Genome-wide identif ication and cold stress-induced expression analysis of the CBF gene family in Liriodendron chinense
2021-12-24YuanlinGuanSiqinLiuWeihuangWuKaiyueHongRongxueLiLimingZhuYangLiuYeLuJinhuiChenLimingYangJisenShi
Yuanlin Guan · Siqin Liu · Weihuang Wu ·Kaiyue Hong · Rongxue Li · Liming Zhu · Yang Liu ·Ye Lu · Jinhui Chen · Liming Yang · Jisen Shi
Abstract Cold-resistance pathways that operate in model plants such as Arabidopsis thaliana and Oryza sativa have been studied extensively. It has been found that CBF genes play an important role in plant cold resistance. Liriodendron chinense, a tree known for its graceful tree shape and widely spread in south China, has weak cold tolerance. However,little is known about its response to cold. To further study the function of L. chinense CBF gene family, we started by characterizing all members of this gene family in the L. chinense genome and their expression prof iling. Phylogenetic analysis found that 14 CBF genes in L. chinense are more closely related to their homologues in woody plants and A.thaliana than those in O. sativa. Cis-acting elements and GO analysis showed that some LcCBF genes participated in the biological process of cold stress response. The transcriptomic and RT-qPCR data showed that most of LcCBF genes displayed an initially increasing and subsequently decreasing trend during cold stress course and the expression prof ile of each member was dif ferent. Some LcCBF genes exhibited a dif ferent abundance in callus, root, stem and leaf tissues.The structure and expression characteristics of LcCBF genes imply that they may have similar and dif ferent functions in response to cold stress conditions. The identif ication and analysis of LcCBF gene family have laid the foundation for future studies into L. chinense cold stress mechanisms and for the cultivation of cold-resistance cultivars.
Keywords Liriodendron chinense · CBF genes ·Phylogenetic relation · Expression prof ile · Cold stress
Introduction
Low-temperature is an important ecological factor that restricts plant growth and geographical distribution, and is also a major natural disaster that af fects agricultural and forestry production (Beck et al. 2007). Low-temperature stress elicits morphological changes in plants, also causes metabolic disorders (Devacht et al. 2011; Erdal et al. 2015).Plant resistance to the low-temperature is the result of longterm evolution and is controlled by genetic factors. In the process of plant evolution, complex and ef ficient response mechanisms have been formed (Zhou et al. 2018; Jiang et al.2020). Plants can survive in the low-temperature environment through the coordination of morphology, physiology and biochemistry (Zhao et al. 2013; Barrero-Gil and Salinas 2017).
When plants were subjected to low-temperature stress, a series of resistance-related genes were induced, for exampleCBFandCOR(Theocharis et al. 2012). It was reported thatArabidopsis thalianacould rapidly initiate the expression of the AP2-type transcription factors CBFs (C-regenerationbinding factors) under cold stress (Azar et al. 2011). CBF,as a transcription activator, specif ically recognizes and binds to CRT/DRE cis-acting elements in promoters to activate the expression of stress response genes, thereby improving plant resistance to low-temperature (Hiraki et al. 2019).
One suchCBFgene, CBF/DREB1, binds to the cis-acting element CRT/DRE to activate the expression of the downstream cold-responsive COR (cold-regulated) gene, which leads to improve plant cold resistance (Gilmour et al. 1998).CBF/DREB1 is regulated by upstream transcription factors,including Inducer of CBF Expression 1 (ICE1). The ICE1-CBF-cold response pathway was also regulated by the transcription factors CAMTA3 and ICE1 respectively (Goldman et al. 2006).ICE1is inactive at normal temperatures inA.thaliana. When plants are exposed to low-temperatures, the expression ofICE1is activated and the protein of theICEgene binds to the ICE box, an associated cis-acting element in theCBFgene promoter, will induce the expression of theCBFgene (Ma et al. 2014; Bai et al. 2018).CBF1is also a transcriptional activator of theCORcold response gene,which can specif ically bind to the core element CRT/DRE and promote the expression of downstreamCORgene. It leads to activate the mechanism of plant resistance to abiotic stress (Zhou et al. 2018; Artlip et al. 2019).
TheCBFgenes are highly conserved within the plant kingdom. There are four members of theA. thaliana CBFgene family:AtCBF1(AT4G25490),AtCBF2(AT4G25470),AtCBF3(AT4G25480) andAtCBF4(AT5G51990), each of which acts on separate pathways (Park et al. 2015).AtCBF1,AtCBF2andAtCBF3all have the same DNA domain AP2 element and are highly homologous (Zhao et al. 2013; Yuasa et al. 2014; Yamasaki and Randall 2016). The three genes share a common ancestor and are closely related to each other; they all reside on chromosome 4 ofA. thaliana(Tang et al. 2020). These three CBF members have dif ferent roles in response to cold treatments through dif ferent pathways.AtCBF4shares relatively low homology and is located on chromosome 5 ofA. thaliana. It cannot be induced by low-temperature, but can be induced by drought and ABA.The tomatoCBFfamily is arranged on chromosome 3 in the orderLeCBF3,LeCBF1andLeCBF2. The similarity is 70–84% between these proteins, and their homology to AtCBF1, AtCBF2 and AtCBF3 proteins is 51–59% (Zhang et al. 2004). In Mangrove (Peng et al. 2020a, b), Wheat(Badawi et al. 2007),Populus simonii(Li et al. 2015),Populus euphratica(Tian et al. 2017),Camellia sinensis(Hu et al. 2020), the identif ication and functional characteristics of theCBFgene family have also been studied, and the regulatory ef fect of CBF on the cold resistance of plants has been conf irmed in these species. Overexpression of theCBFgene signif icantly improved the frost resistance ofBirch(Lv et al.2020) andEucalyptus grandis(Azar et al. 2011).
Liriodendron chinense(Hemsl.) Sarg. is a plant within the genusLiriodendron, Magnoliaceae, a uniquely rare and relic plant in China (Yang et al. 2016; Wu et al. 2020), and is an important wood, landscape and medicinal tree (Chen et al. 2019; Hao et al. 2019; Yang et al. 2019; Li et al. 2020).L. chinenseis mainly distributed naturally within mountainous forests at a 1000-m altitude in southern China (Xu et al.2017; Chen et al. 2018; Ma et al. 2018; Wu et al. 2018).Given the weak cold resistance, the cultivation and promotion ofL. chinensein northern areas of China has thus far been limited (Li et al. 2014). With the completion of theL.chinensegenome, it is possible to explore and study important functional genes ofL. chinenseat a genome-wide level.Cloning and expression analysis of genes related to stress resistance have theoretical and practical value for the mining and application of stress resistance genes inL. chinense. In this study we characterize theL. chinense CBFgene family,analyze gene structure, cis-acting elements, and collinearity,and investigate their expression prof iles under low-temperature stress and dif ferent tissues. These results will lay the foundation for further study ofCBFgene structure and function inL. chinenseresponse to cold resistance.
Materials and methods
Identif ication of CBF family genes
A. thalianaandOryza sativaCBF proteins were acquired from TAIR ( http://www.arabi dopsi s.org/) and Phytozome ( https ://phyto zome.jgi.doe.gov/pz/porta l.html). TheAmborella trichopoda(A. trichopoda),Populus trichocarpa(P. trichocarpa) andSalix purpurea(S. purpurea) similar CBF protein sequences were identif ied from Phytozome( https://phyto zome.jgi.doe.gov/pz/porta l.html) through the program BLASTN 2.2.26 + (E-value<1.0E-10) with theA.thalianaandO. sativaCBF proteins. The CBFs ofC. sinensiswere downloaded from NCBI ( https://www.ncbi.nlm.nih.gov/).L. chinenseprotein sequences were downloaded from Genome Database ( https://hardw oodge nomic s.org).The CBF Pfam number (PF03547) was queried to search forLcCBFprotein sequences using HMMER3.0 software.LcCBFsequences were further authenticated based on the conserved domains using SMART ( http://smart .emblh eidel berg.de). Biochemical properties, such as the molecular weight (Da) and isoelectric point (pI) of each protein, were determined using the Compute pI/Mw tool on the Expasy website ( https://web.expas y.org/protp aram/).
Phylogenetic and GO enrichment analysis
The phylogenetic relationships between LcCBFs and the CBF proteins fromA. thalianaandO. sativawere determined. Evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987). Evolutionary distances were computed using the Poisson correction method, using as a unit the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016). GO (Gene Ontology) annotation enrichment prediction analysis was carried out on the biological function ofCBFgenes by using blast2GO 5.2.5 software.
Chromosomal location and collinearity analysis of the L. chinense CBF genes
The chromosomal locations of theLcCBFgenes were determined based on theL. chinensegenome. Their exon/intron structure was determined with Tbtools (v1.049) using mRNA and theL. chinensegenome. The genomic and coding sequences ofCBFgenes, together with their exon/intron structures, were extracted from the general feature format(GFF3) f ile ofL. chinensesequences. Collinearity analysis of theL. chinense CBFgenes was conducted by BLASTbased method (Bi-direction best hit) to verify the putative orthologous genes (E-value cutof f < 1E-20, identity > 70%)withA. thalianaas well asO. sativa, and the result was visualized by using R package Circlize ( https://cran.r-proje ct.org/web/packa ges/circl ize/).
Gene structure and promoter characteristics analysis of the L. chinense CBF genes
The exon and intron structures of individualCBFgenes were determined using the Gene Structure Display Server (GSDS;http://gsds.cbi.pku.edu.cn/) via alignment of the CDS with their corresponding genomic DNA sequences. The sequence alignment was performed using MEGA 7.0. Sequence relationships were inferred using the NJ method. Conserved motifs were checked using the online Multiple Expectation Maximization for Motif Elicitation (MEME) program. The repetition was set as any number with an optimal width of 6–100 residues and the maximum number of motifs as 20.
The gene structure of theCBFfamily ofL. chinensewas analyzed and the gene structure map was drawn based on the gene location information of the GFF annotation f ile of the tree whole genome. Promoter sequences (2000 bp) ofL.chinense CBFgene were used to predict Cis-acting elements by using online tools PlantCare ( http://bioin forma tics.psb.ugent.be/webto ols/plant care/html/).
Transcriptome analysis of L. chinense response to low-temperature stress
Plants ofL. chinensewith uniform growth were selected and incubated in a 4 °C light incubator (16/8 h light/dark culture)to induce cold stress (Yamaguchi-Shinozaki and Shinozaki 1994). After treatment, leaves were removed (three plants were selected for each treatment, and three leaf sections were selected to mix from each plant) and stored in liquid nitrogen for transcriptome measurement. The gene expression ofLcCBFsbased on transcriptome were taken from the original transcript data ofL. chinenseleaves treated with low-temperature stress, and the obtained data was used to draw heat maps with Tbtools.
RT-qPCR analysis of LcCBF genes
Total RNA from dif ferent tissues was extracted using a FastPure® Plant Total RNA Isolation Kit (Polysaccharides & Polyphenolics-rich) (Vazyme, Nanjing, China).cDNA was synthesized using a HiScript® III 1st Strand cDNA Synthesis Kit (+ gDNA wiper) (Vazyme, Nanjing,China). RT-qPCR (Real Time-Quantitative PCR) was performed on a LightCycler480 II(Roche, Switzerland) using the AceQ®qPCR SYBR Green Master Mix (without ROX)(Vazyme, Nanjing, China). TheL. chinense Actin(LcActin) gene was used as internal reference. 2−ΔΔCtmethod was used to determine the relative abundance of record. Each PCR was performed using three biological replicates. The primer sequences used for qPCR were listed in supplementary Table S1. All primers were designed by Primer 5.0.
Results
Identif ication and phylogenetic analyses of L. chinense CBF gene family
Using the CBF sequences ofA. thalianaandO. sativaas indexes, BLASTP program was performed to search for similar protein sequences in the whole genome database ofL. chinense. A total of fourteenL. chinense CBFgenes were identified, and designated asLcCBF1–LcCBF14(Table 1). The length of their CDS was varied between 675 bp (LcCBF2) and 2940 bp (LcCBF11), which was indicating a large variation in sequence length. Their molecular weight ranged from 24.71 to 110.52 kDa. Each gene contains at least 2 exons, with a maximum of 7 numbers of exons (LcCBF3andLcCBF11), while most genes have one intron, yet with a maximum of 6 numbers of introns ( LcCBF3 and LcCBF11). PI values ranged from 4.89 to 9.56, and PI values of 4 proteins were greater than 7, and that of 10 proteins were less than 7, indicating that the most transcription factors of LcCBF were weakly acidic.
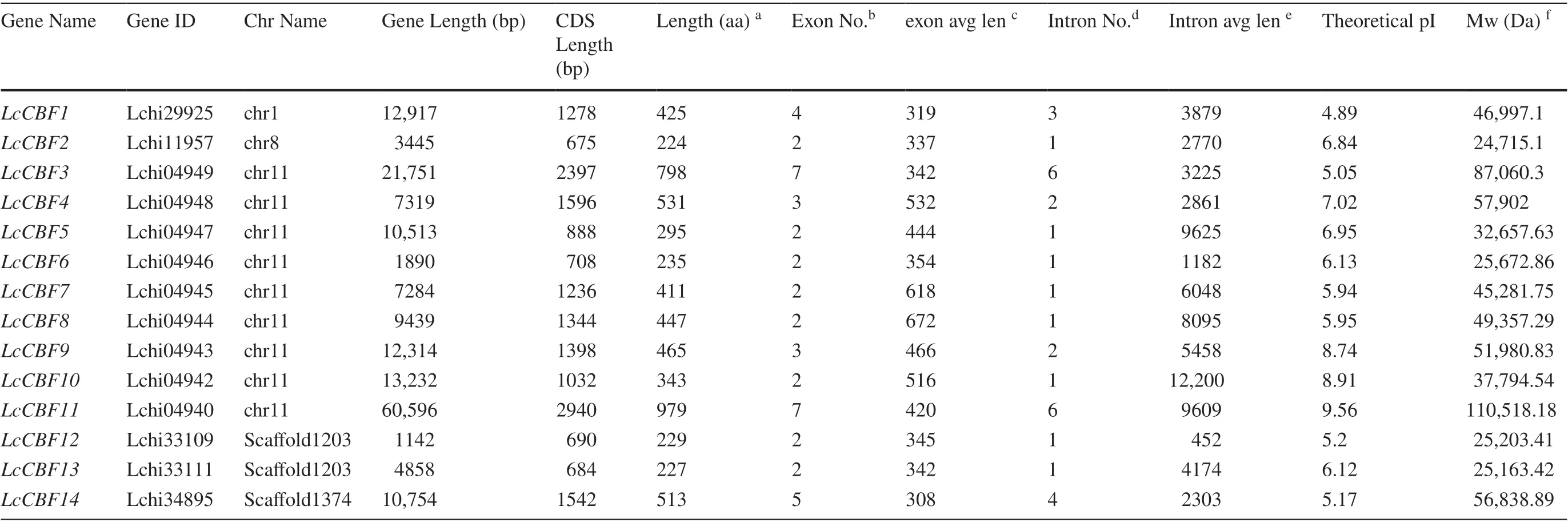
Table 1 Fourteen identifi ed members of the L. chinense CBF gene family
To study the phylogenetic relationship of the CBF family between theL. chinenseand other plants, the members of CBF family fromA. thaliana,O. sativaand f ive woody plants (A. trichopoda,C. sinensis,P. trichocarpaandS. purpurea) were collected to construct the phylogenetic tree. 52 CBF members were divided into four Groups (Fig. 1, Table 2): Group I has 20 genes, and can be divided into two Subgroups: Subgroup I contains 4AtCBFs, 4CsCBFs, 2P. trichocarpagenes (Potri.012G134100, Potri. 015G136400) and 2S. purpureagenes (SapurV1A.0068 s0560, SapurV1A.0400 s0160);Subgroup II contains 8LcCBFs. Group II has 19 genes,including 2LcCBFs, 1OsDREB1Fand other members mainly fromA. trichopoda,P. trichocarpaandS. purpurea. Group III only hasLcCBFs. Group IV has 9 genes that areOsDREBs. The members ofOsDREBsmainly distribute in the Group IV, only one of them distributed in the Group II.AtCBFsare only distributed in Group I.The members ofL. chinenseCBF family and other woody plants have distributed in three Groups, except GroupIV,which implied the dif ference in evolution withA. thalianaandO. sativa.

Table 2 Distributions of the members of CBF gene family from seven species in phylogenetic tree
Chromosome location and collinearity of L. chinense CBF genes
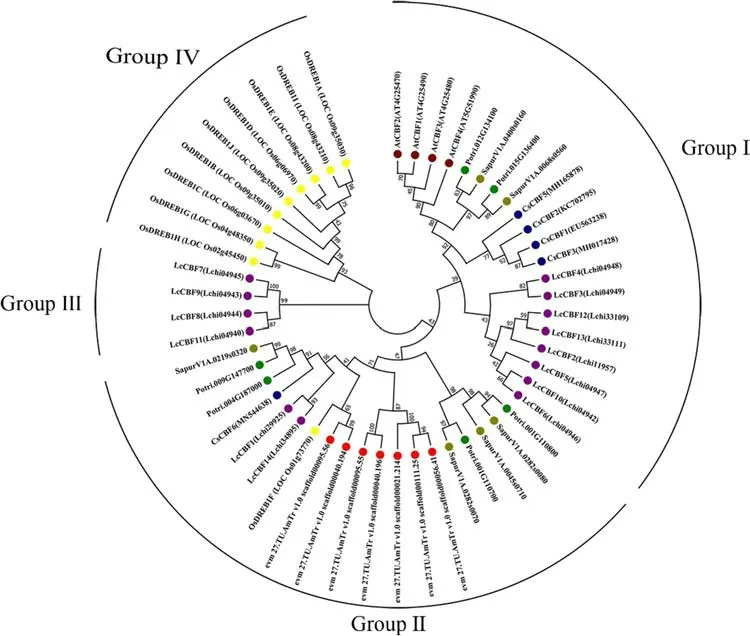
Fig. 1 Phylogenetic analysis of CBF proteins in seven species.The number at the branch of the evolutionary tree indicates the conf idence of the branch.The higher the value, the higher the reliability. Each member of the CBF family was shown the same colour. Lchi: Liriodendron chinense; At: Arabidopsis thaliana; Os: Oryza sativa; AmTr:Amborella trichopoda; Potri:Populus trichocarpa; Sapur:Salix purpurea; Cs: Camellia sinensis
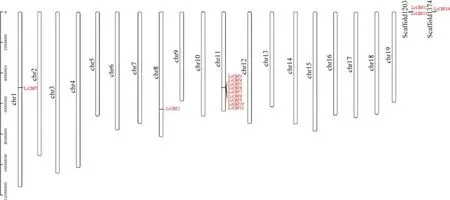
Fig. 2 Chromosomal distribution of the CBF genes of L. chinense. The scale located on the left panel, is in bases indicated chromosome sizes.The chromosome number is indicated at the left of each chromosome
The chromosome location analysis showed that the members of theCBFgene family are not uniformly distributed across all 13L. chinensechromosomes (Fig. 2). 11CBFgenes are distributed on 3 chromosomes: chr1, 8 and 11,and 3 genes are located on still unmapped scaf folds, 9CBFgenes (LcCBF3,4,5,6,7,8,9,10,11) repeat in tandem in a cluster on chromosome 11. Thus, these results suggest that multipleCBFgenes on chromosome 11 in theL. chinenselineage have evolved through tandem repeats. On both chromosome 1 and 8 have a singleCBFgene and this unequal distribution provides clues to their evolution.
To further explore key evolutionary events that may have happened forL. chinenseCBFs, sequence information from LcCBF, OsCBF, and AtCBF, were used to analyze collinearity between the three species (Fig. 3). The results showed that eightLcCBFgenes fromL. chinenseexist linear relationship withO. sativa, and oneLcCBFgene have collinearity withA. thaliana.Lchi_chr11 ofL. chinenseis collinear withOs_chr1,Os_chr2,Os_chr6,Os_chr8,Os_chr9 ofO.sativaand collinear withAt_chr4. There is a collinear relationship betweenLchi_chr1 andOs_chr9.
CBF gene structure, conserved motifs and Cis-acting elements in the promoter region of L. chinense
Diversity of gene structure is the driving force of the evolution of multi-gene families. AllCBFgenes contain exons and introns, and the number of exons varies from 2 to 7 (Fig. 4 a). This data indicates that exon loss and gain occurred in theCBFgene family during evolution, which may explain the functional diversity of closely relatedCBFgenes. In addition to theCBFexon/intron structure, other conserved motifs may also be important for the diverse functions of CBF proteins.
The 10 motifs found were named Motif 1–Motif 10, as shown in Fig. 4 b. AllCBFscontain Motif 2 and Motif 3.Specif ically, allCBFmembers exceptLcCBF11contain Motif 6 at their C-terminus. Motif 2, Motif 3, and Motif 6 are core motifs. Conservative motifs have a high degree of similarity, so genes containing that same motif are likely to be functionally similar.
To understand the transcriptional regulation mechanism of theCBFtranscription factors fromL. chinense, Cisacting elements were predicted in the promoters of the 14LcCBFgenes, as shown in Fig. 5. The common cis-acting elements included those responsive to distinct plant hormones (ABA, auxin, ethylene, and MeJA) and stress factors(anaerobic, drought, light, and low-temperature) (supplementary Table S2), besides the promoters of 8LcCBFgenes(LcCBF1,4,6,7,8,9,10,12) containing low-temperature responsive element. These f indings suggest that theLcCBFgenes could be responsive to a variety of signaling inputs.
Expression patterns of LcCBF genes in response to cold stress treatments
We analyzed the expression levels ofLcCBFgenes in leaves under the dif ferent periods of low-temperature stress(Fig. 6), andLcCBFgenes showed a similar expression pattern in response to cold stress. 5LcCBFgenes (LcCBF6–10)increased their expression at 1 h, 3 h, and 6 h, then decreased at 12 h, after that the expression increased again at 1d and decreased again at 3 days.LcCBF7,8,9,10reached the peak expression level at 6 h, whileLcCBF6peaked at 1 day.LcCBF7andLcCBF9responded to the cold stress faster and reached a higher level of expression at 3 h and 6 h than threeother genes, which implied that theLcCBF7andLcCBF9might be more sensitive to cold. The expression level ofLcCBF6was lower at 1 h and higher at 1 day than four other genes, which indicated theLcCBF6might be delayed in response to low-temperatures.
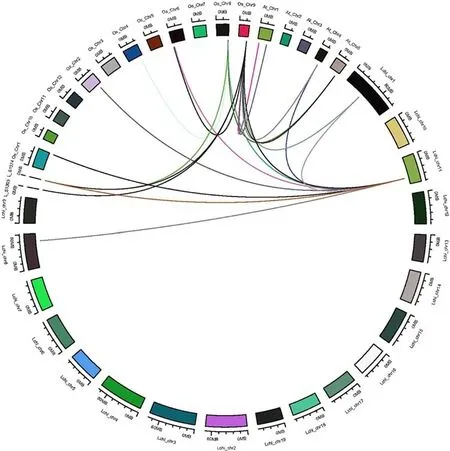
Fig. 3 Syntenic relationships of CBF genes from L. chinense, A. thaliana and O. sativa.Extensive microseconds of CBF regions across L. chinense, A.thaliana and O. sativa. The L.chinense, A. thaliana, and O.sativa chromosomes, shown in dif ferent colors, are labeled Lchi, At, and Os, respectively.Numbers along each chromosome box indicate sequence lengths in megabases. The whole chromosomes of these three species, harboring CBF regions, are shown in a circle
In addition, RT-qPCR was performed to conf irm the response ofLcCBFgenes to cold stress those we had previously detected using RNA-seq. Expression of these genes was analyzed at six consecutive time points after cold treatment, being 0 h, 1 h, 3 h, 6 h, 12 h, and 24 h (Fig. 7).LcCBF4andLcCBF5showed a trend of increasing expression first and then decreasing, and reached the highest expression level at 24 h.LcCBF7, andLcCBF9showed a trend of f irst increasing and then decreasing, and reached the peak expression at 6 h. It is consistent with transcriptome results as shown in Fig. 6.
GO analysis was conducted to understand the potential biological function ofLcCBFgenes detected above (Fig. 8), and foundCBFgenes were mainly related to stress response in biological processes (BP), especially to cold stress response(GO: 0,009,409).LcCBFgenes were found response to cold exceptLcCBF9(supplementary Table S3). The results implied that theCBFgenes ofL. chinenseplay an important role in the cold response mechanism.
Tissue-specif ic expression patterns of LcCBF genes
Next, we used RT-qPCR to analyze the expression of the sevenLcCBFgenes found to be responsive to cold stress in dif ferent tissues ofL. chinense: callus, leaf, root and stem (Fig. 9).The results showed that the expression abundance ofLcCBF10andLcCBF14was the highest in all four tissues, and that ofLcCBF11was very low in four tissues.LcCBF9had the lowest expression level in roots, stems and leaves, but high abundance in callus. The expression level ofLcCBF4was the lowest in callus but the highest in other three tissues.

Fig. 4 Gene structure and conserved motifs of LcCBFs. a Phylogenetic relationships (left), conserved motifs (middle), and gene structure (right) among CBF genes of L. chinense. Dif ferent colored boxes represent dif ferent themes and their positions in each LcCBF sequence. b Motif logo, the amino acid composition of each motif
Discussion
Genome-wide identif ication and characteristics of LcCBF gene family
As a key transcription factor for low-temperature domestication,CBFgene was f irst identif ied in model plantA.thaliana. Since then,CBFhomology and its contribution to improved stress resistance have been reported in several plants. In this study, we conducted a genome-wide search for theCBFhomologous genes inL. chinense, and identif ied fourteenCBFhomologous genes. The number of family members was higher than that ofCBFgenes inO.sativaandA. thaliana. It can be inferred that the number ofCBFfamily members has been expanded inL. chinense(Chen et al. 2019).
In the constructed phylogenetic tree, CBFs ofL. chinensewere found to be more similar to that of dicotyledonousA. thaliana, and theLcCBFswere signif icantly dif ferent between dicotyledons and monocotyledons. In the course of phylogenetic evolution, the evolutionary branches of dicotyledons and monocotyledons are dif ferent. There was no oneto-one correlation betweenCBFgenes ofL. chinenseandA.thaliana, indicating that theCBFgene ofL. chinensewas dif ferentiated during the long evolutionary selection history,which may be the result of adaptation to dif ferent climate environments.
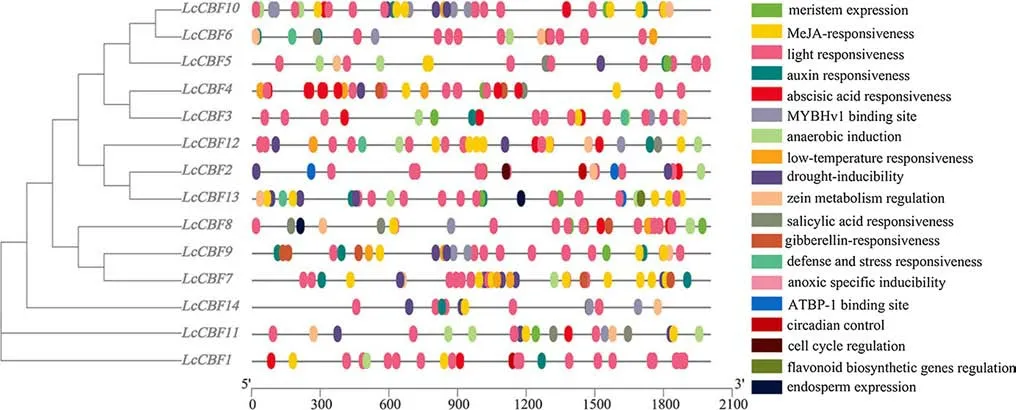
Fig. 5 Various Cis-acting elements in the promoter of LcCBF genes. Dif ferent colored boxes represent dif ferent elements and their positions in each LcCBF promoter

Fig. 6 Expression patterns of CBF genes in L. chinense. Compare and analyze transcriptome of dif ferent time. C-0 h, C-1 h, C-3 h,C-6 h, C-12 h, C-1d, C-3d denote the dif ferent treatment time, and each treatment has 3 replicates. Fold changes of LcCBFs expressions were log 2 transformed. The color histogram on the right of the heat map indicates the level of the expression; red indicates high transcript abundance while green indicates low abundance
Members of theCBFgene family were not uniformly distributed on 13 chromosomes of theL. chinense, and 9LcCBFswere on 11 chromosomes with tandem cluster,indicating that these genes evolved to repeat the lineage of theL. chinense. These tandem genes are more similar in protein sequence, which further supports the idea that the tandem gene is 68%–98% consistent with otherLcCBFs.The tandem repeats ofCBFgenes have also been observed in other species, including Arabidopsis (Cao et al. 2015),tomato (Zhu et al. 2016), barley (Mareri et al. 2020),and eucalyptus (Cao et al. 2020). These results indicate that tandem repeats play an important role in the expansion of theCBFgene family across species. Collinearity analysis results showed that there was a large amount of collinearity betweenCBFfamily members ofL. chinenseandO. sativaof monocotyledons, with gene doubling at the chromosome level, which verif ied tandem duplication on the chromosome.

Fig. 7 Dif ferential expressions of CBF genes under cold stress treatments. RT-qPCR expression levels of selected CBF genes following Low-temperature treatment exposed to 4 °C. The Y-axis indicates the relative expression levels; 0 h, 1 h, 3 h, 6 h, 9 h, 12 h, and 24 h(X-axis) indicate hours of Low-temperature treatment, and error bars denote SE
Fig. 8 GO annotation of LcCBF genes
There are anatomical and physiological differences among species, which may be ref lected in the diversity ofCBFgene structure and conserved motifs. The diversity of exon/intron gene structure plays an important role in the evolution of gene family members. 14LcCBFgenes were identif ied to contain a dif ferent number of exons, indicating that there is a certain diversity in this species. For example,LcCBF11has 7 exons, while other genes in the same phylogenetic branch have 4 exons. However, the characteristics of exon/intron structure and sequence composition are relatively conserved among the closest homologous genes. Most closely related genes in the same lineage have similar gene structures, both in terms of intron number and exon length.Some closely related members have similar structures, which may mean that these CBF proteins function similarly. The presence of specif ic sequences in each branch may have specif ic functions. The similarity of most CBF proteins in gene structure and motif composition was consistent with the phylogenetic analysis of theCBFgene family. The dif ferences in these features between dif ferent evolutionary branches indicate the diversif ied function ofCBFmembers.
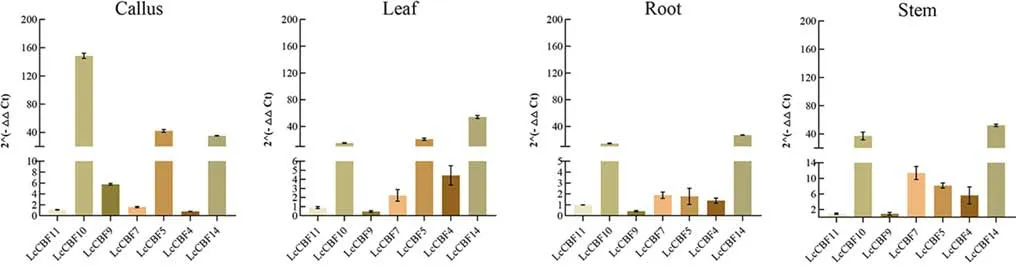
Fig. 9 Tissue-specif ic expression analysis of CBF genes with RT-qPCR in L. chinense. RT-qPCR expression levels of selected CBF genes following dif ferent tissues. The Y-axis indicates the relative expression levels; X-axis indicates seven CBF genes, and error bars denote SE
Plant gene promoter is an important Cis-acting element located in the upstream of the promoter codon and the control center of gene transcription (Doelling and Pikaard 1995). Promoter Cis-acting element analysis showed thatCBFgene promoters had many Cis-acting elements related to growth and stress response, such as stress response elements: sixteen ARE (Cis-acting elements involved in ABA response), eight WUN-motif1 TC enrichment and repetition,and eight LTR indicating thatCBFgene promoters may be involved in the growth, development and stress resistance ofL. chinense. This is similar to the previous report on mangrove (Peng et al. 2020a, b). The promoter region inLcCBFshas a variety of response elements, including low-temperature, drought, high temperature, as well as osmotic stress response elements, light response elements, plant hormones,abscisic acid, gibberellin, MeJA, salicylic acid, and ethylene response element. It is regulated by many factors, including abiotic stress, biological stress and growth development,and so on. The results of gene structure and characteristics analysis implied that 14LcCBFgenes may have dif ferent biological functions in response to abiotic stress.
The response pattern of LcCBF genes to cold
treatments indicates their potential role in cold stress
CBFgene family members are ubiquitous in all kinds of organisms and play a pivotal role in plant response to low-temperature stress by regulating the downstream cold response gene COR to participate in the growth and development of abiotic stress (Akhtar et al. 2012). Dif ferentCBFgenes are believed to have dif ferent functions and can participate in plant growth metabolism, stress resistance and other processes through various signal transduction pathways. For example,A. thaliana CBF1–CBF3andO. sativa DREB1Acan respond to low-temperature stress rapidly (Gong et al.2020).
In this study, it was found that the members ofLcCBFgene family had dif ferent expression dynamics characteristics in cold stress treatment and in dif ferent tissues. MostLcCBFshad the highest expression levels at 3 h and 6 h at low temperature. The overall trend was upward before downward, similar to other plants. The expression ofAtCBF1–3gene was induced at a low-temperature for 15 min, which reached its peak after 2 h and then decreased (Gilmour et al. 1998). ForLeCBF1in tomato plants andGhDREB1in cotton plants, the expression level peaked at 2 h (Zhu et al.2016).
Expression characteristics analyzed in four tissues ofL.chinenseshowed thatLcCBF10was highly expressed in callus.LcCBF14was highly expressed in roots, stems and leaves. The expression ofLcCBF5andLcCBF7in callus and leaf tissue was higher than that in root and stem.CBFgene was expressed tissue- specif ic inL. chinense. This result is consistent with other studies, for example,PtCBF1–4in poplar leaves are signif icantly induced by low-temperature,but onlyPtCBF1andPtCBF3are signif icantly induced in stems (Xie et al. 2019). Under the condition of cold treatment, most of theCBFgenes increased with the time of cold stress, showing a trend of increasing before decreasing, and their expression reached a peak at 6 h, similar to other plants(Jiang et al. 2011).
Conclusions
A genome-wide identif ication and characteristics analysis of 14LcCBFgenes were performed, including gene structures, conserved motifs, and expression patterns. The results showed thatLcCBFgenes were relatively conservative and theLcCBFgenes were strongly induced by cold stress to highly abundant expression. These results will contribute to further understanding the function ofLcCBFsin plant stress- response.
Authors contributionJS and LY designed the experiment; SL and WW collected samples and conducted the analysis of CBF gene family.SL, RL, KH, LZ, YL and YL performed the experiment. YG and SL analyzed the data and wrote the manuscript; JS, LY and JC edited and conf irmed the manuscript, YG and SL contributed equally to this work.All authors have read and approved the f inal manuscript.
杂志排行
Journal of Forestry Research的其它文章
- Characterization and expression analysis of genes encoding Taxol biosynthetic enzymes in Taxus spp.
- Decay rate of Larix gmelinii coarse woody debris on burned patches in the Greater Khingan Mountains
- Characterizing conservative and protective needs of the aridland forests of Sudan
- Point-cloud segmentation of individual trees in complex natural forest scenes based on a trunk-growth method
- Accuracy of common stem volume formulae using terrestrial photogrammetric point clouds: a case study with savanna trees in Benin
- Appropriate search techniques to estimate Weibull function parameters in a Pinus spp. plantation
