and 353 genes were significantly up- or down-regulated (Fig.5A; Table S3).A set of 1043 DEGs were identified in the comparison of OE (LP) vs.OE (HP),among which 793 and 250 genes were significantly up- or downregulated(Fig.5B;Table S4).As represented by Fig.5C,many genes that were highly expressed in WT(HP)roots were repressed in WT(LP),OE(HP)and OE(LP)roots.The WT(LP)and OE(LP)roots had the highest number of genes induced by LP treatment, but the OE(LP)roots contained a group of genes uniquely and highly induced by low-Pi(G1 in Fig.5C).Some of the highly expressed genes in OE(HP) roots overlapped with the low-Pi induced genes in WT (LP)and OE (LP) roots, but again a small group of genes (G2 in Fig.5C) were uniquely and highly expressed in OE (HP) roots.These results indicate that PSR genes are normally regulated in WT (LP) roots, many PSR genes are similarly regulated in OE (LP)and WT (LP) roots, and some PSR genes are misregulated in the OE(HP)roots,given that they are highly expressed in the presence of sufficient Pi.Overexpression ofTaMADS2-3Dactivated the expression of a group of non-PSR genes in OE (HP) roots (G2 in Fig.5C),and increased the expression of a group of weakly induced PSR genes in OE(LP)roots(G1 in Fig.5C).Thus,in both OE(HP)and OE(LP)roots,there were uniquely and highly expressed genes not found in the WT(HP)or the WT(LP)roots.These unique genes may have been directly or indirectly activated by the overexpressedTaMADS2-3Dunder HP or LP conditions.
The DEGs obtained by comparing OE (HP) vs.WT (HP) were subjected to GO analysis.Five biological processes were markedly enriched, such as secondary metabolic process, response to oxidative stress,response to karrikin,phosphate ion transport,and inorganic anion transport(Fig.5D).This GO enrichment result suggests the activation of PSR genes and processes inTaMADS2-3DOElines under HP conditions, in agreement with the morphological and physiological characteristics ofTaMADS2-3DOEplants under HP conditions, which are similar to those of WT plants subjected to LP treatment(Fig.4).In agreement with this suggestion,five genes functioning in Pi transport and RSA modification under low-Pi conditions:AtPHT1;8,AtPHT2;1,AtETC1,AtLPR1, andAtZAT6, were highly expressed inTaMADS2-3DOEplants cultured with high Pi supply (Fig.5E), supporting the occurrence of constitutive activation of PSRs byTaMADS2-3Doverexpression inArabidopsis.
3.8.Additional gene expression changes caused by overexpressing TaMADS2-3D in Arabidopsis
In addition to the PSR genes, the DEGs identified by comparing OE (HP) vs.WT (HP) root transcriptomes included genes involved in the processes of ion transport and balance and of sugar and malate transport inArabidopsis(Table S5).The expression levels of genes involved in redox process and flavonoid biosynthesis were also changed (Fig.6A, B).Thus,TaMADS2-3Dactivated manyArabidopsisgenes when overexpressed in transgenic plants.

Fig.4.Overexpressing TaMADS2-3D affects the growth and Pi content of Arabidopsis.(A-C)Comparisons of primary root length(A),lateral root number(B),and lateral root density (C) of Arabidopsis TaMADS2-3DOE (SM109, SM110) and WT plants.The measurement was made after seedlings were transferred to solid MS media containing 625 μmol L-1 Pi (HP) or 10 μmol L-1 Pi (LP) for 7 days.(D) Phenotypes of Arabidopsis TaMADS2-3DOE and WT plants.The plots shown were made 7 days after transfer of seedlings to HP or LP media.Scale bars, 1 cm.(E, F) Fresh weight of shoots and roots in Arabidopsis TaMADS2-3DOE and WT plants under different Pi conditions.(G, H) Pi contents in shoots and roots of Arabidopsis TaMADS2-3DOE and WT plants under HP and LP conditions.Error bars indicate SD (three biological replicates, with at least 15 seedlings analyzed per genotype per experiment) and asterisks indicate means statistically different at P ≤0.05 (one-way ANOVA followed by LSD and Duncan tests).
Several genes involved in redox process were upregulated in the roots ofArabidopsis TaMADS2-3DOEplants under the HP conditions(Fig.6A).These genes may directly or indirectly affect ROS homeostasis, which influences root development.For example, the expressions ofAtTH8andAtTH7, encoding thioredoxins involved in ROS homeostasis, were upregulated (Fig.6A).Likewise,AtGSTU17andAtGSTU20, encoding glutathione S-transferases (GSTs),were induced inTaMADS2-3DOEplants under HP conditions(Fig.6A).
Six genes involved in flavonoid biosynthesis showed increased expression levels in the roots ofTaMADS2-3DOEplants under HP conditions (Fig.6B).Among them,AtTT4,AtTT5,AtTT7, andAtFLSencode key enzymes governing flavonoid accumulation in plant cells.Their function increases cellular flavonoid content, thereby affecting ROS homeostasis and plant stress responses.It can be also observed that the anthocyanin content was markedly increased inTaMADS2-3DOEplants grown with high Pi supply (Fig.S1).
3.9.Overexpressing TaMADS2-3D increased H2O2 and O2- production in Arabidopsis roots
Levels of hydrogen peroxide (H2O2) and superoxide (O2-) in roots of WTArabidopsisandTaMADS2-3DOEplants were measured under HP or LP conditions.H2O2and O2-were increased in primary root tips ofTaMADS2-3DOEplants under HP conditions, but this increase was not evident under LP conditions (Fig.6C).The level of H2O2in LR primordia showed no significant changes, and the level of O2-was slightly decreased in LR primordia ofTaMADS2-3DOEplants.These observations suggest an association between the inhibition of PR growth byTaMADS2-3Doverexpression and an increased accumulation of H2O2and O2-in PR root tips under HP conditions.
3.10.Overexpression of TaMADS2-3D in wheat also inhibited plant growth
Transgenic wheat overexpressingTaMADS2-3Dusing the maize ubiquitin gene promoter was generated,and the growth dynamics of three independent overexpression lines (CM5, 29, and 15) in hydroponic culture were observed.TaMADS2-3Dexpression level was strongly increased in the three lines (Fig.7A).Under either HP or LP conditions, the growth parameters ofTaMADS2-3D-overexpression plants,including plant height,root length,and biomass of root and shoot, were all significantly decreased in comparison with those of the WT controls(P< 0.05)(Fig.7B-E).The total root number of overexpressing lines did not markedly differ from that of the WT control (Fig.7F).No significant difference in Pi content was found among theTaMADS2-3D-overexpression plants and the WT plants under either HP or LP conditions (Fig.7G, H).However, Pi contents in the shoot tissues of overexpressing plants tended to be higher than those determined for the WT controls in the HP environment (Fig.7G, H).Thus, overexpression ofTaMADS2-3Din wheat also resulted in growth inhibition and some increase in Pi uptake under HP conditions, a tendency similar to that inArabidopsislines overexpressingTaMADS2-3Dcultured with sufficient Pi.
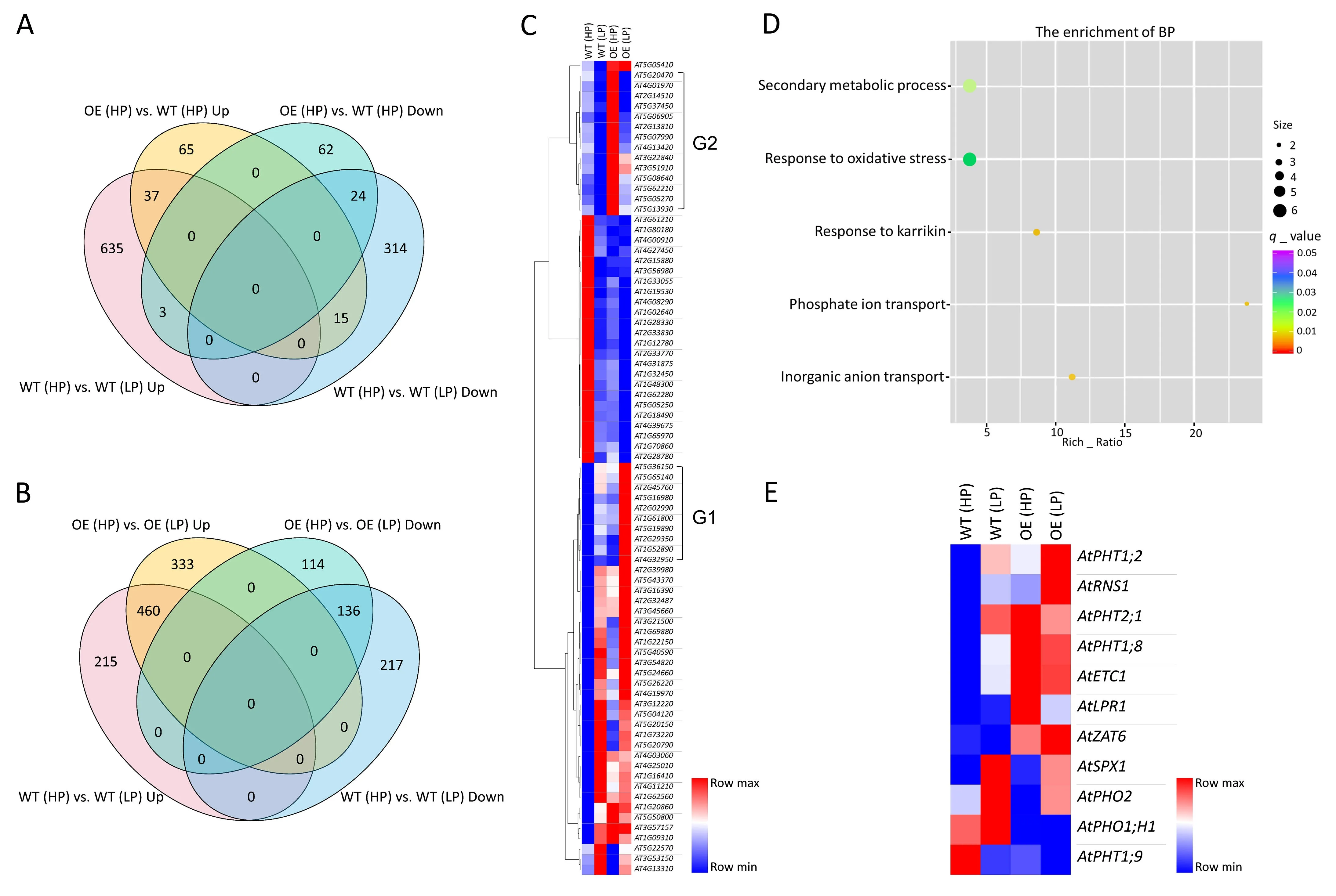
Fig.5.Transcriptomic analysis and expression heat map of differentially expressed genes(DEGs)in Arabidopsis TaMADS2-3DOE and WT roots.(A,B)Venn diagrams showing the DEGs obtained in the comparison of TaMADS2-3DOE(OE)(HP)vs.WT(HP)or WT(LP)vs.WT(HP)(A)and the DEGs computed for OE(LP)vs.OE(HP)or WT(LP)vs.WT(HP)(B).The words‘‘Up”and‘‘Down”refer to the numbers of up-or down-regulated DEGs.(C)Overlap of up-or down-regulated genes among the roots of WT(HP),WT(LP),OE (HP), and OE (LP) seedlings.G1 and G2 indicate the two groups of genes highly up-regulated in only OE (LP) roots or in the roots of OE (HP).(D) Significantly enriched biological processes computed for the DEGs obtained in the comparison of OE(HP)vs.WT(HP).(E)Expression heat map of known PSR genes among the roots of WT(HP),WT(LP), OE (HP), and OE (LP) seedlings.
4.Discussion
4.1.TaMADS2-3D regulates plant PSRs
The information on the genetic and molecular mechanisms controlling PSRs is still limited in crop plants, especially in wheat,although it has been reported that plant PSRs were modulated by multiple genes via different pathway [38,39].We suggest thatTaMADS2-3Dacts in the regulation of PSRs.The most compelling evidence for this suggestion is that overexpression ofTaMADS2-3DinArabidopsisled to constitutive activation of PSRs under HP conditions.Arabidopsis TaMADS2-3DOEplants not only showed morphological and developmental phenotypes (shortened PR,increased LR number and density, retarded plant growth, and increased anthocyanin content) resembling those of PSRs but also accumulated more Pi in their root cells.Furthermore, many PSR genes were activated inArabidopsis TaMADS2-3DOEplants under HP conditions.These concurrent changes suggest that PSRs are fully activated inArabidopsis TaMADS2-3DOEplants cultured with HP supply.OverexpressingTaMADS2-3Dalso activated wheat PSRs constitutively, as the transgenic lines showed inhibited shoot and root growth and increased Pi accumulation in its cells under HP conditions.Pi contents increased in shoot tissues of transgenic lines under HP conditions.Though not significant, they were consistent across the two experiments.Thus,TaMADS2-3Dis likely a functional regulator of plant PSRs and could act at an upstream node of the Pi signaling pathway.Otherwise it may not be able to activate PSRs at a large scale when overexpressed on its own.
The expression profiles ofTaMADS2-3DandTaMADS2in wheat under LP conditions appear consistent with the above suggestion thatTaMADS2-3Dacts at an upstream position of the plant Pi signaling pathway.TaMADS2-3D, and probably its homoeologsTaMADS2-3AandTaMADS2-3B, may be required for the initial transduction of low-Pi signal and the early phase of Pi signalling,given thatTaMADS2expression was up-regulated at the early stage of low-Pi stress:3 h post-low-Pi treatment (Fig.1C).TaMADS2expression needs to be down-regulated at the later stages of low-Pi stress, possibly because excessive function ofTaMADS2may lead to excessive low-Pi signalling and uncontrolled abnormal PSRs.In this context, it is understandable why overexpression ofTaMADS2-3DinArabidopsisand wheat results in constitutive PSRs and inhibition of plant growth in the absence of low-Pi stress.However, further work is needed to determine whether overexpression ofTaMADS2-3AorTaMADS2-3Bcould also lead to constitutive PSRs in plants and whether the three homoeologs ofTaMADS2differ in their regulation of plant PSRs.

Fig.6.Altered expression of genes involved in redox process and flavonoid biosynthesis and levels of hydrogen peroxide (H2O2)and superoxide(O2-)in roots of Arabidopsis TaMADS2-3DOE plants and WT controls.(A)Expression heat map of genes involved in redox process among the roots of WT(HP),WT(LP),OE(HP),and OE(LP)seedlings.(B)Expression heat map of genes involved in flavonoid biosynthesis among the roots of WT(HP),WT(LP),OE(HP),and OE(LP)seedlings.(C)Results of DAB and NBT staining in LRs and PRs of TaMADS2-3DOE lines(SM109 and SM110)and the WT control cultured on HP(625 μmol L-1)or LP(10 μmol L-1)media for 7 days.Intensities of DAB and NBT staining reflect levels of H2O2 and O2-, respectively.Scale bars, 100 μm.
Although many genes have been reported to regulate PSRs in model and crop plants [40-43], to our knowledge, no other genes have been shown to activate PSRs when overexpressed alone inArabidopsisand wheat.However, the effects of overexpressingTaMADS2-3Don PSR phenotypes, especially the changes in root system parameters and Pi content, were similar but not identical inArabidopsisand wheat, indicating that the function ofTaMADS2-3Din PSR regulation differs to some extent among species.This difference may be associated with the differing root systems:Arabidopsishas a taproot and wheat a fibrous root system.Still,TaMADS2-3Dis likely unique among PSR regulators in being able to confer a range of PSR phenotypes when overexpressed on its own in both the dicot speciesArabidopsisand the monocot plant wheat.
4.2.Potential molecular mechanism underlying TaMADS2-3D function
The transcriptome comparisons generated in this study lead to several suggestions about the molecular mechanism involved inTaMADS2-3Dfunction.First, TaMADS2-3D acts as a nucleuslocated TF and directs the expression of a large number of target genes including both PSR and non-PSR genes.This is evident from a comparison of genes highly expressed among the roots of OE(HP), WT (LP) and OE (LP)Arabidopsisplants (Fig.5).Second, ROS homoeostasis may contribute to the inhibition of PR growth by overexpressedTaMADS2-3D.This proposition is supported by the association among shortening of PR inArabidopsis TaMADS2-3DOEplants, elevated hydrogen peroxide and superoxide levels,and increased expression of genes functioning in ROS metabolism in the tip region of PR on HP medium.Several studies[44-47]have shown that disrupting ROS homoeostasis exerts profound effects onArabidopsisroot development under either normal or stress conditions.Of particular interest is theUBP1gene encoding a bHLH TF,whose overexpression up-regulates hydrogen peroxide and superoxide accumulation in root tips and results in strong inhibition ofArabidopsisPR growth under normal growth conditions [48].WhetherTaMADS2-3Doverexpression inArabidopsisleads to increased expression and function ofUBP1awaits further research.Finally, increasedAtZAT6expression is probably linked to the elevated flavonoid and anthocyanin levels inArabidopsis TaMADS2-3DOElines, given thatAtZAT6, induced during Pi deprivation, is a repressor of PR growth[12],and has been shown[13]to be essential for ROS-stimulated accumulation of flavonoids and anthocyanins inArabidopsis.
Considering that the gene expression profiles ofArabidopsis TaMADS2-3DOEplants differed substantially from those of WT plants under either HP or LP conditions and that PSRs were partially impaired inTaMADS2-3DOEplants under LP supply, the molecular and biochemical events followingTaMADS2-3Doverexpression may be more complicated than suggested above.The gene expression changes, as well as the phenotypes, ofArabidopsis TaMADS2-3DOEplants may not be simply attributed to PSRs,given that overexpressingTaMADS2-3Dcould increase the expression of non-PSR genes(G2)as seen in OE(HP)roots(Fig.5C).A full understanding of the biological processes activated byTaMADS2-3Doverexpression will depend on systematic identification of genes regulated by TaMADS2-3D transcriptional activity.

Fig.7.Analyses of transgenic wheat lines overexpressing TaMADS2-3D.(A)Expression levels of TaMADS2-3D in three independent transgenic wheat lines(CM5,29,and 15)and the WT control(cv.Fielder)measured by qRT-PCR.Values are typical of three biological replicates.(B-F)Comparisons of plant height(B),root length(C),fresh weight of shoots (D) and roots (E) and total number of roots (F) between transgenic wheat lines and the WT control.(G, H) Pi contents of shoots (G) and roots (H) for the transgenic wheat and WT plants grown under HP(200 μmol L-1)or LP(10 μmol L-1)conditions.(I)Phenotypes of transgenic wheat and WT plants hydroponically cultured for 21 days under HP or LP conditions.Wheat seedlings were germinated for 7 days before being used for hydroponic culture.The hydroponic culture trial was repeated twice.Scale bars,5 cm.In (B) to (H), error bars indicate SD (n = 3, with at least 10 seedlings used per genotype per experiment in each analysis), and asterisks indicate means statistically different at P ≤0.05 (one-way ANOVA followed by LSD and Duncan tests).
4.3.Implications for future research
We suggest the following directions for future research.First,identification of the target genes of TaMADS2-3D could be achieved by chromatin immunoprecipitation sequencing.Second,the mechanism behind the elicitation of ROS accumulation in plant roots by overexpressionTaMADS2-3Dand its involvement in the activation of PSRs should be clarified.This will require the preparation and analysis of more genetic materials, such as knockout mutants ofTaMADS2-3D.The use ofArabidopsismay facilitate such research, given that the function ofTaMADS2-3Dappears to be conserved to some extent betweenArabidopsisand wheat, based on the observation that overexpressedTaMADS2-3Dconfers a similar set of PSR phenotypes.Finally,the value ofTaMADS2-3Din improving the P use efficiency of wheat plants could be studied.Although overexpression ofTaMADS2-3Dusing a strong promoter (pUbi) inhibited plant growth, it enabled wheatTaMADS2-3DOEplants to absorb more Pi from the environment, a trait valuable for maintaining wheat growth under LP conditions.To ameliorate the undesirable effects ofTaMADS2-3Doverexpression, a low-Pi inducible promoter could be employed to driveTaMADS2-3Dexpression in transgenic plants.Alternatively, allelic variants ofTaMADS2-3Dmay be sought in wheat and related species.Allele(s) permitting more Pi absorption but not growth inhibition could be identified and used in breeding P-efficient wheat lines.
5.Conclusions
In summary, we have identified a MADS-box gene,TaMADS2-3D, whose ectopic expression leads to constitutive PSRs.This gene is likely to be valuable for further elucidating the molecular mechanism controlling plant PSRs and may represent a useful target for improving the P-use efficiency of wheat cultivars.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Yingchun Hanperformed acquisition of data as well as analysis and interpretation of data and drafted the manuscript.Na Liuperformed data analysis and drafted the manuscript.Chuang Li,Shuaiwu Wang,Lihua Jia,Rui Zhang,and Hui Lihelped in preparing materials and discussions,Wenming Zheng, Jinfang Tan, and Hongwei Xuewere responsible for conception and design as well as analysis and interpretation of data, and revised the manuscript.
Acknowledgments
This study was supported by the National Key Research and Development Program of China (2018YFD0200600), the Natural Science Foundation of Henan Province (182300410023), and the State Key Laboratory of Wheat and Maize Crop Science of China.We thank Prof.Daowen Wang for constructive suggestions and proofreading this manuscript.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.03.020.
杂志排行
The Crop Journal的其它文章
- Brief Guide for Authors
- Heavy soil drying during mid-to-late grain filling stage of the main crop to reduce yield loss of the ratoon crop in a mechanized rice ratooning system
- GmNMHC5 may promote nodulation via interaction with GmGAI in soybean
- H2O2 mediates transcriptome reprogramming during Soybean mosaic virus-induced callose deposition in soybean
- Contrasting patterns of accumulation,partitioning,and remobilization of biomass and phosphorus in a maize cultivar
- Calcineurin B-like protein 5 (SiCBL5) in Setaria italica enhances salt tolerance by regulating Na+ homeostasis
