Inf luence of soil microorganisms and physicochemical properties on plant diversity in an arid desert of Western China
2021-12-24XiaodongYangYanxinLongBinoySarkarYanLiGuanghuiArshadAliJianjunYangYueCao
Xiaodong Yang · Yanxin Long · Binoy Sarkar ·Yan Li · Guanghui Lü · Arshad Ali ·Jianjun Yang · Yue-E. Cao
Abstract Soil microorganisms and physicochemical properties are considered the two most inf luencing factors for maintaining plant diversity. However, the operational mechanisms and which factor is the most inf luential manipulator remain poorly understood. In this study, we examine the collaborative inf luences of soil physicochemical properties(i.e., soil water, soil organic matter (SOM), salinity, total phosphorus and nitrogen, pH, soil bulk density and f ine root biomass) and soil microorganisms (fungi and bacteria)on plant diversity across two types of tree patches dominated by big and small trees (big trees: height ≥ 7 m and DBH ≥ 60 cm; small trees: height ≤ 4.5 m and DBH ≤ 20 cm)in an arid desert region. Tree patch is consists of a single tree or group of trees and their accompanying shrubs and herbs.It was hypothesized that soil physicochemical properties and microorganisms af fect plant diversity but their inf luence dif fer. The results show that plant and soil microbial diversity increased with increasing distances from big trees.SOM, salinity, f ine root biomass, soil water, total phosphorus and total nitrogen contents decreased with increasing distance from big trees, while pH and soil bulk density did not change. Plant and soil microbial diversity were higher in areas close to big trees compared with small trees, whereas soil physicochemical properties were opposite. The average contribution of soil physicochemical properties (12.2%–13.5%) to plant diversity was higher than microbial diversity (4.8%– 6.7%). Salinity had the largest negative af fect on plant diversity (24.7%–27.4%). This study suggests that soil fungi constrain plant diversity while bacteria improve it in tree patches. Soil physicochemical properties are the most important factor modulating plant diversity in arid desert tree patches.
Keywords Arid ecosystem · Soil microbial diversity ·Soil physicochemical properties · Plant diversity · Soil salinity
Introduction
Arid deserts account for approximately one-fourth of the global land surface (Polis 1991). Over the past decades, arid deserts have been recognized as one of the most vulnerable ecosystems for biodiversity loss, possibly due to global climate changes and increases in deforestation, extensive agriculture and urbanization (Elkeblawy 2014; Wang et al. 2017;Yang et al. 2019). Expanding maintenance mechanisms of plant diversity in arid desert ecosystems has become a major concern in government, industry and academic sectors (Li et al. 2018b; Saiz et al. 2018; Berdugo et al. 2019). Previous studies have suggested that arid desert ecosystems are composed of a continuous shrub-grass layer and a discontinuous tree patch (Ludwig et al. 2004). A tree patch consists of a single tree or group of trees and their accompanying shrubs and herbs (Fig. 1), which is the main contributor to biodiversity conservation, productivity maintenance and bio physiochemical functioning of arid desert ecosystems (Belsky 1994; Li et al. 2019). However, the mechanism of plant diversity maintenance at the level of tree patches remains unclear.
Soil microorganisms are important biotic factors for the maintenance of plant diversity under dif ferent climates(Janzen 1970; Liu et al. 2012; Peng et al. 2019). Soil microorganisms can change the spatial distribution and availability of nutrients, thus af fecting plant diversity via resource differentiation among species (Chaparro et al. 2012; Hodge and Storer 2015; Lou et al. 2016). Changes in soil microbial community composition, especially the increasing abundance of microorganisms closely related to plant growth and survival, can also improve plant diversity (Bulgarelli et al. 2015; Simões 2015). However, studies linking plant diversity and soil microorganisms are lacking for arid desert ecosystems (Bagchi et al. 2011; Peng et al. 2019).
The ef fect of soil microorganisms on plants varies in direction and magnitude (Tian et al. 2018; Huangfu et al.2019). The shift in direction might be determined by the type of microorganisms present in a particular ecosystem (Packer and Clay 2000; Peng et al. 2019). Bacteria may improve diversity through their positive improvement to environmental conditions and plant adaptation (Bulgarelli et al. 2015;Laforest-Lapointe et al. 2017). For example, bacteria reduce environmental stress (e.g., salt stress) and increase the activity of roots to resist oligotrophic stress via their secretions(Asaf et al. 2018; Cho et al. 2018). In contrast, as suggested by the Janzen-Connell hypothesis, pathogens, like a strain of fungus, may maintain alpha diversity in plant communities by reducing the survival rates of conspecif ic seeds and seedlings located close to reproductive adults or in areas of high conspecif ic density (Packer and Clay 2000). However,our understanding of the dif ferent functions of soil fungi and bacteria on plant diversity has yet to be validated in arid desert tree patches.
Soil physicochemical properties may also play an important role in the maintenance of plant diversity at a tree-patch scale due to their regulation of nutritional supplies (Chaparro et al. 2012; Silva et al. 2014; Gong et al. 2018). In arid desert forest, several studies consider that soil nutrients,salinity, pH and soil water are the most important abiotic factors af fecting plant diversity (Li et al. 2008; Yang et al. 2009;Leuzinger et al. 2015; Yan et al. 2015). Salt stress inhibits seed germination, plant survival, distribution through high osmotic pressure and imbalanced ion metabolism (Leuzinger et al. 2015; Yan et al. 2015; Zeng et al. 2020). Soil water is closely related to plant physiological metabolism, photosynthesis, reproduction and electrolyte balance (Li et al. 2008,2018b; Zhou et al. 2010). Water shortages result in plant mortality and reduced abundance (Gong et al. 2018; Zhang et al. 2018). pH af fects plant diversity through changing soil enzyme activities and root nutrient absorption (Sang 2009;Cambrollé et al. 2015). The improvement of soil nutrients,especially nitrogen and phosphorus, increases species abundance and richness in nutrient-poor desert ecosystems (Yang et al. 2009; Li et al. 2018b). However, these studies are generally carried out at a large scale, such as river basins, landscape transects and natural reserves (Li et al. 2008, 2018c;Zhou et al. 2010; Zeng et al. 2020).Whether these factors inf luence plant diversity at a micro-scale like tree patches is unclear. In addition, as described above, soil physicochemical factors and microorganisms af fect plant diversity;whether their contributions are dif ferent is not known.

Fig. 1 Dif ferences in abundance and richness between areas around large (big) and small trees in the study region;control is the area that does not support tree growth
Populus euphraticaOliv. is a dominant tree species in arid desert regions of Midwestern Asia, North Africa to southern Europe. The species has its largest range and highest population in China, especially in the northwestern regions.P.euphraticashows excellent tolerance to high temperatures and salinity, and is considered as a model species for studying plant adaptability to the environment (Yang et al. 2014).In the Ebinur Lake National Nature Wetland Reserve of the Xinjiang Uygur Autonomous Region in northwestern China,shrubs and herbaceous plants may grow sparsely around a singleP. euphraticaindividual (Fig. 1). Plant abundance and richness around a single individual dif fers between trees of dif ferent stem sizes as well as dif ference in distance from the trunk (Fig. 1). However, only a few studies related to plant diversity at tree patch levels have been conducted in this area(Yang et al. 2017; Wu et al. 2019). The purpose of this study is to explore the mechanisms of plant diversity maintenance in arid desert tree patches. We hypothesize that both soil physicochemical properties and microorganisms af fect plant diversity but their inf luences may be dif ferent. In order to verify this hypothesis, there are three main objectives in this study: (1) to understand the variability of plant diversity; (2)to determine the relationships of soil microorganisms and physicochemical properties to plant diversity; and, (3) to compare the relative contributions of soil microorganisms and physicochemical properties to plant diversity.
Materials and methods
Study site
The study site is located in the Ebinur Lake National Nature Wetland Reserve (82°36′–83°50′ E, 44°30′–45°09′ N) in the southern part of the Gurbantünggüt Desert, Xinjiang Uygur Autonomous Region, western China. The site has a typical arid temperate continental climate, with annual hours of sunshine approximately 2800 h; the mean annual precipitation is less than 100 mm. Annual potential evaporation ranges from 1500 to 2000 mm and the annual average temperature ranges from 6 to 8. The soil is part of the Arenosols according to the World Reference Base for Soil Resources (Yang et al.2019), and the surface salt content ranges from 4 to 8%. The zonal vegetation is xerophytic and composed of desert plants(e.g.,P. euphratica,Haloxylon ammodendronC.A.Mey.,Halocnemum strobilaceumPall.,Reaumuria soongonicaPall.,Nitraria tangutorumBobr., Halimodendron halodendronPall., Suaeda glaucaBunge, Alhagi sparsifoliaShap.andSalsola chinensisGand.The f irst six species are woody plants while the last three are herbaceous). Total vegetation cover is < 10% (Yang et al. 2014).
Experimental plot and plant community investigation
A 100 × 100 m long-term experimental plot (approximately 20 years-old) of naturalP. euphraticaforest was chosen as the study plot; 76P. euphraticatrees had 30% canopy cover.Three small trees and three large (big) ones were randomly selected in mid-July, 2018. Big and small trees were selected as the study subjects as plant abundance and distribution were different under their canopies (Fig. 1). The space between the selected trees was greater than 15 m. Height and diameter at breast height (DBH) def ined the two types of trees: small trees (height ≤ 4.5 m and DBH ≤ 20 cm),and large trees (height ≥ 7 m and DBH ≥ 60 cm). Height,DBH and crown area were measured with a Vertex-IV meter(Haglöf, Dalarna, Sweden), diameter tape and a meter stick,respectively. Based on the distance from the trunk, one ringed plot and one cambered plot were established around each big tree, while one ringed plot was established around each small tree. The radii of each ringed and cambered plot around the big tree were 3 m and 6 m, respectively, (i.e.,Big tree0−3 m, and Big tree3−6 m), whereas the radius of the ringed plot around the small tree was 3 m (i.e., Small tree0−3 m). The maximum radii of the ringed plots were 6 and 3 m for the big and small trees because their crown lengths were less than 12 m and 6 m, respectively. Outside the canopies of both, three circular plots were established as the controls. Therefore, the four experimental sites were:Big tree0−3 m, Big tree3−6 m,Small tree0−3 m,and control.In each plot, plant community characteristics, i.e., species name, abundance, richness, canopy cover and height, were recorded (Fig. 2). In order to remove the inf luence of dif ferent sampling areas on plant characteristics, all plots were 28.26 m 2 because this was close to the minimum area of the local plant community (25–30 m 2 ) (Yang et al. 2010a).
Soil sample collection and physicochemical property measurements
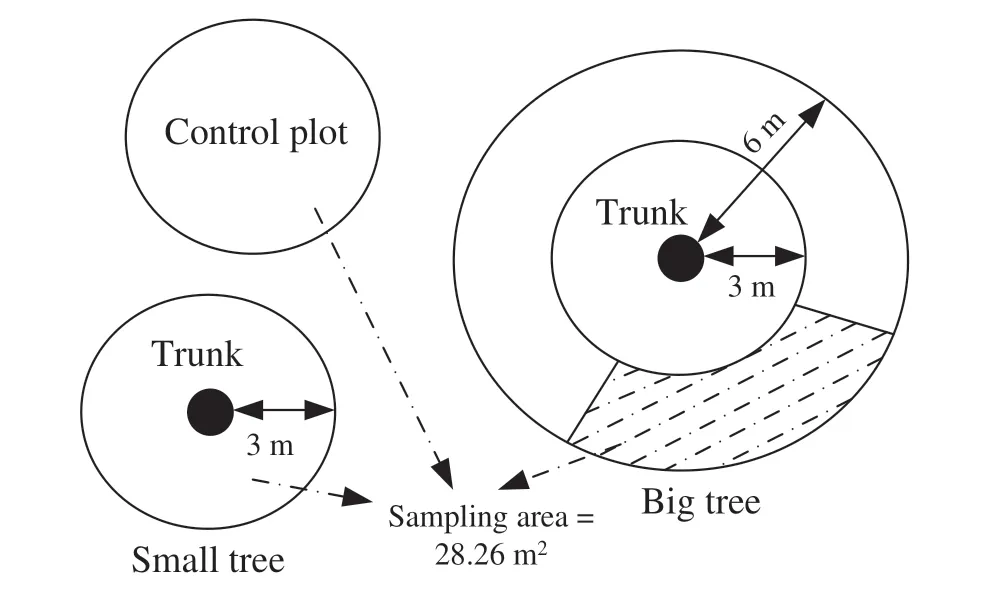
Fig. 2 Diagram of the ringed community around and outside the trunks
The ring-knife method, in which a ring-knife was f illed with soil, was used to measure soil bulk density (upper 20 cm layer) in three randomly selected points in each plot. Five soil samples from the upper 20 cm layer were randomly collected using a core sampler with an inner diameter of 2.5 cm.The sampler was sterilized with 75% ethanol before collecting each sample. In order to reduce the inf luence of spatial heterogeneity on microbial diversity and physiochemical property analysis, f ive soil samples were mixed into one composite sample in each plot. Therefore, each plot (Big tree 0−3 m, Big tree3−6 m,Small tree0−3 m,and control) included three representative composite soil samples. The mixed soil sample was divided into two parts, one packed in self-sealing plastic bags to determine physicochemical properties at the Plant Physiology Laboratory of Xinjiang University. The second part was placed into a 5 mL sterile freezing tube and stored at 4 °C for the determination of soil microbial community composition and diversity at the Noah Source Gene Sequencing Company in Beijing.
A wet sieve was used to separate f ine roots from other organic material (Gao et al. 2014). Based on the degree of cohesion between cortex and periderm and color and tissue resilience, living and dead roots were separated by hand. Live roots ≤ 2 mm in diameter were selected and oven dried at 80 °C for 48 h (Gao et al. 2014; Yang et al. 2019).Fine root biomass was calculated using the dry weight of root biomass and sampling area (i.e., (dry weight × 10 6 )/[π × D 2 /4]; g/m 2 ; where,Dis the inner diameter of soil cores as suggested by Yang et al. ( 2019). Soil water, organic matter (SOM), total nitrogen and total phosphorus contents were measured using oven drying, oil bath-K2CrO4titration, semi-micro Kjeldahl and acid-soluble-molybdenumantimony colorimetric methods, respectively (Cha 2017).Soil salinity was measured using a conductivity meter in 1:5 (w/v) soil suspension in deionized water (DDS-12A,Hongyi, Inc., Shanghai, China). The pH was determined in a 1:2.5 (w: v) soil suspension in water using a pH meter(PH3-3C, LEICI, Shanghai, China). All physiochemical measurements were repeated three times for each composite soil sample. Fine root biomass was included in physicochemical properties because it was regarded as an inf luential factor of microbial community composition and plant diversity, and also because it was equivalent to the other physicochemical factors such as SOM and total phosphorus in mathematical analysis. Some researchers have classif ied f ine root biomass as an organic part of the soil and considered it an important soil property (Zhu and He 1992). In this study,it was included under physicochemical properties in order to increase the convenience of data presentation and analysis.
Measurement of soil microbial community composition and diversity
The composition of soil microorganisms was measured using 16S rDNA amplifier sequencing. Soil DNA was extracted from 0.30 g fresh soil according to manufacturer’s instructions using the MoBio PowerSoil Extraction Kit(MoBio Laboratories Inc., Carlsbad, CA, USA). The quality of DNA was examined using a Nanodrop ND2000 spectrophotometer (Thermo Scientif ic, Wilmington, DE, USA).The V3-V4 regions of 16S rRNA gene of bacteria and ITS1 gene of fungi were amplif ied and sequenced for analysis.The bacterial 16S rRNA genes were amplif ied by the primer pairs Arch338F and 806R (Caporaso et al. 2012) and the fungal ITS1 gene amplif ied by the primer pair ITS5-1737F/ITS2-2043R. All samples were amplif ied in triplicate and no-template controls were included in all steps of the process. PCR products were visualized using electrophoresis on 1.5% agarose gels and purif ied using a GeneJET Gel Extraction Kit (Thermo Scientif ic). The purif ied PCR amplicons products were sequenced on the Illumina MiSeq (300-bp paired-end reads) platform (Illumina Inc., San Diego, CA,USA) at the Novogene Bioinformatics Technology Co., Ltd.(Beijing, China).
The acquired sequences were f iltered for quality according to Edgar ( 2010) and Haas et al. ( 2011). All ef fective tags of samples were clustered into operational taxonomic units (OTUs) using UPARSE (Version 7.0.1001) based on the 97% identity of sequences. Sequences with the highest occurrence in OTUs were selected as representative. Annotation analysis of representative sequences was conducted using the MOTHUR (Version 1.30.1) to obtain the community composition in each taxonomic level (i.e., kingdom,phylum, class, order, family and genus). PyNAST (Version 1.2) and “Core Set” of GreenGene database were used to obtain the phylogenetic relationship among OTUs and a normalized database. The results of a rarefaction curve of the gene sequence showed that OTU numbers of bacteria and fungi in each sample tended to be f lat with the increase of sequence number. This indicated that DNA sequencing had a high rationality and the measured sequence included the majority of species information on bacteria and fungi (Fig.S1). Good coverage of bacteria and fungi reached 99.1%to 99.8%, respectively, also indicating that most of the soil microorganisms were captured in the DNA sequence.
Soil microbial diversity was taken as the α-diversity,i.e., Shannon—Wiener diversity and abundance-based coverage estimator (ACE) richness indices, which was calculated using QIIME 1.8.0 software based on a normalized database of OTUs. Venn diagram, β-diversity and a principal coordinate analysis (PCoA) were conducted to test the dif ference in the composition of the soil microbial community among the four sites. β-diversity was calculated by both taxonomic and phylogenetic measurements.Taxonomic measurements were based on the normalized database of OTUs using the Bray—Curtis distance matrix,while phylogenetic measurements were based on both the normalized database of OTUs and community phylogeny using the Unweight Unifrac distance matrix. The calculation of PCoA was based on the weighted Unifrac distance matrix. The weighted, unweighted UniFrac and Bray–Curtis distance matrices were calculated by QIIME 1.8.0 software based on a normalized database of OTUs.
Statistical analysis
One-way analysis of variance (ANOVA) examined the dif ferences in soil physicochemical properties, plant and microbial diversity among Big tree0−3 m, Big tree3−6 m,Small tree0−3 mand the control. Individual tree (or experimental replicates) was set as the random ef fect. If the variance of the indicators was homogeneous, the least-square mean separation with Duncan’s correction was used to test the dif ferences among the four treatments. Alternatively, if the variance was heterogeneous, Tamhane’s T3 test was used to test the dif ferences. Plant diversity indices included Shannon–Wiener and Simpson diversity which were calculated using the data of plant community characteristics (Lakhani 1989).
At the genus level, the Bray—Curtis distance-based redundancy analysis (db-RDA) with forward selection was used to explore the relationships between physicochemical properties and microbial composition. Hierarchical partitioning analysis (HPA) was used to obtain the relative contributions of soil physicochemical properties and microbial diversity to the variability of plant diversity among the four sites. Contribution is the proportion of each independent variable from the goodness-of-f it measures across all variable combinations in a hierarchy(Chevan and Sutherland 1991). Prior to HPA, Pearson correlation analysis was used to reduce the number of independent variables. Our results showed that pH and soil bulk density were not relate signif icantly with plant diversity (p> 0.05) and these two factors were not included in the HPA (Table S1). PCoA, db-RDA, HPA and Pearson correlation analysis were conducted using R. 3.4.3 software. The PCoA and db-RDA were evaluated in theveganpackage. Hierarchical partitioning analysis was evaluated with therdacca.hppackage (Lai and Peres-neto 2020).
Results
Dif ferences in plant diversity across the four sites
One-way AVONA results show that Shannon–Wiener and Simpson diversity were highest in control relative to other sites (Fig. 3 a, b). The ef fect of plant size on plant diversity indicated that Shannon–Wiener and Simpson diversity at Small tree0−3 mwere not signif icantly dif ferent from that at Big tree3−6 m(p> 0.05) but were signif icantly dif ferent from Big tree0−3 m(p< 0.05). The ef fect of distance from tree trunk on diversity indicated that Big tree3−6 mhad higher Shannon–Wiener and Simpson diversity compared with Big tree0−3 m(Fig. 3 a, b).
Dif ferences in soil physicochemistry, microbial community composition and diversity among the four sites
One-way ANOVA showed that f ine root biomass, SOM,salinity, soil water, and total nitrogen and phosphorus contents signif icantly changed over the four sites (Fig. 4), while pH and soil bulk density did not change (p> 0.05) (Table S1 and Fig. S2). Pairwise comparison indicated that these were signif icantly highest in the Big tree0−3 mthan other sites(p< 0.05) (Fig. 4). Salinity was signif icantly higher in Big tree 3−6 m than Small tree 0−3 m and control (p< 0.05) (Fig. 4).Fine root biomass, SOM, soil water, total nitrogen and phosphorus contents showed no signif icant dif ference among Big tree3−6 m, Small tree0−3 mand control (Fig. 4). The control had similar soil physicochemical properties as the Small tree 0−3 m . .
At dif ferent taxonomic levels, the four sites were distinguished from each other in their bacterial compositions (Fig. S3 − S7). At the phylum level of bacteria,Bacteroidetes was more abundant in Big tree3−6 m,while Proteobacteria was more abundant in Big tree0−3 m. The abundance of Actinobacteria and Chlorof lexi followed the order: Small tree 0−3 m > control > Big tree 0−3 m > Big tree 3−6 m (Fig. S3). At the genus level, members ofGillisiawere higher in Big tree3−6 mthan that in other three sites, while members of the generaAliifodinibiusandSalinimicrobiumwere higher in Big tree0−3 mand Big tree3−6 mthan that in the other two sites. Abundance ofSphingomonasandAdhaeribacterspecies followed the order: Small tree0−3 m> control > Big tree0−3 m> Big tree 3−6 m(Fig. S7). Similarly, fungal community compositions in the four sites were also distinguished at the levels of phylum, class, order, family, and genus. At the phylum level, members of Ascomycota were highest in Big tree 0−3 mwhile Mortierellomycota followed the order: Small tree 0−3 m > control > Big tree 0−3 m > Big tree 3−6 m (Fig.S3). At the genus level,unidentif ied_ SordariomycetesandRussulawere more abundant in Big tree 0−3 m , whilePenicillium,MortierellaandAmauroascuswere highest in the Small tree0−3 m. The abundance ofAlternariafollowed the order: control > Big tree3−6 m> Small tree0−3 m> Big tree 0−3 m (Fig. S7). The term “unidentif ied” means that a microbe ofSordariomycetesis currently named only at the class level but not named and classif ied at the subtaxonomic level due to limited information.
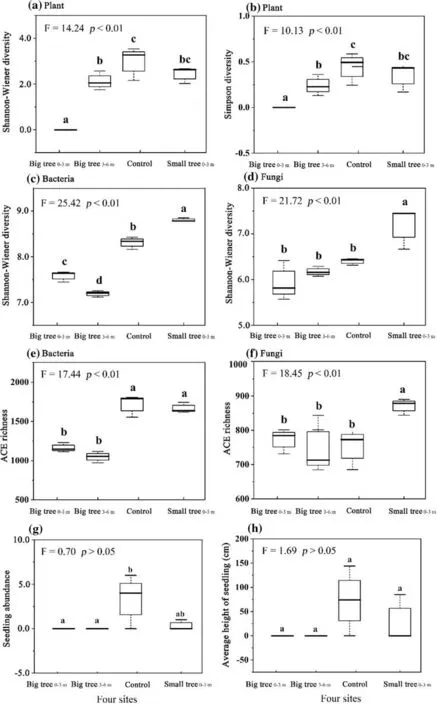
Fig. 3 Dif ferences in Shannon–Wiener and Simpson diversities,microbial diversity (i.e., Shannon–Wiener diversity and ACE richness), and seedling height and abundance of dominant species among the four sites;Big tree 0−3 m and Big tree 3−6 m are the sampling areas 0 − 3 m and 3 − 6 m away from the big tree trunk; Small tree 0−3 m is located 0 − 3 m away from the small tree trunk. Control is the area without any dominant tree species
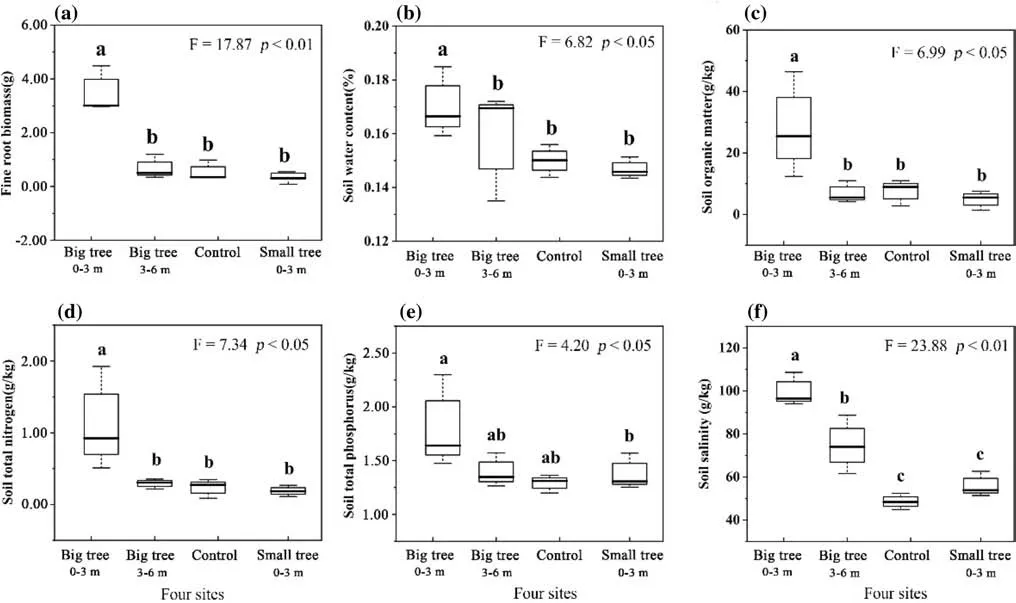
Fig. 4 Dif ferences in soil physicochemical properties among the four sites
A Venn diagram demonstrated that operational taxonomic units (OTUs) dif fered among the four sites (Fig. 5 a). In the bacterial community, there were 216, 39, 430 and 313 sitespecif ic OTUs associated with Big tree0−3 m, Big tree3–6 m,Small tree0−3 mand control, respectively. Only 479 in 3011 OTUs were shared in all four sites (15.9% in total OTUs)(Fig. 5 a). In the fungal community, the numbers of site-specif ic OTUs were 252, 218, 218, and 123 for Big tree0−3 m,Big tree3−6 m, Small tree0−3 m, and control, respectively.Only 423 in 2063 OTUs were shared by all four sites (20.5%in total OTUs) (Fig. 5 b).
The β-diversity of microbial communities demonstrates a signif icant variation among the four sites (Fig. 5 c, d).Higher Unweight Unifrac distances indicate larger dissimilarities of microbial community composition between two paired sites. In the bacterial community, there were small Unweight Unifrac distances between Big tree0−3 mand Big tree3−6 m(0.386), while a larger distance between Big tree 0−3 mand control (0.568) was observed. Small tree0−3 mhad a smaller distance to control (0.373), whereas there was a larger distance between Big tree0−3 m(0.641) and control(0.579) (Fig. 5 c). In the fungal community, Big tree 0−3 m had a small distance to Big tree 3−6 m (0.567), while there was a larger distance between Small tree0−3 m(0.571) and control(0.560). Small tree0−3 mhad a smaller distance to control(0.389), whereas there was a larger distance to Big tree0−3 m(0.571) and Big tree3−6 m(0.546) (Fig. 5 d).
Principle coordinate analysis showed that the composition of microbial communities signif icantly dif fered among the four sites because of clear clustering (p< 0.01) (Fig. 6).Both in the bacterial and fungal communities, small distance between Small tree0−3 mand control suggest that both had similar microbial community composition. In contrast, larger distances between Small tree0−3 m, Big tree0−3 mand Big tree 3−6 m , as well as control, demonstrates that the microbial community composition in Small tree 0−3 m and control were dissimilar to Big tree0−3 mand Big tree3−6 m(Fig. 6).
Shannon–Wiener diversity and ACE richness of microbial communities also showed a signif icant dif ference among the four sites (Fig. 3 c–f). Specif ically, in the bacterial community, both were the highest in control, the intermediate in Big tree0−3 m, and the lowest in Big tree3−6 m. These indices were not signif icantly dif ferent between control and Small tree0−3 m(Fig. 3 c, e). In the fungal community,Shannon—Wiener diversity and ACE richness followed the order: Small tree0−3 m> control ≥ Big tree3−6 m≥ Big tree 0−3 m.The Shannon–Wiener diversity and ACE richness were signif icantly dif ferent in Small tree 0−3 m relative to other sites, while no signif icant dif ference was observed among Big tree0−3 m, Big tree3−6 mand control (Fig. 3 d, f).

Fig. 5 Dif ferences in community composition of soil microorganisms among the four sites; Fig. 5 a, b display the degree of overlap of microbial OTUs among the four sites; Fig. 5c, d show dif ferences in Unweight Unifrac distances among the four sites (beta diversity).Higher Unweight Unifrac distances indicate lager dif ference in community composition of soil microorganisms between two paired sites
Relationships among soil physicochemistry,and microbial and plant diversity
The Bray–Curtis distance-based redundancy analysis (db-RDA) with forward selection demonstrated that soil physicochemical properties were signif icantly connected with microbial composition at the genus level (p< 0.01), which explained 90.9% and 87.9% of variations for bacterial and fungal communities, respectively (Fig. 7). Among soil physicochemical properties, f ine root biomass and salinity were the most important variables that correlated with bacterial community composition. Salinity had a larger inf luence on bacterial community composition than f ine root biomass.However, other soil physicochemical properties, including total nitrogen, total phosphorus, soil water and SOM, were not reserved in db-RDA, indicating that these parameters were not correlated with bacterial community composition (Fig. 7). For the fungal community, salinity, f ine root biomass, SOM and total nitrogen had signif icant correlation (p< 0.01) with community composition, whereas total soil phosphorus and water were not correlated. The inf luence of physicochemical properties on fungal community composition followed the order: salinity > f ine root biomass > SOM > total nitrogen (Fig. 7).
Hierarchical positioning analysis (HPA) showed that physicochemical properties and microbial diversity af fected the alpha diversity of tree patches (Fig. 8). The average inf luence of physicochemical properties to plant Shannon—Wiener (12.2%) and Simpson (13.5%) diversities were higher than microbial diversity (i.e., Shannon—Wiener diversity = 6.7%; and ACE richness = 4.8%) (Fig. 8 a, b). Fungi diversity had a smaller contribution to plant diversity compared with the diversity of bacteria (Fig. 8 a, b). Soil salinity had the greatest ef fect on plant diversity compared with other properties (Shannon–Wiener diversity = 27.4%; and Simpson diversity = 24.7%). The contributions of f ine root biomass, SOM, soil water, total phosphorus, total nitrogen to Shannon–Wiener diversity were 13.6%, 6.8%, 12.1%, 5.3%,and 7.8%, while Simpson diversity values were 20.7%, 8.7%,9.7%, 7.4%, and 9.4%.

Fig. 6 Principal Coordinate Analysis (PCoA) prof iles for soil microbial communities displayed using Weighted UniFrac distances
To further clarify the dif ferent inf luence of bacteria and fungi on plant diversity, HPA was re-used to obtain the connection between plant diversity and the most dominant genera of soil microorganisms. The f ive most dominant genera were selected from each bacterial and fungal community to construct the hierarchical partitioning analysis as they may play major roles in the relationship between microorganisms and plants (Na et al. 2018). The results show that the average contribution of these f ive dominant bacteria genera to plant Shannon–Wiener (11.5%)and Simpson (11.8%) diversities were higher than that of fungi (Shannon–Wiener 8.5%; Simpson 8.2%) (Fig. 8 c and 8d). However, among all the dominant genera, a genus of fungi, unidentif ied_Sordariomycetescontributed the most to Shannon–Wiener (30.4%) and Simpson (20.9%) diversities than other dominant genera (Fig. 8 c and 8d).

Fig. 7 Bray–Curtis distance-based redundancy analysis (db-RDA)with forward selection showing correlations between bacterial communities and soil physicochemical properties; arrows are physicochemical properties; BN, BF, C and S are sampling codes of 12 samples representing Big tree 0−3 m , Big tree 3−6 m , control and Small tree 0−3 m , respectively; data analyzed at the genus level of soil microorganisms; only ten most dominant genera of soil microorganisms are presented
Discussion
Variabilities of soil physicochemistry and microbial diversity in tree patches
Fine root biomass, soil organic matter, salinity, water, total nitrogen and phosphorus differed among the four sites(Fig. 4), indicating that tree size and the distance to the tree trunk af fected mineral nutrients, organics and water. This might be inf luenced by the “island of fertility” ef fect and hydraulic lift (Vessey 2003; Yang et al. 2017; Ochoa-Hueso et al. 2018). The “island of fertility” ef fect is the phenomenon where trees act as nutrient pumps, taking up nutrients from deep soil layers and soil outside the tree’s canopy and deposit the nutrients under the tree canopy through litter fall and/or leaching (Ludwig et al. 2004). Another explanation of the “island of fertility” is that trees are an ef fective trap for nutrient-rich dust f loating in the atmosphere or to attract mammals which deposit their dung under the tree canopy (Vessey 2003; Ochoa-Hueso et al. 2018). In the patches, mineral nutrients and organic accumulations in close proximity to the trunk are likely to be higher than in more distant places due to the reductions of radial roots and crown depth (Vessey 2003; O’Brien et al. 2017). Therefore,salinity, SOM, total nitrogen and total phosphorus contents decreased with increasing distance from the trunk (Big tree 0−3 m> Big tree3−6 m> control). Similarly, compared with small trees, big trees had greater inf luence on the “island of fertility” ef fect on physicochemical properties because of larger root systems and crown areas. Hence concentrations of mineral nutrients and organic materials were higher in areas close to big trees. In arid deserts, Yang et al. ( 2017)and Li et al. ( 2018a) found that deep-rooted species such asP. euphraticaandH. ammodendronhave hydraulic lift properties that transport water from moist layers to drier layers via their roots. Due to the positive relationship between the water amounts of the hydraulic lift and root distribution, soil water decreased with an increasing distance from the trunk and was higher in areas closest to large trees (Yang et al.2017). As tree species were not in the control group, the“island of fertility” ef fect and hydraulic lift properties were not present, and thus the contents of soil minerals, organic matter and water were lower in controls than in areas around big and small trees (Vessey 2003; Zhu et al. 2017). The pH and soil bulk density were not signif icantly dif ference among the four sites (Fig. S2), indicating that tree size and distance to the trunk did not af fected pH and soil bulk density. This is related to the nutrient-poor and highly salinized soils. In arid deserts, soil is mainly constituted of sand, highly subjected to salinization and alkalization (pH > 7.6) (Zhang et al.2018). Organic inputs, such as litter fall and root exudates,can reduce soil bulk density and pH but on a spatial microscale such as a tree patch, they do not result in a signif icant reduction of these two properties because of limited inputs.
Soil microbial composition and diversity were also affected by tree size and distance from trunk (Fig. 3,5– 6 ). Previous studies have shown that soil microbial composition and diversity were mainly affected by physicochemical properties, plant litter and root exudates (el Zahar et al. 2014; Huangfu et al. 2019). In tree patches,due to the influence of the “island of fertility” effect and the changes of mass distribution of foliage and roots, the magnitudes of soil physicochemical properties, plant litter and root exudates decreased with increasing distance from the trunk, and higher values were expected in the areas close to big trees (Chaparro et al. 2012; Huangfu et al. 2019). In this study, microbial composition and diversity showed differences among the four sites. This result was also shown by the Venn diagram and the variation of β-diversity of microbial communities (Fig. 5).The similarity in plant-environment relationships, such as the “island of fertility” effect, resulted in Small tree0–3 mhaving the largest number of mutually shared OTUs, and the smallest Unweight Unifrac distances with the control group, while displaying the largest differences in site-specific OTUs and distance with the Big tree0−3 m(Fig. 5).
Fine root biomass and salinity have greater influence on soil microbial composition and diversity compared with other soil properties (Fig. 8). In arid desert areas,high soil salinity leads to excessive water loss and death of microbial cells, thus changing microbial community composition and diversity (Gong et al. 2018; Zhang et al.2018). It is well-known that all microorganisms consume organic matter to sustain life (Yang et al. 2010b). Fine root biomass correlated positively with the amount of root exudates and organic matter volume (Tian et al. 2018;Huangfu et al. 2019). Changes in fine root biomass results in an obvious difference in microbial composition and diversity. In addition, our results show that soil organic matter and total nitrogen affected fungal community composition but not bacterial composition, indicating the influence of these two properties differs. This may be related to the different living properties between these two organisms (Peng et al. 2019). Compared with bacteria, fungi have higher demands on soil environmental conditions, especially nutrition (Chaparro et al. 2012),therefore, changes of SOM and total nitrogen would alter fungal community composition and diversity.
Comparative ef fect of soil physicochemistry and microbial diversity on plant diversity
There was signif icant plant diversity over the four sites(Fig. 3). Soil physicochemical properties had a greater contribution to plant diversity than microbial diversity (Fig. 8).This was because the soil was a nutrient source to the plants and acted as an intermediator regulating the inf luence of soil microorganisms on plant diversity (Bever et al. 1997; Lou et al. 2016; Lozano et al. 2017). The inf luence of soil physicochemistry on microbial community composition amplif ies the interrelationship between soil microorganisms and plant diversity (Ochoa-Hueso et al. 2018; Tian et al. 2018).As noted previously, these results indicate that soil physicochemical properties were signif icantly correlated with microbial composition at the genus level (Fig. 7).
Salinity had the greatest ef fect on plant diversity (Fig. 8 a,b), because it is one of the main limiting environmental factors in arid deserts (Giuseppe et al. 2008; Gong et al.2018 ). Salt stress in soils was considered the most important obstructive reason for seed germination and seedling survival (Hegarty 1978; Giuseppe et al. 2008). Thus, a decrease in salinity with increasing distance from the trunk increases plant diversity simultaneously through improved seed germination and seedling survival (Fig. 3 a, b) (Giuseppe et al.2008; Huangfu et al. 2019). Our results also suggest that f ine root biomass was the second largest contributor to plant diversity (Fig. 7, Fig. 8 a, b). This was possibly due to the physiological property ofP. euphratica.As a typical secretohalophyte or salt tolerant species,P. euphraticaexcretes large amounts of electrolytes from its f ine roots into soils(Hu and Tsen-Li 1993). Since such species have radial root systems that extend well beyond the trunk, a high abundance of f ine roots in close proximity of trunks would accumulate more salts. Thus, f ine root biomass has a negative relationship with plant diversity through its inf luence on the distribution of salinity. Additionally, variability in f ine root biomass changes nutrient distributions via root release(Leuzinger et al. 2015). As a result, it af fects soil microbial community composition and diversity. Since there was signif icant correlation between soil microorganisms and plant survival and abundance, the variability of f ine root biomass af fects plant diversity.
Soil organic matter, soil water, total phosphorus and total nitrogen also negatively af fected plant diversity at the tree patch (Table S1). This result dif fers from previous studies that showed a positive af fected (Yang et al. 2009; Zhang et al. 2018; Zeng et al. 2020). Since water is a limiting factor of forest ecosystems in arid areas, the increase in soil water was benef icial to the improvement of plant diversity(Li et al. 2008). However, there was a negative correlation between soil water and plant diversity at the tree patch. This may be due to the synergic transport between soil salinity and water (Zhang et al. 2018). In our study area, soil salinity and water content were highly correlated because salt was transported from deep soil layers to the surface for evaporation (Ma et al. 2018). Plants in soils with high water levels demonstrate that they also suf fer from high salt stress. This was also conf irmed by Gong et al. ( 2018) and Zhang et al.( 2018) who found that plant diversity in habitats with high water and salt levels was lower than in ecosystems with less water and salt. Soil organic matter, total nitrogen and phosphorus were negatively correlated with plant diversity,which may be the cause of the negative impact of pathogen attack on plant survival (Liu et al. 2012). As noted previously, this study found that soil nutrition is benef icial for the abundance and diversity of fungi. In this case, the number of dead seedlings and ungerminated seeds caused by pathogen attack would increase with increased nutrient levels, thus reducing plant diversity. In addition, allelopathy, shading and resource competition of maturePopulus euphraticaon herbs and shrubs would negatively af fect their growth and survival at a tree patch (Sher et al. 2011; Yang et al. 2017).Allelopathy is a biological process by which mature trees produce one or more chemicals that inf luence the growth,survival, and reproduction of other plant species (Sher et al.2011). However, our study did not consider these factors and more research are needed.
Soil fungi and bacteria af fect plant diversity dif ferently
Unidentif ied_Sordariomycetescontributed the most to plant diversity compared with other dominant fungal genera(Fig. 8 c, d). Its relative abundance followed the order: Big tree 0−3 m > Big tree 3−6 m > Small tree 0−3 m > control (Fig.S6). These results indicate that fungi were negatively correlated with plant diversity. Previous studies have reported thatSordariomycetesinhibited seedling survival because this genus includes many pathogenic fungal species (Yang et al. 2018). As suggested by the Janzen-Connell hypothesis, with the increase in plant size, pathogen attack might reduce survival rates and density, and constrain height growth of conspecif ic seedlings located close to big trees(Liu et al. 2012; LaManna et al. 2016). In this study, both the abundance and tree height ofP. euphraticaseedlings increased with increasing distance from the big tree trunk,and also were higher around small trees than around big trees (Fig. 3 g, h), indicating that pathogen attack reduced plant diversity (Janzen 1970; Liu et al. 2012). However, our results dif fered from previous studies on the Janzen-Connell hypothesis showing that the abundance of heterogenic plants located close to mature trees did not change with distance from the tree trunk (Liu et al. 2012). Our results show that the abundance of heterogenic plants increased with increasing distance from the trunk (Table S2). Heterogenic plants are genetically and taxonomically dif ferent from matureP.euphratica, i.e., they are dif ferent from mature trees and not the result of seed germination from trees. The dif ference of our f inding from that of Liu et al. ( 2012) could be ascribed to the variability of soil physicochemical properties. More specif ically, the inf luence of soil physicochemical properties on plant diversity, especially the constraint of soil salinity on plant diversity, increased the abundance of heterogenic plants with increasing distance from the tree trunk.
Except for theunidentif ied_Sordariomycetes, the contributions from other dominant fungal genera on plant diversity were lower than bacteria (Fig. 8 c, d). The average contribution of the f ive most dominant genera of bacteria to plant diversity was also higher than that of fungi (Fig. 8 c,d). These results suggest that soil bacteria had a greater impact on plant diversity than fungi at tree patches. This was possibly because plants may have a closer relationship with bacteria than fungi under nutrient-poor environments(Ingham et al. 1985; Yang et al. 2010b; Asaf et al. 2018).Additionally, previous studies have shown that the inf luencing mechanism of bacteria to regulate plant diversity dif fers from that of fungi (Slabbert et al. 2010; Cho et al. 2018).Bacteria may improve plant diversity through the distribution of functionally identical strains along dif ferent sampling positions (Bulgarelli et al. 2015). For example, among the f ive most dominant genera of bacteria, two halophilic strains,AliifodinibiusandSalimicrobium, were positively related to plant diversity due to their ability to decrease salt stress (Cho et al. 2018). The other strain among the f ive most dominant bacterial genera,Sphingomonas, also was positively related to plant diversity because it helped plants resist oligotrophic stress and enhanced root activity in nutrient-poor soils (Asaf et al. 2018). In addition, sinceSphingomonassecrets growth hormones during metabolism, an increase in its abundance would promote plant growth and improve plant diversity in nutrient-poor environments (Asaf et al. 2018).
Conclusions
The collective inf luences of soil physicochemical properties and soil microorganisms on plant diversity in arid desert tree patches were examined. The results show that plant diversity, physicochemical properties, and microbial community composition and diversity dif fered with distance from the tree trunks, as well as between areas around large and small trees. Salinity had the largest ef fect on plant diversity compared with other soil properties. These results are in agreement with the limiting factor hypothesis, suggesting soil salinity as one of the most important factors for determining plant diversity in arid desert regions. The results also show that soil fungi and bacteria may shape plant diversity in dissimilar ways. Bacteria may enhance diversity through environmental improvements via functional strains, whereas fungi may constrain diversity through pathogen attack. However, microbial inoculation was not used to verify the different inf luences between fungi and bacteria, especially for dominant microorganisms (e.g.,unidentif ied_Sordariomycetes,Sordariomycetes, Aliifodinibius, SalimicrobiumandSphingomonas) on seed germination and seedling survival in tree patches. Allelopathy, shading and resource competition by mature trees over herbs and shrubs may af fect plant diversity but these were not examined in this study. Further study is needed to fully reveal plant diversity maintenance in tree patches. This study provides a new insight for explaining the maintenance of plant diversity in tree patches in arid desert regions.
AcknowledgementsWe are highly grateful to anonymous reviewers,handling editor (Yanhui Wang) and Corresponding editor (Zhu Hong)for their insightful comments which greatly improved an earlier version of this manuscript.Author contributions X.D. Yang, Y.X. Long, Y. Cao, A. Ali and J.J.Yang developed the original idea and the protocol, abstracted and analyzed data, wrote the manuscript. B. Sarkar, Y. Li and G.H. Lv contributed to the development of the protocol and prepared the manuscript.
Compliance with ethical standards
Conf lict of interestThe authors declare that the research was conducted in the absence of any commercial or f inancial relationships that could be construed as a potential conf lict of interest.
杂志排行
Journal of Forestry Research的其它文章
- Genome-wide identif ication and cold stress-induced expression analysis of the CBF gene family in Liriodendron chinense
- Decay rate of Larix gmelinii coarse woody debris on burned patches in the Greater Khingan Mountains
- Characterizing conservative and protective needs of the aridland forests of Sudan
- Point-cloud segmentation of individual trees in complex natural forest scenes based on a trunk-growth method
- Accuracy of common stem volume formulae using terrestrial photogrammetric point clouds: a case study with savanna trees in Benin
- Appropriate search techniques to estimate Weibull function parameters in a Pinus spp. plantation
