Calcineurin B-like protein 5 (SiCBL5) in Setaria italica enhances salt tolerance by regulating Na+ homeostasis
2022-02-19JingweiYnLnYngLiuYingdiZhoTongHnXingfenMioAyingZhng
Jingwei Yn, Ln Yng, Y Liu, Yingdi Zho, Tong Hn, Xingfen Mio, Aying Zhng,b,*
a College of Life Sciences, Nanjing Agricultural University, Nanjing 210095, Jiangsu, China
b State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing 210095, Jiangsu, China
c College of Agriculture, Heilongjiang Bayi Agricultural University, Daqing 163319, Heilongjiang, China
Keywords:Setaria italica SiCBL5 Salt tolerance Na+ homeostasis
ABSTRACT Salinity,a major abiotic stress,reduces plant growth and severely limits agricultural productivity.Plants regulate salt uptake via calcineurin B-like proteins(CBLs).Although extensive studies of the functions of CBLs in response to salt stress have been conducted in Arabidopsis,their functions in Setaria italica are still poorly understood.The foxtail millet genome encodes seven CBLs,of which only SiCBL4 was shown to be involved in salt response.Overexpression of SiCBL5 in Arabidopsis thaliana sos3-1 mutant rescued its salt hypersensitivity phenotype, but that of other SiCBLs (SiCBL1, SiCBL2, SiCBL3, SiCBL6, and SiCBL7) did not rescue the salt hypersensitivity of the Atsos3-1 mutant.SiCBL5 harbors an N-myristoylation motif and is located in the plasma membrane.Overexpression of SiCBL5 in foxtail millet increased its salt tolerance,but its knockdown increased salt hypersensitivity.Yeast two-hybrid and firefly luciferase complementation imaging assays showed that SiCBL5 physically interacted with SiCIPK24 in vitro and in vivo.Cooverexpression of SiCBL5, SiCIPK24, and SiSOS1 in yeast conferred a high-salt-tolerance phenotype.Compared to wild-type plants under salt stress conditions,SiCBL5 overexpressors showed lower accumulations of Na+ and stronger Na+ efflux, whereas RNAi-SiCBL5 plants showed higher accumulations of Na+and weaker Na+ efflux.These results indicate that SiCBL5 confers salt tolerance in foxtail millet by modulating Na+ homeostasis.
1.Introduction
Salinity is an abiotic stress that reduces plant growth and severely limits agricultural productivity [1].Na+is the predominant ion in most saline soils.Excess cytosolic Na+absorbed by plants from saline soil is toxic to several plant metabolic processes.Plants have evolved various adaptation strategies to prevent or reduce cytosolic Na+accumulation in response to salt stress [1,2].The calcium (Ca2+)-dependentsalt overly sensitive(SOS) pathway is a central player in modulating ion homeostasis, as established by molecular and genetic characterization of several salt hypersensitivity mutants inArabidopsis.Salt stress causes a rapid increase in cytosolic Ca2+that can be perceived by calcium-binding proteins such as SOS3.SOS3 recruits the downstream kinase SOS2 to the plasma membrane and increases its kinase activity.The SOS2-SOS3 complex then activates a Na+/H+exchanger protein in the plasma membrane,SOS1.Finally,SOS1 pumps Na+out of the plant cell to prevent the damage caused by excess cytosolic Na+[2-5].
SOS3 belongs to the family of calcineurin B-like proteins (CBL),which harbor EF-hand motifs that sense Ca2+[6].Several CBLs have been implicated in regulation of salt stress in species includingArabidopsis, rice, maize, poplar, and sorghum [7-12].In response to salt stress inArabidopsis, AtCBL4/AtSOS3 functions mainly in roots and AtCBL10/AtSCaBP8 in shoots [13-16].Loss of function ofAtCBL1andAtCBL9increases the salt hypersensitivity ofArabidopsisseedlings, whereas overexpression ofAtCBL1andAtCBL5increases their salt tolerance [17-20].AtCBL10 is vital for reproductive development under salt stress [21].In poplar, PtSOS3 shows functional similarity to AtCBL4/AtSOS3 [22].PtCBL10a and PtCBL10b regulate shoot salt tolerance by modulating ion homeostasis [23].These studies suggest the complexity and diversity of CBLs in multiple plant species under salt stress.
Foxtail millet (Setaria italica) is a diploid C4grass and has a small genome (of about 500 MB) that makes it attractive as a model system [24,25].It is widely grown in northern China and some Asian countries.Despite comprehensive studies of CBLs inArabidopsis,little is known about the roles of CBLs in foxtail millet.Recently [26], overexpression ofSiCBL4inAtsos3-1mutant was shown to rescue the hypersensitivity of anAtsos3-1mutant.Although the foxtail millet genome encodes seven CBLs [27], the functions of additional CBLs in response to salt stress remain to be determined.In this study,we searched for additional SiCBLs that could rescue the salt hypersensitivity of theAtsos3-1mutant.
2.Materials and methods
2.1.Plant materials and growth conditions
The foxtail millet inbred line Yugu 1 and tobacco(Nicotiana benthamianaL.)were used.Seeds were sown in pots containing a mixture of 1/2 soil and 1/2 vermiculite (v/v) and grown in a growth chamber under conditions of 25 °C, 200 μmol m-2s-1, 14 h light/10 h dark,and 60%humidity.One-week-old seedlings grown in pots were treated with 350 mmol L-1NaCl for various times,and samples were collected and immediately frozen in liquid nitrogen.
Arabidopsis thaliana(L.) Heynh.ecotype Columbia (Col-0) and theAtsos3-1mutant (CS3864) were used.Seeds were sterilized and sown on solid medium containing 1/2 Murashige and Skoog salts including vitamins and 1%(w/v)sucrose at 4°C for two days,and then grown in a growth chamber (22 °C, 100-200 μmol m-2s-1, 14 h light/10 h dark, 60% humidity).
2.2.Isolation of total RNA and real-time PCR analysis
Total RNA was isolated from plant materials as described by Zhang et al.[28].Real-time PCR was performed using EvaGreen 2× qPCR MasterMix-No Dye (Applied Biological Materials (abm)Inc.,Canada)according to the manufacturer’s instructions.Expression levels of candidate genes were measured using the 2-ΔΔCT method withSiActin 7orAtActin 2as described previously [29].Data were collected from three repeats.
2.3.Foxtail millet and Arabidopsis transformation and regeneration
To generateSiCBL5overexpressors, full-lengthSiCBL5was amplified by PCR using specific primers (Table S1) and was introduced into the plant expression vector pCUN-NHF driven by theZea mays polyubiquitin-1promoter.To obtainSiCBL5-knockdown plants,the fragment ofSiCBL5from 1 to 250 bp was cloned in both the sense and antisense directions in the vector RNAi vector driven by theZea mays polyubiquitin-1promoter using specific primers(Table S1).Yugu 1 was used as the plant receptor.Agrobacteriummediated foxtail millet shoot-tip transformation [30] was used.Positive transformants were selected by spraying with 25 μg mL-1DL-phosphinothricin (Sigma-Aldrich, USA), and then further confirmed by PCR amplification.The T2seedlings showed a resistantto-susceptible segregation of 3:1, and resistant seedlings were transferred to pots for continuous cultivation until homozygous T3seeds from individual lines were obtained.
The full-lengthSiCBLswere amplified by PCR using specific primers (Table S1) and cloned into the pEarleyGate 101 vector.The recombinant plasmid was introduced toAtsos3-1mutant byAgrobacterium tumefaciensstrain GV3101-mediated transformation as described [31].Positive transformants were selected on 1/2 MS medium containing 25 μg mL-1DL-phosphinothricin.The T2seedlings showed resistant-to-susceptible segregation of 3:1,and the resistant seedlings were transferred to pots for continuous cultivation until homozygous T3seeds were obtained from individual lines.
2.4.Subcellular localization of SiCBLs
Four-week-oldN.benthamianaleaves were coinfiltrated withA.tumefaciensstrain GV3101 carrying the35S-SiCBLs-YFPfused construct and theAtPIP2A-mCherrymarker (pm-rk) or γ-TIP-mCherrymarker (vac-rk) as described [32,33].The samples were mounted on glass slides and examined under Leica TCS SP8 confocal laser scanning microscope (TCS SP8, Leica (Microsystem), Germany)with the following parameters:YFP, excitation at 513 nm and emission at 518-582 nm;mCherry,excitation at 587 nm and emission at 598-630 nm.
2.5.Yeast test
To test the function of SiCBL5, theSaccharomyces cerevisiaemutant strain AXT3K, which lacks the main plasma membrane Na+transporters, was used [16,34].The full-length ofSiCBL5andSiCIPK24was separately cloned into pGADT7 and p414-GPD and the full-lengthSiSOS1was cloned into p416-GPD using specific primers (Table S1).These constructs were introduced into AXT3K using the PEG/LiAc method.Dilutions of various positive transformed cells and untransformed cell were then spotted onto AP medium with or without 100 or 150 mmol L-1NaCl [16].
2.6.Firefly luciferase complementation imaging (LCI) and yeast twohybrid assays
Full-lengthSiCBL5andSiCIPK24were cloned into pCAMBIA1300-cLUC and pCAMBIA1300-nLUC vectors, respectively.Agrobacteriumstrain GV3101 carrying these vectors was infiltrated into four-week-old tobacco leaves.Three days after infiltration,1 mmol L-1D-luciferin(Sigma-Aldrich)was sprayed on the leaves, which were held for 20 min in the dark, and then the LUC image was captured with a low-light cooled CCD camera (Tanon 5200 Multi, Tanon Biomart, China).
For yeast two-hybrid assay (Y2H), full-lengthSiCBL5andSiCIPK24were separately introduced into the pGADT7 AD vector and pGBKT7 using specific primers (Table S1).These constructs were transformed into the Y2H strain and cultured for three days on 40 μg mL-1X-α-gal (Clontech, USA) synthetic dropout/-Leu/-Trp/-His/-Ade plates.
2.7.Phenotype analysis
Four-day-oldArabidopsisseedlings growing on 1/2 MS medium were exposed to salt stress by addition of 100 mmol L-1NaCl.Root length and fresh weight were recorded seven days after treatment.Shoot fresh weight was recorded for three repeats and root length was recorded for more than ten repeats.
One-week-old foxtail millet seedlings were grown in pots and treated with 350 mmol L-1NaCl for nine days (at three-day intervals), 45 °C for two days, and normal temperature for three days,and no irrigation for 12 days to simulate salt, heat, and drought stress, respectively.The fresh weights of WT,SiCBL5overexpressors and RNAi-SiCBL5plants were recorded after treatment.Shoot fresh weight, root fresh weight, and chlorophyll content were recorded for three biological replicates with 10 plants per replicate.
2.8.Determination of Na+ content, K+ content, and Na+ flux
Foxtail millet seedlings were separated into shoots and roots.The dry samples were digested in nitric acid at 80 °C for 6 h.The contents of Na+and K+were determined using an inductively coupled plasma-optical emission spectrometry instrument (Perkin Elmer, USA) [35].Values were recorded for six repeats.
For measurement of Na+flux, three-day-old seedlings of WT,SiCBL5overexpressors, and RNAi-SiCBL5plants were treated with 100 mmol L-1NaCl for 24 h.Na+flux was measured in the primary root zone (-600 μm from the root tip) using non-invasive microtest technology (NMT, Younger, Amherst, MA, USA) following Yin et al.[16].Values were recorded for six repeats.
3.Results
3.1.Subcellular localization of SiCBLs
The locations of SiCBL1-YFP, SiCBL2-YFP and SiCBL3-YFP proteins partially overlapped with that of the tonoplast marker protein (vac-rk), suggesting that the three proteins were present in the tonoplast (Fig.1A-C).However, distinct distributions of SiCBL1-YFP, SiCBL2-YFP, and SiCBL3-YFP that did not colocalize with the tonoplast marker were also observed [32].SiCBL4 and SiCBL5 were shown to be localized only in the plasma membrane along with a plasma membrane-localized marker (pm-rk)(Fig.1F-I).Signals of SiCBL6-YFP and SiCBL7-YFP were detected in the cell membrane, cytosol, and nucleus (Fig.1D-E).
3.2.SiCBL5 and SiCBL4 rescue the salt-sensitive phenotype of the Atsos3-1 mutant
To determine whether other SiCBLs respond to salt stress,SiCBLsdriven by the35Spromoter were transformed into theAtsos3-1mutant.All seven SiCBL genes (SiCBL1,SiCBL2,SiCBL3,SiCBL4,SiCBL5,SiCBL6, andSiCBL7) were separately expressed in the mutant lines and were detected by RT-PCR and qRT-PCR assays(Fig.S1).No difference was observed in the primary root lengths and fresh weights of the wild type and these genotypes in the absence of NaCl.In the presence of NaCl, the primary root lengths of these lines separately overexpressingSiCBL1,SiCBL2,SiCBL3,SiCBL6, orSiCBL7inAtsos3-1mutant were shorter than those of the wild type but similar to that of theAtsos3-1mutant.However,the primary roots of these lines separately overexpressingSiCBL4orSiCBL5inAtsos3-1mutant were longer than those of theAtsos3-1mutant and similar to that of the wild type.The reduction in fresh weight was correlated with the suppression of primary root growth in wild type and different genotypes (Fig.2, Fig.S2).These results suggest both SiCBL4 and SiCBL5 were found to be functional orthologs of AtCBL4.
3.3.SiCBL5 increased salt tolerance in foxtail millet
SiCBL5was preferentially expressed in roots and young leaves,and expressed at a lower level in mature leaves, seeds, stem, and inflorescence (Fig.3A).NaCl did not affectSiCBL5expression(Fig.3B).
Two independentSiCBL5overexpressors (OE-SiCBL5#1 and OESiCBL5#2) and two independentSiCBL5-knockdown lines (RNAi-SiCBL5#1 and RNAi-SiCBL5#2) were confirmed by qRT-PCR(Fig.S3).AsSiCBLs genes are highly similar (Fig.S4), the expressions of the otherSiCBLswere also detected and onlySiCBL5expression was reduced by ~ 60% in RNAi-SiCBL5lines compared with the wild type(Figs.S3-S5).Thus,SiCBL5expression was suppressed by RNAi.

Fig.1.Subcellular localization of SiCBLs.(A-C) Tobacco leaf cells co-expressing fusion proteins SiCBL1-YFP or SiCBL2-YFP or SiCBL3-YFP, tonoplast marker (vac-rk), brightfield image and with a merged image in a tobacco cell.Images of (a-c) represent the enlarged square in (A-C).(D and E) Tobacco leaf cells co-expressing fusion proteins SiCBL6-YFP or SiCBL7-YFP,plasma membrane marker(pm-rk),bright-field image and with a merged image in a tobacco cell.(F and H)Tobacco leaf cells co-expressing fusion protein SiCBL4-YFP or SiCBL5-YFP, plasma membrane marker (pm-rk), bright-field image and with a merged image in a tobacco cell.(G and I) The fluorescence intensity profiles along the direction of white arrows in (F) and (H) are shown on the right panel.Scale bars, 10 μm.Experiments were repeated three times.

Fig.2.Overexpression of SiCBL5 in Atsos3-1 mutants rescued its salt sensitivity phenotype.(A-F) Salt sensitivity associated with overexpression of SiCBL1 (A), SiCBL2 (B),SiCBL3(C),SiCBL5(D), SiCBL6(E),or SiCBL7(F)in Atsos3-1 mutant.Four-day-old Arabidopsis seedlings growing on 1/2 MS medium were exposed to salt stress by addition of 100 mmol L-1 NaCl for an additional seven days, and representative photographs are shown.Values for seedling fresh weight (three seedlings) are means±SD (n = 3) and values for root length are means±SD(n>10).Scale bars,1 cm.Asterisk shows significant difference between various genotypes and wild type using the unpaired Student’s ttest (*, P < 0.05).
Salt sensitivity assays in the wild-type,SiCBL5overexpressors,and RNAi-SiCBL5revealed no significant difference in the growth of foxtail millet seedlings between wild-type and transgenic lines in the absence of NaCl.However, in the presence of NaCl,SiCBL5overexpressors showed less severe wilting and chlorosis than wild-type plants, whereas RNAi-SiCBL5lines showed more severe wilting and chlorosis (Fig.3C).Under NaCl treatment, the shoot and root fresh weight ofSiCBL5overexpressors were greater than those of the wild type, while the shoot and root fresh weight of RNAi-SiCBL5lines were lower than those of the wild type(Fig.3D, F, and G).Under NaCl treatment, chlorophyll contents were significantly higher inSiCBL5overexpressors but markedly lower in RNAi-SiCBL5lines than in the wild type.However,no significant difference in chlorophyll content was observed between the wild-type plants and any transgenic plant in the absence of NaCl (Fig.3E).Clearly, SiCBL5 promotes salt tolerance in foxtail millet.However, no such positive effect was observed under heat stress (Fig.S6A).Drought tolerance of foxtail millet was increased in theSiCBL5transgenic plant but returned to the WT level in the RNAi-SiCBL5plants (Fig.S6B), suggesting that SiCBL5 may also function in drought tolerance.

Fig.3.SiCBL5 increases salt tolerance.(A) SiCBL5 expression in various tissues of foxtail millet.(B) SiCBL5 expression in the wild type under NaCl treatment.One-week-old seedlings grown in pots were treated with 350 mmol L-1 NaCl for various times as indicated and results of qRT-PCR analysis of their SiCBL5 expressions.(C) Seedling phenotype,(D)shoot fresh weight,(E)chlorophyll contents,(F)root phenotype,and(G)root fresh weight of the wild type,SiCBL5 overexpressors and RNAi-SiCBL5.One-weekold seedlings of wild type,SiCBL5 overexpressors,and RNAi-SiCBL5 grown on pots were irrigated with or without 350 mmol L-1 NaCl for nine(C)or six(D-G)days,after which the above photographs and indexes were recorded.Scale bars,2 cm in(C)and(F).Values in(D),(E)and(G)are means±SD from three biological replicates with 10 plants in each replicate.Asterisk shows significant difference between various genotypes and wild type using the unpaired Student’s t-test (**, P < 0.01).
3.4.SiCBL5 interacts with SiCIPK24 in vitro and in vivo
Y2H assay was used to confirm the interaction between SiCBL5 and SiCIPK24, a homolog of AtSOS2/AtCIPK24 in foxtail millet.SiCBL5 interacted with SiCIPK24 in yeast cells (Fig.4A).In the LCI assay, the signal was strongly detected only in tobacco leaves injected with SiCBL5-nLUC and cLUC-SiCIPK24 (Fig.4B).Thus,SiCBL5 interacted with SiCIPK24 in vitro and in vivo.
3.5.SiCBL5-SiCIPK24 affects the function of SiSOS1 in yeast cells
A putative SOS1 in foxtail millet was identified by BLAST search against AtSOS1 protein sequence.The protein sequence of SiSOS1 showed 57.2% identity with that of AtSOS1 (Fig.S7).The yeast mutant strain AXT3K was very sensitive to NaCl and could not survive in AP medium supplement with 100 mmol L-1NaCl and 150 mmol L-1NaCl (Fig.5).Expression ofSiSOS1or coexpression ofSiSOS1andSiCIPK24in yeast mutant strain AXT3K did not rescue the salt hypersensitivity of the AXT3K mutant strain.However, yeast cells co-expressingSiSOS1,SiCIPK24,andSiCBL5were more tolerant to NaCl than the others.Thus, SiCBL5-SiCIPK24 affected the function of SiSOS1 in yeast cells.

Fig.4.SiCBL5 physically interacts with SiCIPK24 in vitro and in vivo.(A) Yeast two-hybrid assay of the interaction between SiCBL5 and SiCIPK24.Y2H strains were cotransformed with various combinations as indicated.SD/-Leu/-Trp/-His/-Ade+X-α-gal medium was used for interaction test.The combinations of BD-P53/AD-SV40 and BDLam/AD-SV40 were used as positive and negative controls,respectively.(B)LCI assay of SiCBL5 interaction with SiCIPK24.Tobacco leaves were co-transformed with various combinations as indicated.Luciferase signals were captured using the Tanon-5200 image system.Scale bar, 1 cm in (B).All experiments were repeated three times.

Fig.5.SiCBL5-SiCIPK24 affects the function of SiSOS1 in yeast cells.Yeast AXT3K cells transformed with the indicated combinations of SiSOS1, SiCIPK24, or SiCBL5 were spotted on AP plates with or without NaCl.Plates were cultured at 28 °C for 3 days.Experiment was repeated three times.
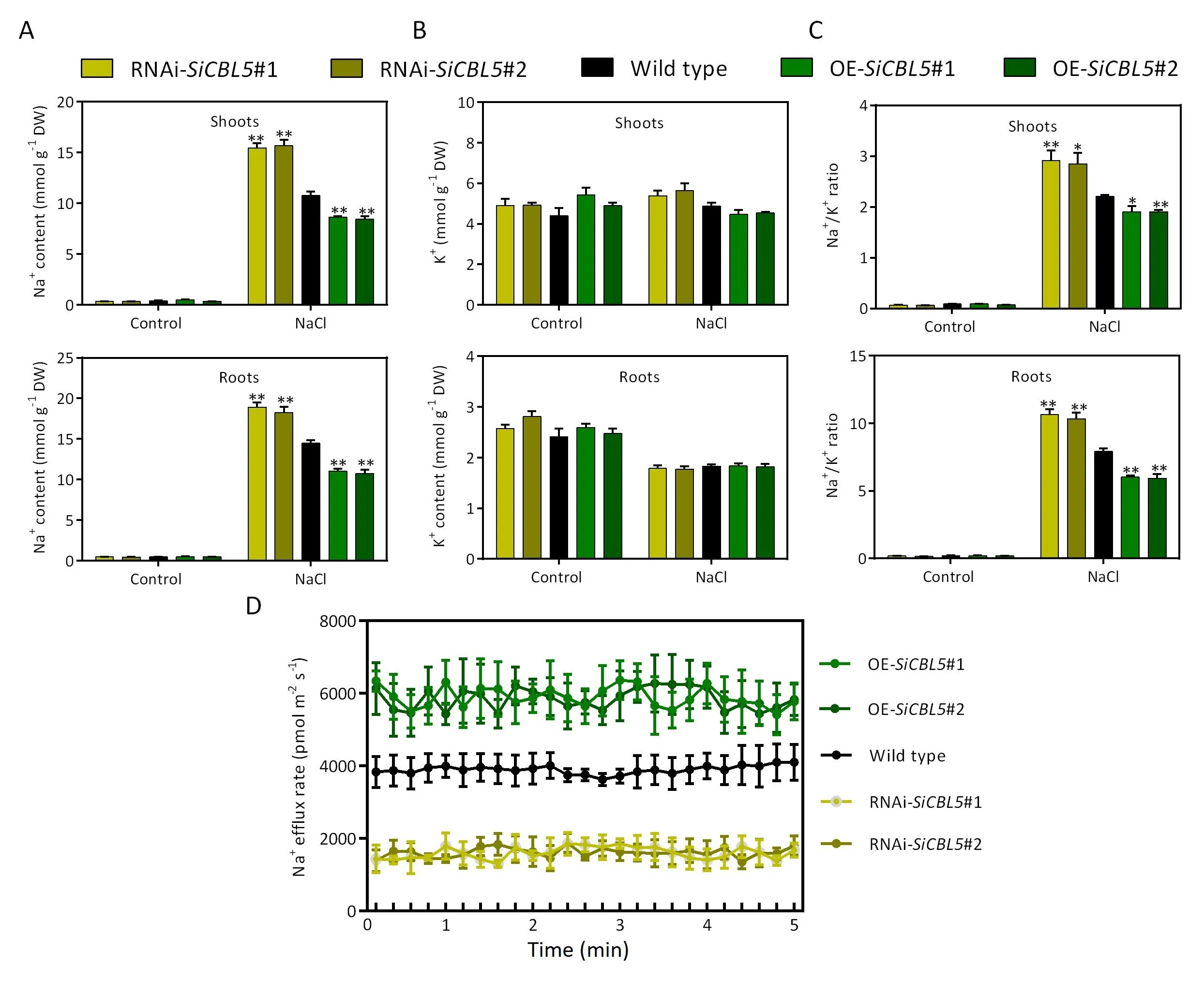
Fig.6.Effects of SiCBL5 on Na+and K+contents and Na+efflux in foxtail millet.(A)Na+contents,(B)K+contents,and(C)Na+/K+ratio of shoots and roots in SiCBL5 transgenic lines exposed to NaCl treatment.One-week-old seedlings were treated with 350 mmol L-1 NaCl for 6 days,after which the shoots and roots were harvested.(D)Na+efflux in wild type,SiCBL5 overexpressors,and RNAi-SiCBL5 in the presence of NaCl.Seedlings were treated with 100 mmol L-1 NaCl for 24 h,after which Na+flux was measured in the primary root zone(-600 μm from the root tip)using NMT.Values are means±SE(n=6).Asterisks indicate significant differences between various genotypes and wild type using the unpaired Student’s t-test (*, P < 0.05; **, P < 0.01).
3.6.SiCBL5 positively maintains Na+ homeostasis in foxtail millet
To investigate the effect of SiCBL5 on Na+homeostasis in foxtail millet, the Na+contents in shoots and roots of wild-type plants,SiCBL5overexpressors, and RNAi-SiCBL5transgenic lines were investigated.In the presence of NaCl, the Na+contents in both shoots and roots were significantly higher in RNAi-SiCBL5lines but markedly lower inSiCBL5overexpressors.However,no significant difference in Na+content was observed between the wild-type plants and all transgenic plants in the absence of NaCl (Fig.6A).Shoots and roots of wild-type and transgenic lines showed approximately equal K+contents in both the absence and presence of NaCl(Fig.6B).Under NaCl treatments,the Na+/K+ratio was significantly lower inSiCBL5overexpressors, while the Na+/K+ratio was obviously higher in RNAi-SiCBL5lines than in the wild type (Fig.6C).Because the high expression ofSiCBL5in roots suggested that SiCBL5 might affect Na+efflux in roots, the NMT test was used to monitor Na+efflux in roots.All roots from wild-type andSiCBL5transgenic plants pretreated with NaCl showed Na+efflux characteristics.The Na+efflux rate in roots was significantly weaker in RNAi-SiCBL5lines but markedly stronger inSiCBL5overexpressors than in the wild type (Fig.6D).Thus, SiCBL5 maintains Na+homeostasis in foxtail millet by promoting Na+export from cells.
4.Discussion
In plants, Ca2+acts as a second messenger in regulating plant development,plant growth,and abiotic and biotic stress responses.Cellular Ca2+is perceived and transduced to cellular response by various Ca2+sensors including calcineurin B-like protein (CBL),calmodulin protein (CaM), calmodulin-like protein (CML),calcium-dependent protein kinase (CDPK), and calcium/calmodulin-dependent protein kinase (CCaMK) [36,37].Among these Ca2+sensors,CBL is believed to be a key regulatory node linking the perception of stimuli to cellular responses[7].Many studies have shown that CBL functions in plant response to abiotic and biotic stresses.InArabidopsis, 10 CBLs have similar and distinct functions.For example, AtCBL1, AtCBL4, AtCBL5, AtCBL9, and AtCBL10 are involved in salt stress response [15,18-20,38].AtCBL1 and AtCBL9 modulate cold stress response [19,39,40].AtCBL3 is involved in low K+stress response [41].AtCBL2 is involved in membrane H+transport [42].AtCBL5 increases drought tolerance[20].However,the functions of CBLs in foxtail millet remain largely unknown.
We infer that SiCBL5 increased salt tolerance, based on four findings.(1)Heterologous expression ofSiCBL5inArabidopsiscompletely rescued the salt hypersensitivity of theAtsos3-1mutant(Fig.2).(2)Overexpression or knockdown ofSiCBL5in foxtail millet increased or decreased salt tolerance(Fig.3).(3)SiCBL5 interacted with SiCIPK24 to influence the function of SiSOS1 in yeast cell(Figs.4, 5).(4) SiCBL5 positively affected Na+homeostasis by promoting Na+export from cells in roots of foxtail millet(Fig.6).These findings suggest that SiCBL5 plays a regulatory role in modulating root Na+exclusion, but also affected shoot Na+accumulation(Fig.6A).In a previous study, PaSOS3 was involved in longdistance transport of Na+in poplar [23].We speculate that SiCBL5 affects the Na+transportation system from root to shoot in foxtail millet.
In a study [26] that investigated the role of the SiCBL4 in salt stress tolerance in foxtail millet, overexpression ofSiCBL4inAtsos3-1mutant rescued the hypersensitivity ofAtsos3-1mutant.However,that study did not elucidate the ion homeostasis regulatory mechanism of SiCBL4 in this process.In our study,SiCBL5 positively modulated Na+homeostasis by promoting Na+export from cells to increase salt tolerance.Our study also revealed vital differences between SiCBL5 and SiCBL4 in stress response.In the previous study [26], expression ofSiCBL4was induced by salt stress,whereas in the present studySiCBL5expression was not affected by NaCl treatment.This is consistent with the finding of a previous study [14] that the expression ofAtCBL4/AtSOS3was not induced by NaCl treatment.Our results indicate that these two calcium sensors,SiCBL4 and SiCBL5,in foxtail millet are differentially regulated by salt stress at the transcriptional level.These sensors also display differing expression patterns in foxtail millet,implying that different tissues are involved in salt tolerance.The finding thatSiCBL4was highly expressed in leaves andSiCBL5preferentially expressed in roots suggests that they are involved in salt tolerance in different tissues.Both SiCBL4 and SiCBL5 completely rescued the salt hypersensitivity of theAtsos3-1mutant (Figs.2, S2) [26].These findings suggest that both sensors are functional homologs of AtCBL4 in response to salt stress.Both SiCBL4 and SiCBL5 showed high identify with AtCBL4[27].The double copies of CBL4 in foxtail millet might have resulted from evolutionary adaptation of higher plants to survive in severe adverse environment.Poplar and the halophyteThellungiella parvulathat have shown strong tolerance to salinity carry two or more copies of CBL10 genes in their genomes [23,43].The double copies may help explain foxtail millet’s excellent tolerance to salt stress.
The N-myristoylation motif in some CBL proteins is important for their subcellular localization in plants and fulfillment of their biological functions [26,44,45].InArabidopsis, AtCBL1, AtCBL4,AtCBL5,and AtCBL9,all carrying an N-myristoylation motif in their N-terminus, were shown to be localized in the plasma membrane,whereas AtCBL2, AtCBL3, AtCBL6, and AtCBL10 were localized mainly in the vacuolar membrane.AtCBL7 and AtCBL8 were localized in the plasma membrane, cytoplasm and nucleus [36].AtCBL4/SOS3 targets AtCIPK24/SOS2 to the plasma membrane and thus activates SOS1 to pump Na+out of the cells[2].In foxtail millet, SiCBL4 and SiCBL5, harboring an N-myristoylation motif,were localized in the plasma membrane(Figs.1,S8).SiCBL5 interacted with SiCIPK24 to affect the function of SiSOS1 in yeast cells(Figs.4, 5).Although AtCBL10 protein lacks an N-myristoylation motif, overexpression ofAtCBL10inAtsos3-1mutant could partly suppress salt hypersensitivity ofAtsos3-1mutant [14].Surprisingly, SiCBL1, SiCBL2, SiCBL3, SiCBL6, and SiCBL7, all lacking an N-myristoylation motif, could not rescue the salt hypersensitivity ofAtsos3-1mutant.Two reasonable explanations to this observation could be(1)distinct functions of CBL family in various species and (2) a defect of heterologous expression.Thus, there may be other CBLs in foxtail millet that respond to salt stress.Future studies should investigate the functions of foxtail millet CBLs in response to salt stress.
5.Conclusions
In foxtail millet, overexpression ofSiCBL5increased its salt tolerance, but knockdown ofSiCBL5increased salt hypersensitivity.SiCBL5 increased salt tolerance by regulating Na+homeostasis.This study sheds light on the function of CBLs in plants and on salt stress-tolerance mechanisms in plants.
Accession numbers
Sequence data from this article can be found under accession numbersSiCBL1(Seita.9G164200),SiCBL2(Seita.7G294700),SiCBL3(Seita.3G375800),SiCBL4(Seita.3G166700),SiCBL5(Seita.5G214600),SiCBL6(Seita.5G291900),SiCBL7(Seita.5G259100),SiActin 7(Seita.7G294000),SiCIPK24(Seita.4G221700),SiSOS1(Seita.3G409000),AtCIPK24/AtSOS2(AT5G35410),AtActin2(AT3G18780),AtSOS1(At2G01980),AtCBL1(AT4G17615),AtCBL4/AtSOS3(AT5G24270),AtCBL5(AT4G01420), andAtCBL9(AT5G47100).
CRediT authorship contribution statement
Jingwei Yan:Methodology, Writing - original draft, Funding acquisition,Project administration,Data curation,Formal analysis.Lan Yang:Data curation, Formal analysis, Methodology.Ya Liu:Writing-review &editing,Methodology.Yingdi Zhao:Methodology.Tong Han:Writing-review&editing.Xingfen Miao:Writing-review&editing.Aying Zhang:Writing-review&editing,Funding acquisition, Project administration, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the National Natural Science Foundation of China(32001445 and 31871534)and the Natural Science Foundation of Jiangsu Province (BK20200557).We are grateful to Prof.Yuanqing Jiang (Northwest A&F University) for providing theAtsos3-1mutant, Prof.Caifu Jiang (China Agricultural University) for providing yeast strain AXT3K, Dr.Yang Zhou (Hainan University) for providing p414-GPD and p416-GPD vectors, and Dr.Weijuan Liu (Yangtze University) and Dr.Zhiyong Li (Hebei Academy of Agriculture and Forestry Sciences) for revising the manuscript.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.06.006.
杂志排行
The Crop Journal的其它文章
- Brief Guide for Authors
- Heavy soil drying during mid-to-late grain filling stage of the main crop to reduce yield loss of the ratoon crop in a mechanized rice ratooning system
- GmNMHC5 may promote nodulation via interaction with GmGAI in soybean
- H2O2 mediates transcriptome reprogramming during Soybean mosaic virus-induced callose deposition in soybean
- Contrasting patterns of accumulation,partitioning,and remobilization of biomass and phosphorus in a maize cultivar
- TaMADS2-3D, a MADS transcription factor gene, regulates phosphate starvation responses in plants
