Epigenetic regulation by long noncoding RNAs in osteo-/adipogenic differentiation of mesenchymal stromal cells and degenerative bone diseases
2022-02-12KaiXiaLiYuanYuXinQiHuangZhiHeZhaoJunLiu
Kai Xia, Li-Yuan Yu, Xin-Qi Huang, Zhi-He Zhao, Jun Liu
Kai Xia, Li-Yuan Yu, Xin-Qi Huang, Zhi-He Zhao, Jun Liu, State Key Laboratory of Oral Diseases & National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, Sichuan Province, China
Kai Xia, Li-Yuan Yu, Xin-Qi Huang, Zhi-He Zhao, Jun Liu, Department of Orthodontics, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, Sichuan Province, China
Abstract Bone is a complex tissue that undergoes constant remodeling to maintain homeostasis, which requires coordinated multilineage differentiation and proper proliferation of mesenchymal stromal cells (MSCs).Mounting evidence indicates that a disturbance of bone homeostasis can trigger degenerative bone diseases, including osteoporosis and osteoarthritis.In addition to conventional genetic modifications, epigenetic modifications (i.e., DNA methylation, histone modifications, and the expression of noncoding RNAs) are considered to be contributing factors that affect bone homeostasis.Long noncoding RNAs (lncRNAs) were previously regarded as ‘transcriptional noise’ with no biological functions.However, substantial evidence suggests that lncRNAs have roles in the epigenetic regulation of biological processes in MSCs and related diseases.In this review, we summarized the interactions between lncRNAs and epigenetic modifiers associated with osteo-/adipogenic differentiation of MSCs and the pathogenesis of degenerative bone diseases and highlighted promising lncRNA-based diagnostic and therapeutic targets for bone diseases.
Key Words: Long noncoding RNA; Epigenetics; DNA methylation; Histones; Cell differentiation; Bone diseases
INTRODUCTION
The skeletal system contains bones, joints, and ligaments that function together as a locomotive organ and provide structural support.Originating from mesenchymal progenitors during embryogenesis, the skeletal system undergoes modeling and remodeling throughout life[1].Mesenchymal stromal cells (MSCs) refer to a heterogeneous unfractionated population of cells, which include fibroblasts, myofibroblasts, and progenitor cells[2,3].MSCs are able to differentiate into chondrocytes or osteoblasts to comply with bone formation and regeneration needs[4].It is worth mentioning that adipocytes, as well as osteoblasts, derive from the same population of MSCs.A shift in the osteoadipogenic differentiation balance may lead to bone diseases, such as osteoporosis, which typically manifests as a shift toward adipogenesis[5,6].Likewise, osteoarthritis is usually characterized by impairment of cartilage regeneration due to the attenuated chondrogenic capacity of MSCs[7,8].Therefore, the differentiation of MSCs, which proceeds under the control of various transcription factors, influences the pathogenesis of common bone diseases[9-11].
In addition to conventional genetic and environmental factors, epigenetic modifications can influence the bone phenotype and the development of skeletal diseases[12,13].Epigenetic mechanisms alter gene expression patterns without changing the DNA sequence by three major mechanisms, including DNA methylation, histone modifications, and altered expression of noncoding RNAs[14].With the rapid development of next-generation sequencing (NGS) and advanced bioinformatic tools, the crucial roles of epigenetic mechanisms in the differentiation of MSCs and the pathogenesis of bone diseases have begun to be elucidated[15-17].
Long noncoding RNAs (lncRNAs) are defined as a set of noncoding RNAs longer than 200bp that have no protein-coding ability.Evidence is rapidly accumulating on the functions of lncRNAs in epigenetic regulation in the differentiation of MSCs and the occurrence of many diseases[18-21].In this review, we revisit the epigenetic regulatory mechanisms of lncRNAs involved in DNA methylation and histone modifications and summarize the biological functions of lncRNAs in regulation crucial differentiation- and bone disease-related genes by interacting with key epigenetic modifiers.It is our hope that this review may provide an updated summary that sheds light on the lncRNA-based precise regulation of the MSC differentiation process and highlights possible therapeutic targets of degenerative bone diseases.
DNA METHYLATION
DNA methylation functions as a regulator of osteogenesis and adipogenesis of MSCs and is involved in common bone diseases[22-24].In humans, the majority of DNA methylation occurs at cytosines in cytosine-phospho-guanosine (CpG) dinucleotides[25,26].Approximately 75% of all gene promoters are within CpG-rich regions, known as CpG islands, that are mostly unmethylated[27].It is generally accepted that the methylation of these CpG islands is associated with the repression of gene expression[28].Nevertheless, it is worth mentioning that DNA methylation is also associated with upregulated gene expression under certain circumstances[29].
As writer enzymes, DNA methyltransferases (DNMTs) catalyze DNA methylation by transferring a methyl group onto the C5 position of a cytosine at CpG dinucleotide sites to form 5mCpG[30].A member of the DNMT family, DNMT1, which is also called the maintenance DNMT, maintains the original methylation pattern during DNA replication, while DNMT3a and DNMT3b are involved inde novomethylation[30,31].The interaction of lncRNAs with DNMTs is varied and reciprocal.For example, lncRNAs can recruit DNMTs to the promoters of target genes and regulate their expression patterns.In turn, the changes in the methylation level of specific lncRNA gene promoters can alter the expression of lncRNAs, including downstream lncRNA-regulated genes[32,33].In MSCs, lncRNAs, as regulators of DNA methylation, have received increasing attention due to their great importance in the regulation of differentiation and bone-related diseases (Figure 1).
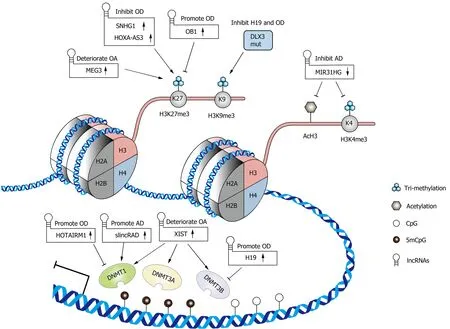
Figure 1 A brief illustration of the interactions between long noncoding RNAs and the epigenetic modification associated with osteo-/adipogenic differentiation of mesenchymal stromal cells and osteoarthritis.
LncRNAs regulate DNA methylation during osteogenic differentiation
H19, a well-known lncRNA, plays a crucial role in embryo development, cell differentiation, and the occurrence and development of bone diseases[34-37].In human dental pulp stromal cells (hDPSCs), H19 positively regulates odontogenic differentiationviahypomethylation of distal-less homeobox 3 (DLX3), a key factor in odontogenic differentiation[32].H19 decreases SAHH and DNMT3B activity, consequently promoting the expression ofDLX3[32].In turn, a mutation ofDLX3identified in dentin hypoplasia patients could increase DNMT3B activity, and the subsequently repressed H19/miR-675 axis impairs the odontoblastic differentiation of hDPSCs[38].Similarly, in valve interstitial cells (VICs), which have a mesenchymal origin[39], the knockdown of H19 attenuated their osteogenic differentiation capacity by increasing the transcription ofNOTCH1and decreasing the levels of RUNX2 and BMP2[40].In mineralized aortic valve tissue, H19 was upregulated as a result of hypomethylation of CpG in its promoter region[40].These results suggest the possibility that H19 forms a positive feedback loop with DNMTs and promotes the osteogenic differentiation of MSCs.
Another study found an inverse association between the methylation level of perinatalCDKN2A, which encodes the lncRNA antisense noncoding RNA in the INK4 Locus (ANRIL), and bone mass at ages 4 and 6 years[41].Considering that transitional hypomethylation ofCDKN2Ahas been identified in human bone marrow stromal cells (hBMSCs) during osteogenic differentiation[42], the authors further verified that the methylation ofCDKN2Adecreased the binding of transcription factors SMAD3/4 and consequently downregulated the expression of ANRIL[41].In terms of the functional mechanism of ANRIL, it has been demonstrated that the knockdown of ANRIL decreased the number of live cells and induced cell apoptosis of SaOS-2 cells[41].
Given the crucial roles ofHOXgenes in development and differentiation, it is reasonable to believe that the lncRNAs encoded by theHOXgene cluster could also exert their function as critical biological regulators (i.e.,HOTAIRin theHOXCcluster andHOTAIRM1in theHOXAcluster)[43-45].In human dental follicle stromal cells (hDFSCs), lncRNA HOTAIRM1 promoted osteogenesis by inhibiting the enrichment of DNMT1 in theHOXA2promoter region and subsequently maintaining two CpG islands in a hypomethylated state, which guaranteed the transcriptional activation ofHOXA2[17].
LncRNAs regulate DNA methylation during adipogenic differentiation
lncRNA HOTAIR, encoded by theHOXCgene cluster as mentioned above, could also inhibit the adipogenic differentiation of hBMSCs[46].In this process, HOTAIR probably directly interacts with DNMTs or is involved in gene regulation by triple helix formation[46].
Peroxisome proliferator-activated receptor-gamma (PPAR-γ) and CCAAT enhancer binding protein-alpha (C/EBP-α) are key transcription factors involved in adipogenesis.They synergistically promote the transcriptional activation of genes that induce the adipocyte phenotype and maintain their expression throughout the entire differentiation process and the entire life of the adipocytes[47,48].In mouse ST-2 cells (bone marrow stromal cells), 3T3-L1 cells (committed preadipocytes derived from MSCs), and C3H10T1/2 cells (embryonic stem cells) as well as in bone marrow stromal cells, lncRNA Plnc1 promotes adipogenesis by increasingPpar-γ2transcription through reducing the DNA methylation level on its promoter[49].
Upregulation of lncRNA slincRAD is also observed in the early stages of adipocyte differentiation in 3T3-L1 cells[50].LncRNA slincRAD guides Dnmt1 to translocate to the perinuclear region in S phase and direct Dnmt1 to the promoter of cell cyclerelated genes, including p21 (Cdkn1a)[50].As p21 is a cyclin-dependent kinase inhibitor that plays an important role in the differentiation of 3T3-L1 cells, this effect facilitates the progression of differentiation[50,51].
HISTONE MODIFICATIONS
The building block of chromatin is the nucleosome, which consists of a complex of DNA and four types of core histone subunits (H2A, H2B, H3, and H4)[52].Histone proteins are subject to a variety of modifications, with most studies focusing on methylation and acetylation.Lysine (K) residues in histone H3 are commonly modified by methylation, which is orchestrated by histone methyltransferases (HMTS) and histone demethylases (HDMs)[53,54].Previous studies have revealed that trimethylation of H3K4 (H3K4me3) promotes transcription, whereas H3K9me3 and H3K27me3 restrict gene expression[53].Likewise, acetylation and deacetylation of lysine residues in histones are regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively.It is believed that the addition of an acetyl group to lysine residues alters the structure and folding of the nucleosome and consequently loosens the chromatin to enable transcription[55].During cellular biological and pathologic processes, including cell differentiation, bone regeneration and disease, histone modifications are dynamically changed[53,56].This process is at least in part mediated by lncRNAs that recruit histone-modifying enzymes to targeted gene promoters and alter histone modification enrichment (Figure 1).
Involvement of lncRNAs in osteogenic differentiation through histone modifications
As mentioned earlier, a mutation ofDLX3identified in dentin hypoplasia patients could increase DNMT3B activity[38].This study also reported that this mutation was capable of repressing H19 expression by increasing the enrichment of H3K9me3 in the promoter region of the H19 gene and retarding the odontoblastic differentiation of hDPSCs[38].
Similar to RUNX2, Osterix (OSX) is considered a master transcription factor that regulates the osteogenic differentiation of MSCs and it is required for the maturation of functional osteoblasts[57].lnc-OB1 promotes osteogenic differentiation of MSCs, probably by upregulating OSXviathe inhibition of H3K27me3 in the OSX promoter region[58].In human osteoblast cells, this regulation might be mediated by an interaction between lnc-OB1 and SUZ12, which is an integral component of polycomb repressive complex 2 (PRC2), responsible for H3K27me3[58,59].
Another core part of PRC2, EZH2[59], was also found to interact with lncRNAs and regulate osteogenic differentiation.It has been shown that lncRNA SNHG1 inhibits the osteogenic differentiation of human periodontal ligament stromal cells by repressing the expression of KLF2, a positive regulator of osteoblast differentiation[60], through EZH2-mediated H3K27me3 of its promoter[61].Likewise, lncRNA HOXA-AS3 inhibits hBMSC osteogenesis, possiblyviaEZH2-dependent H3K27me3, and represses RUNX2 expression[62].
Involvement of lncRNAs in adipogenic differentiation through histone modifications
As a critical transcription factor for adipogenesis, C/EBP-α was found to be upregulatedviathe recruitment of the MLL3/4 complex to its promoter, which is guided by the binding of PA1 (a component of the MLL3/4 complex) to lncRNA ADINR during adipogenic differentiation of human adipose-derived stromal cells (hASCs)[63].It is believed that MLL3/4 complexes are involved in the maintenance of H3K4me3 and the removal of H3K27me3, thereby regulating downstream gene expression[64,65].
Adipocyte fatty acid-binding protein (A-FABP, also known as FABP4 or aP2), a downstream target gene of PPAR-γ and C/EBP-α, is considered a marker of adipogenic differentiation[66,67].The knockdown of lncRNA MIR31HG suppressed FABP4 expression by reducing the enrichment of acetylated histone 3 (AcH3) and H3K4me3 in the FABP4 promoter, leading to the inhibition of adipogenic differentiation of hASCs[16].
H19 and miR-675 (derived from H19) inhibited the adipogenic differentiation of hBMSCs through the miRNA-mediated repression of HDAC4, 5 and 6.In turn, the inhibition of HDACs decreased CCCTC-binding factor (CTCF) occupancy on the imprinting control region (ICR) of H19 and reduced H19 expression[68].This evidence, combined with that mentioned in an earlier section that H19 is considered a positive regulator of osteogenic differentiation, suggests that DNA methylation and histone modifications might be linked together by H19 and shift the osteoadipogenic differentiation balance toward osteogenesis.
ROLE OF LNCRNAS IN DEGENERATIVE BONE DISEASES
More recently, epigenetic regulation of bone homeostasis has been considered as an important factor in the pathogenesis of degenerative bone diseases, such as osteoporosis, arthritis, post menopausal osteoporosis,etc.[69,70].As mentioned above, lncRNAs have attracted considerable attention in the epigenetic regulation of bone homeostasis.The potential link between degenerative bone diseases and lncRNAs at the epigenetic level is also an intriguing area for exploration.
LncRNAs regulate DNA methylation in osteoarthritis and osteoporosis
Osteoarthritis (OA) is a common degenerative joint disease that is associated with the impairment of cartilage regeneration, chondrocyte apoptosis, and the degradation of the cartilage extracellular matrix (ECM)[71,72].In this sophisticated balance between biosynthesis and degradation, lncRNAs play a role in the survival of chondrocytes and the regulation of arthritis-associated factors[73].
It has been reported that the overexpression of lncRNA CTBP1-AS2 downregulates miR-130a by increasing the methylation level of themiR-130agene, which finally leads to a decreased proliferation rate of chondrocytes in OA patients[74].
As a natural inhibitor of matrix metalloproteinases (MMPs), TIMP-3 deficiency can lead to mild cartilage degeneration in patients with OA[75].lncRNA XIST is capable of downregulating the expression of TIMP-3 through the recruitment of DNMT1, DNMT3A, and DNMT3B, which increased the methylation ratio of the CpG island in theTIMP-3promoter region, and consequently increased collagen degradation in OA chondrocytes[76].
Increasing evidence suggests that small nucleolar RNA host gene (SNHG) family members are involved in the pathogenesis of OA[77-79].The overexpression of lncRNA SNHG15 alleviated ECM degradation and promoted chondrocyte formationviacompeting endogenous RNA (ceRNA) SNHG15/miR-7/KLF4 axis[33].In human OA cartilage tissues, however, the promoter region of lncRNA SNHG15 had a higher level of methylation than in normal cartilage tissues, and this might be a promising therapeutic target for OA[33].AnotherSNHGfamily member, lncRNA SNHG9, was found to be downregulated in chondrocytes from OA patients[80].Functional studies indicated that the overexpression of SNHG9 led to a decreased apoptotic rate through increased methylation of themiR-34agene that suppressed the expression of miR-34a[80].
Osteoporosis is characterized by a loss of bone mass and microarchitectural deterioration of the skeletal structure[81].The imbalance of bone homeostasis between osteoblastic bone formation and osteoclastic bone resorption plays a fundamental role in the pathogenesis of osteoporosis[82].Emerging evidence suggests that epigenetic modifications are deeply involved in bone metabolism, which contributes to the development of osteoporosis.
The ERK-MAPK signaling pathway is a well-established pathway with critical roles in immune responses and embryonic development, including the regulation of bone massviacontrolling osteoblast differentiation[83].A previous study suggested that lncRNA H19 promoted tension-induced osteogenesis of hBMSCs through the FAKERK1/2-RUNX2 signaling pathway[84].Likewise, an alteration in H19 methylation may also be involved in the disruption of bone formation in disuse osteoporosis.It has been shown that DNMT1-induced hypermethylation of the H19 promoter results in H19 downregulation and ERK-MAPK signaling inhibition, which leads to osteogenesis impairment bothin vivoandin vitro(rat osteoblast/osteocyte-like UMR-106 cells)[85].
LncRNAs regulate histone modifications in osteoarthritis
An abnormality of cartilage regeneration can be related to attenuated chondrogenic differentiation of MSCs in OA patients[8].Similar to other MSCs derived from other tissues, synovium-derived mesenchymal stromal cells (SMSCs) are multipotent but have the greatest chondrogenesis potential, representing a promising stem cell source for cartilage repair in OA patients[86].lncRNA MEG3 was reported to have the ability to inhibit the chondrogenic differentiation of SMSCs and the expression of cartilageassociated genes (aggrecan and Col2A1) by inhibiting TRIB2 expression through EZH2-mediated H3K27me3[87].
CONCLUSION
LncRNAs are extensively involved in various types of epigenetic modifications, including DNA methylation, histone modifications, and noncoding RNA interactions, during MSC differentiation and the occurrence and progression of degenerative bone diseases.Concerning the large body of available literature and comprehensive reviews on the RNA-RNA interactions of lncRNAs (i.e., ceRNA mechanisms)[88,89], this topic of epigenetics is not discussed in this review, but it is worth mentioning that in some cases, ceRNA mechanisms act as mediators between lncRNAs and epigenetic modifiers.Another potential involvement of lncRNAs in epigenetics is the interaction with the key enzyme of methyl metabolism.It is known that DNMT and HMT utilize S-adenosylmethionine (SAM) as a major methyl-group donor in mammals, which is consumed and regenerated in one-carbon metabolism[90,91].Several studies have shown that lncRNAs play a role in SAM-dependent methylation through regulating enzymes related to the metabolism[92,93].However, similar studies on differentiation and bone diseases are lacking.Further studies are needed to assess the potential importance of lncRNAs on the methyl metabolism.
Although it seems that DNA methylation and histone modification are two different types of epigenetic modification, these two systems can be dependent on and influence one another during organism development[94].However, the underlying molecular mechanisms are complicated and remain vague.Intriguingly, lncRNAs are capable of regulating gene expression either in a cis- or trans- manner by guiding or serving as scaffolds for transcription factors or epigenetic modifiers to specific gene loci[95].This raises the possibility that lncRNAs could be coordinator of these processes.In this review, we summarized the roles of lncRNAs played in MSC differentiation and common degenerative bone diseases through reciprocal interactions between lncRNAs and epigenetic modifiers.A complete list of the epigenetic regulatory mechanisms of lncRNAs discussed in this review is available in Tables 1-3.
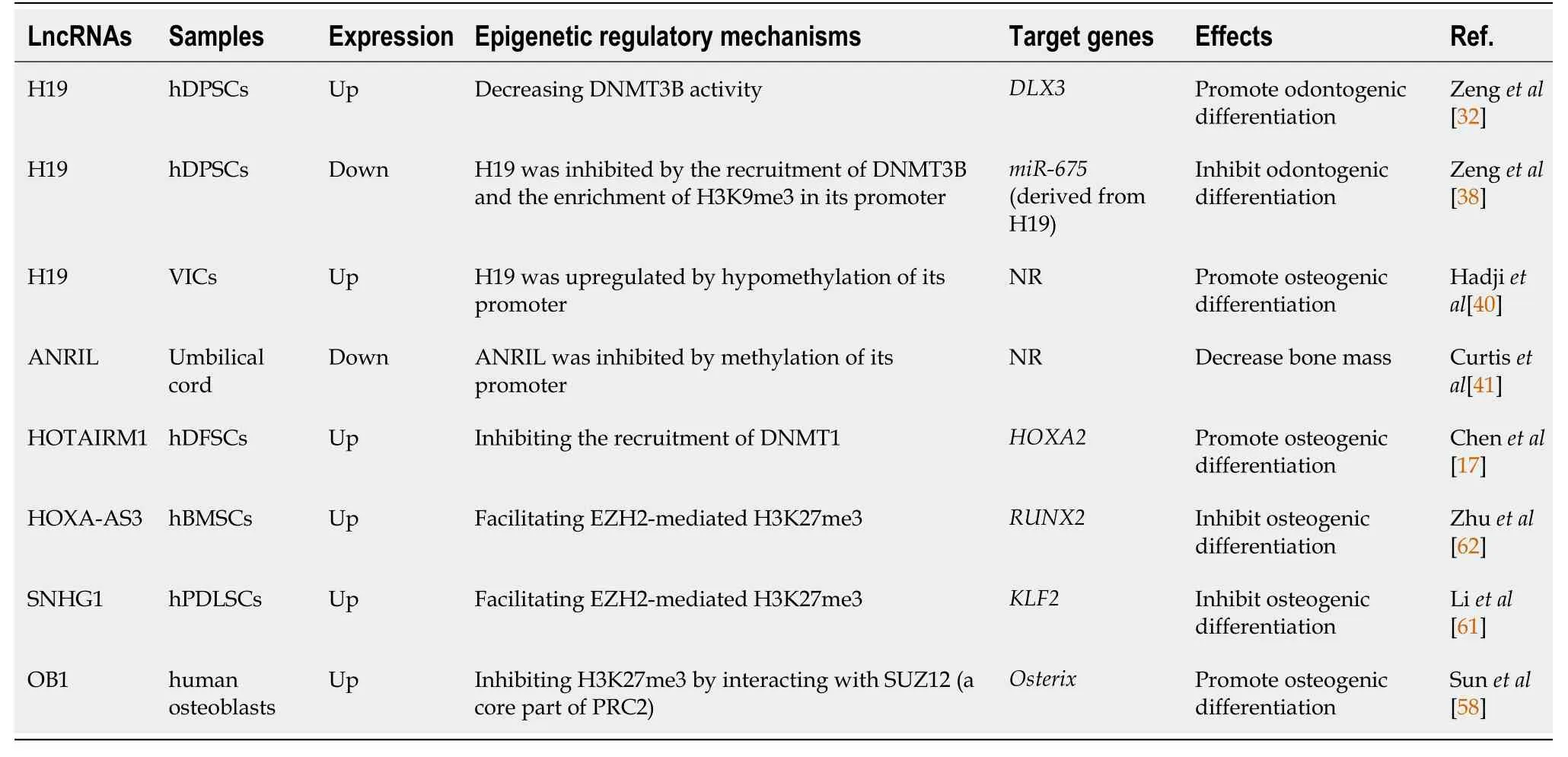
Table 1 Interactions between long noncoding RNAs and epigenetic modifiers during osteogenic differentiation of mesenchymal stromal cells
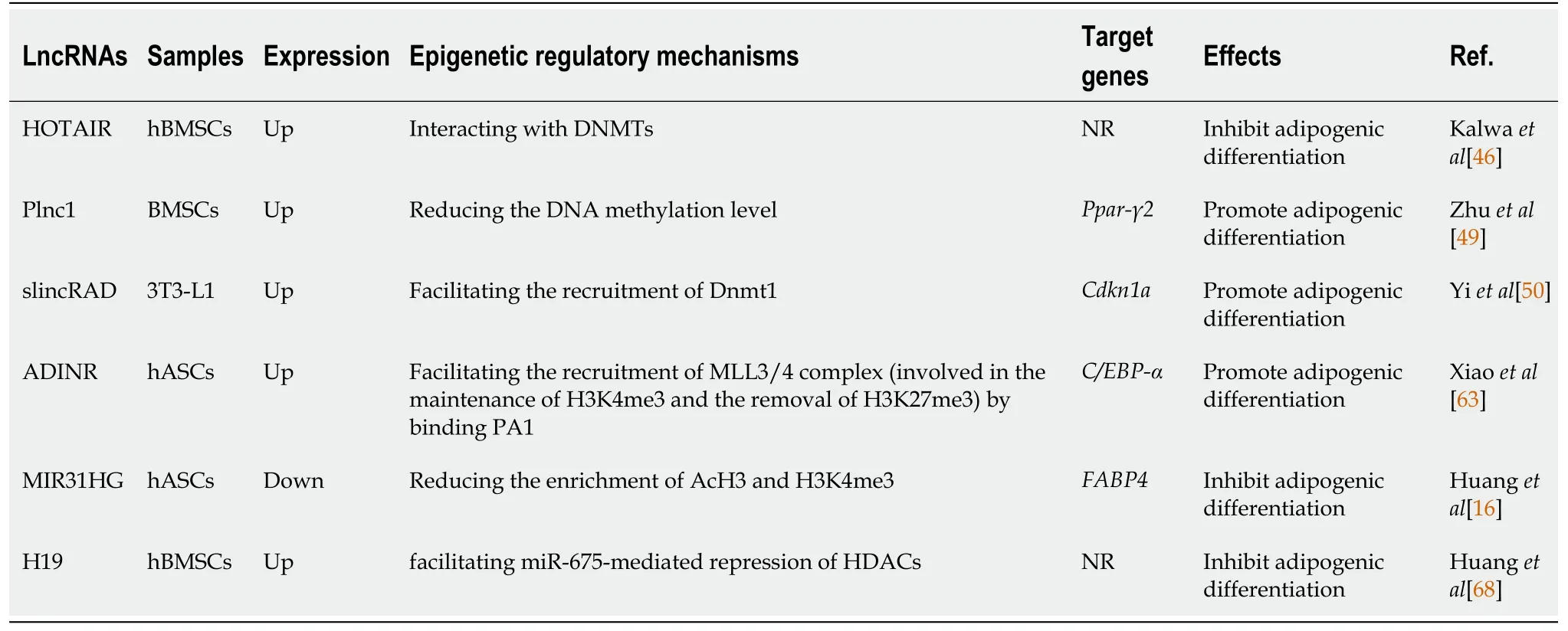
Table 2 Interactions between long noncoding RNAs and epigenetic modifiers during adipogenic differentiation of mesenchymal stromal cells
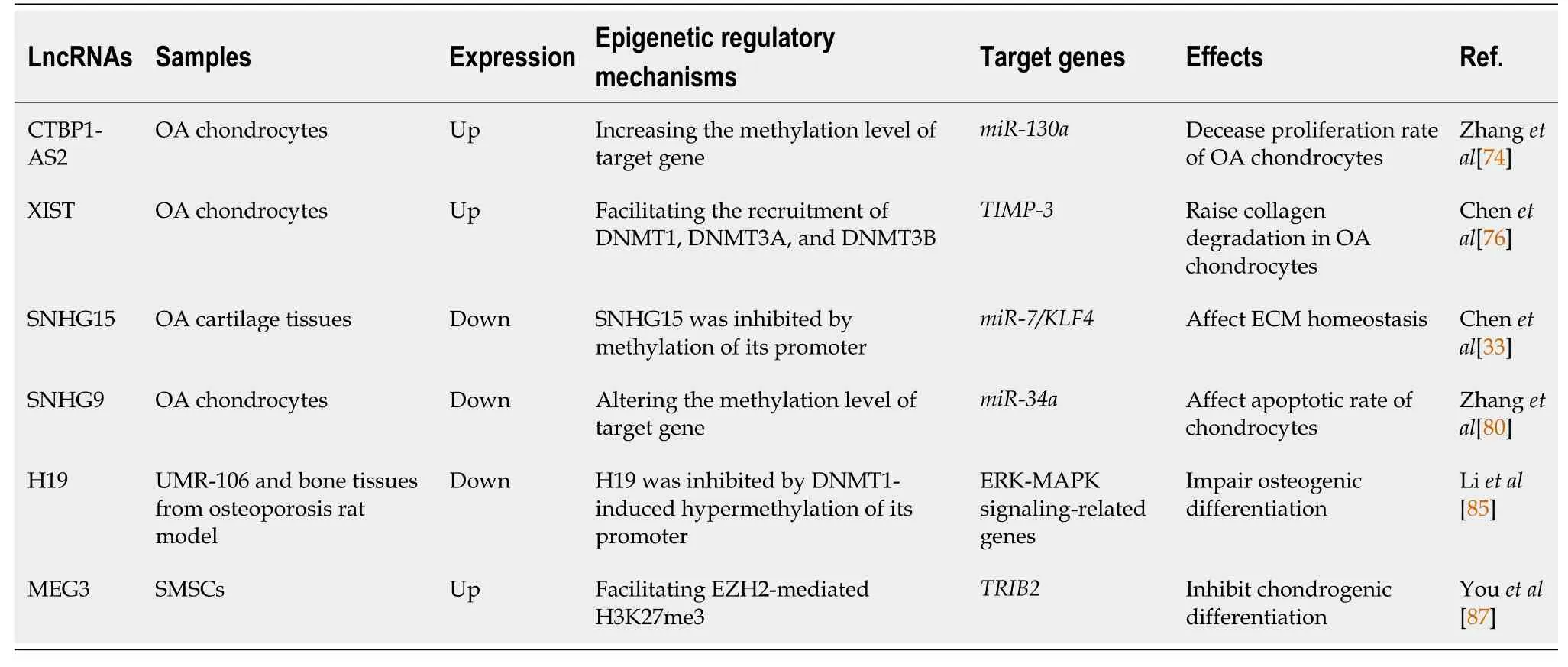
Table 3 Interactions between long noncoding RNAs and epigenetic modifiers in degenerative bone diseases
Taken in combination with previous studies[96-98], the present evidence indicates that lncRNAs could be diagnostic and prognostic biomarkers in degenerative bone diseases.Moreover, as lncRNAs can be manipulated pharmacologically to modulate epigenetic modifications[99], this also opens new avenues for future therapeutic interventions.However, multiple challenges need to be overcome before clinical applications can be achieved.Given that lncRNAs have complex secondary structures, one of the challenges that lies ahead is the off-target possibilities, as a single lncRNA is capable of binding to multiple epigenetic modifiers and targeting several genes.Therefore, more reliable bioinformatic tools in terms ofin silicoalgorithms for comprehensive lncRNA interaction prediction and sequencing technologies are required.Despite these impediments, lncRNA-based epigenetic interventions have shown potential in the regulation of MSC differentiation and therapeutic strategies for bone diseases.
杂志排行
World Journal of Stem Cells的其它文章
- Cardiac stem cells: Current knowledge and future prospects
- Multiple roles of mothers against decapentaplegic homolog 4 in tumorigenesis, stem cells, drug resistance, and cancer therapy
- Modified mesenchymal stem cells in cancer therapy: A smart weapon requiring upgrades for wider clinical applications
- Molecular mechanism of therapeutic approaches for human gastric cancer stem cells
- Therapeutic effects of menstrual blood-derived endometrial stem cells on mouse models of streptozotocin-induced type 1 diabetes
- Stem cell therapy applied for digestive anastomosis: Current state and future perspectives
