Characterization and expression analysis of genes encoding Taxol biosynthetic enzymes in Taxus spp.
2021-12-24LuyuanJiangKaikaiZhangXingLiyingYangShuaiWangDuanfenChenYanfangYangDeyouQiu
Luyuan Jiang · Kaikai Zhang · Xing Lü ·Liying Yang · Shuai Wang · Duanfen Chen ·Yanfang Yang · Deyou Qiu
Abstract Taxol (Paclitaxel), an important anticancer drug,is derived at very low yields from Taxus (yew) species that grow very slowly. In the present study, thirteen genes that encode enzymes involved in Taxol biosynthesis in Taxus spp. were analyzed with bioinformatics methods, and their expression levels in dif ferent tissues and after cold and hormone treatments were also analyzed. The results indicated that many cis -elements related to abiotic stresses and hormones were found in the promoter sequences of the 8 genes involved in Taxol biosynthesis. Moreover, the 13 enzymes encoded by the target genes were located in dif ferent organelles and had many phosphorylation sites in the response proteins. The 13 genes were expressed highly either in roots or in stems, with lower transcripts in needles, and they were highly expressed after treatment with cold, gibberellin,methyl jasmonate or coronatine, consistent with predictions based on the bioinformatics analysis. These results suggest that the factors such as hormones and abiotic stresses stimulate taxane biosynthesis in yews, providing an important way to sustainably generate taxanes from yew trees or their cell cultures to improve Taxol yields.
Keywords Gene expression · Bioinformatics analysis ·Acyltransferase · Cytochrome P450 · Abiotic stress ·Hormone Project funding: This work was supported by National Natural Science Foundation of China (31570675), a Grant from the National Key Research and Development Program of China(2017YFD060070605), and a Grant for National non-prof it Research Institutions of Chinese Academy of Forestry(CAFYBB2018SY009).
Introduction
Taxol, the well-known brand name for the diterpenoid paclitaxel, is an anticancer compound, mainly extracted fromTaxusspecies (yews), with curative ef fects against breast,lung, ovarian, endometrial, and cervical cancers (Hou et al.2019; Khalifa et al. 2019; Zagouri et al. 2019). Since its discovery, Taxol, has been an important research topic globally, as the number of cancer patients continues to increase.Because the content of Taxol is low in yews and the yew grows slowly; the molecular mechanism of Taxol biosynthesis in yews has been well studied. The biosynthetic process mainly includes cyclizations of the taxane skeleton, hydoxylations by cytochrome P450, acylations after yielding the important baccatin III (BAC III) intermediate, and f inally,side chain assembly at the C13-O-position of BAC III to yield Taxol (Croteau et al. 2006; Nasiri et al. 2016).
Thirteen genes involved in the Taxol biosynthesis pathway have been cloned and isolated over the past 20 years,including the genes encoding geranylgeranyl diphosphate synthase (GGPPS), taxadiene synthase (TS), six cytochrome P450s, and three acetyltransferases (Croteau et al. 2006).The formation of GGPP is f irst catalyzed by GGPPS, then GGPP is cyclized by TS to form taxa-4(5),11(12)-diene(Croteau et al. 2006), and then a series of hydroxylation and acylation reactions modify the taxane core. The order of oxygenation of the taxane core has been proposed as follows: C5 and C10, followed by C2 and C9, C13 followed by C7, and f inally C1 is hydroxylated (Floss and Mocek 1995; Guerra-bubb et al. 2012; Jennewein et al. 2001). The construction of a phenylalanine-derived C13-side chain is catalyzed by phenylalanine aminomutase (PAM), then attached tobaccatino (BAC) III by baccatin III-3-amino,3-phenylpropanoyltransferase (BAPT) (Jennewein et al.2004). Previous studies have shown that the conversion of 10-deacetylbaccatin III (10-DAB) into BAC III could be a rate-limiting step in the Taxol biosynthetic pathway (Nasiri et al. 2016), and 10-deacetylbaccatin III-10-O-acetyltransferase (DBAT), BAPT, and 3′-N-debenzoyl-2′-deoxyTaxol-N-benzoyltransferase (DBTNBT) are presumed to be the most important rate-limiting enzymes for Taxol biosynthesis inTaxus baccataL. cell suspension cultures when synthesis is elicited by coronatine (COR) and methyl-β -cyclodextrin(Kashani et al. 2018).
Methyl jasmonate (MeJA) and salicylic acid (SA) are other known elicitors of Taxol synthesis. In response to these elicitors, genes related to Taxol synthesis are upregulated, and Taxol synthesis can be signif icantly increased(Onrubia et al. 2013; Kashani et al. 2018; Sarmadi et al.2018; Sykłowska-Baranek et al. 2019). The functions and properties of seven genes isolated from yews, that encode enzymes catalyzing early and late steps in the pathway, have been described and summarized (Walker and Croteau 2001).However, information on the expression and regulation of the genes related to Taxol biosynthesis is still lacking. Thus,in the present study, 13 genes known to encode the Taxol biosynthetic enzymes were analyzed with bioinformatics methods, including promoter component analysis, protein characterization, and expression analyses in dif ferent tissues and after abiotic stresses. The expression prof iles of these genes were also investigated after drought, salinity, and COR exposure. The biological functions of the key enzymes involved in Taxol biosynthesis and the molecular mechanism of Taxol biosynthesis need to be understood to improve Taxol yields and meet demands for this valuable drug.
Materials and methods
Plant materials and stress treatments
Five-year old plants ofTaxus chinensisvar.maireiwere planted in plastic pots (diameter 23 cm, height 18 cm)with soil (3:1:1, nutritional soil: vermiculite: perlite) in the greenhouse of the Chinese Academy of Forestry. The highsalinity, drought and COR treatments were carried out as described previously (Yang et al. 2018). Plants were watered with 1 L NaCl (1 mol/L) for the high-salinity stress or 20%PEG 6000 for the drought stress. After 0 and 6 h of treatment, leaf samples were collected and immediately frozen in liquid nitrogen, and then stored at − 80 °C. COR was f ilter-sterilized and added to B5 solid medium before solidif ication for a f inal concentration of 1 μmol/L. Cells were harvested at 0 and 8 h after COR treatment and immediately frozen in liquid nitrogen, and then stored at − 80 °C. For RNA-sequence (seq), the GA3dissolved with 95% ethanol and diluted to 200 μmol/L was sprayed to the shoots of the plants until drips, two times a day at 9:00 AM and 3:00 PM,respectively. At the third day 9:00 AM, the leaves were collected and immediately frozen in liquid nitrogen, and then stored at − 80 °C. All treatments were performed with three biological replicates.
RNA isolation and cDNA preparation
Total RNA was isolated using the TRIzol Reagent (Invitrogen) and the supplier’s procedure, then with DNase to remove genomic DNA. The quality and purity of the extracted RNA was checked by measuring A260/A280 and A260/A230 absorption ratios in a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientif ic, Waltham, MA, USA)and by electrophoresis in 1.2% agarose. With the TransScript f irst-strand cDNA synthesis SuperMix (With gDNase)(Transgen, Beijing, China), f irst strand cDNA was synthesized from 0.6 μg total RNA, and then diluted 5 times.
Bioinformatics analysis
The protein and promoter sequences of Taxol biosynthesis were download from GenBank ( https ://www.ncbi.nlm.nih.gov/genba nk/; accession information in Supplementary File 1). Thecis-element analysis was performed using PlantCARE ( http://bioin forma tics.psb.ugent.be/webto ols/plant care/html/). The location of the proteins was analyzed using Plant-mPLoc ( http://www.csbio .sjtu.edu.cn/bioin f/plant-multi/). The signal peptide was analyzed based on the default Hidden Markov model in SignalP 5.0 Server. To analyze whether the enzymes in the Taxol biosynthesis pathway contained a transmembrane region, the proteins were analyzed using the online analysis website TMH MM-2.0. The phosphorylation site of these proteins was also predicted using Netphos 3.1.
Expression analysis
The RNA-Seq datasets for tissues (accession number SRP127697), low-temperature-treated (accession number SRP096539), and MeJA treated accessions of yews (accession number SRP133888 and SRP083793) were downloaded from the SRA database of NCBI. High-quality reads were assembled into contigs using Trinity 2.0.6 (Grabherr et al.2011), and annotated using a Blast search against the Gen-Bank nonredundant protein database.
Quantitative real-time PCR
To determine the expression levels of the genes involved in Taxol biosynthesis after the dif ferent treatments, quantitative real-time PCR (qRT-PCR) as described by Zhang et al. ( 2019) using Roche Light Cycler 480 (Roche, Basel,Switzerland) with KAPA SYBR FAST qPCR Kit Master Mix (2×) (Sigma-Aldrich, St. Louis, MO, USA). The PCR reaction volume (10 μL) contained 5 μL KAPA SYBR FAST qPCR Kit Master Mix (2×), 0.65 μL of each gene-specif ic primer, 1.2 μL of diluted cDNA, and 2.5 μL sterile ddH2 O.TheTBC41orGAPDH1was selected as the reference gene,and primers for the test genes and reference genes are listed in Table S1. The relative expression of target genes was analyzed by using the 2−ΔΔCTmethod (Livak and Schmittgen 2001).
Results
Cis-element analysis
According to the results of the PlantCARE analysis of the promoter sequences of the known key Taxol biosynthetic enzyme genes (Table S2), the eight Taxol biosynthesisrelated gene promoters contained basic components, such as G-box (exceptBAPT), hormone response components,such as auxin response element (AurxRE and AuxRR-core),ABA response element (ABRE), gibberellin (GA) response element (P-box and TATC-box), and SA (TCA-element) and MeJA components (CGTCA-motif and TGACG-motif). In addition, a variety of abiotic response components were also present, including low temperature stress (LTR), drought response components (MBS), and heat stress response elements (HSE). Among these genes,TS,T13OH, andDBTNBThad more than sixcis-elements, indicating that the three genes had complex functions and participated in multiple network pathways.
Subcellular localization
Based on the location predictions for the enzymes, P450 hydroxylase proteins were located in the endoplasmic reticulum and acetyltransferases were located in the cytoplasm(Table 1). The GGPPS and TS proteins were located in the chloroplast or plastid, respectively, and PAM was located in the nucleus.

Table 1 Plant-mPLoc predictions of subcellular locations of the 13 target enzymes involved in Taxol biosynthesis in yews
Signal peptide analysis
For the 13 target proteins analyzed with the online SignalP 5.0 Server, the predication for each was “other”, suggesting that they did not contain any signal peptide (Fig. S1).
Protein transmembrane analysis
The transmembrane region analyses using TMH MM-2.0 showed that hydrolases T2OH, T5OH, T10OH, and T13OH had transmembrane areas at the N terminal. Moreover,T7OH had two transmembrane regions between amino acids 20–42 and 149–168. The other proteins had no transmembrane areas (Fig. 1).
Phosphorylation analysis
According to Netphos 3.1 online software ( http://www.cbs.dtu.dk/servi ces/NetPh os/), all 13 proteins had phosphorylation sites in dif ferent locations and numbers (Fig. 2). The number of serine phosphorylation sites was the highest among all 13 proteins, and the number of tyrosine phosphorylation sites was lowest (Table 2). Comparing with the other proteins, TS and PAM had relatively more serine and threonine phosphorylation sites. Among the hydroxylases,T2OH and T5OH had more serine and threonine phosphorylation sites, respectively, and among the acetyltransferases,BAPT and TAT had more serine and threonine phosphorylation sites, respectively.
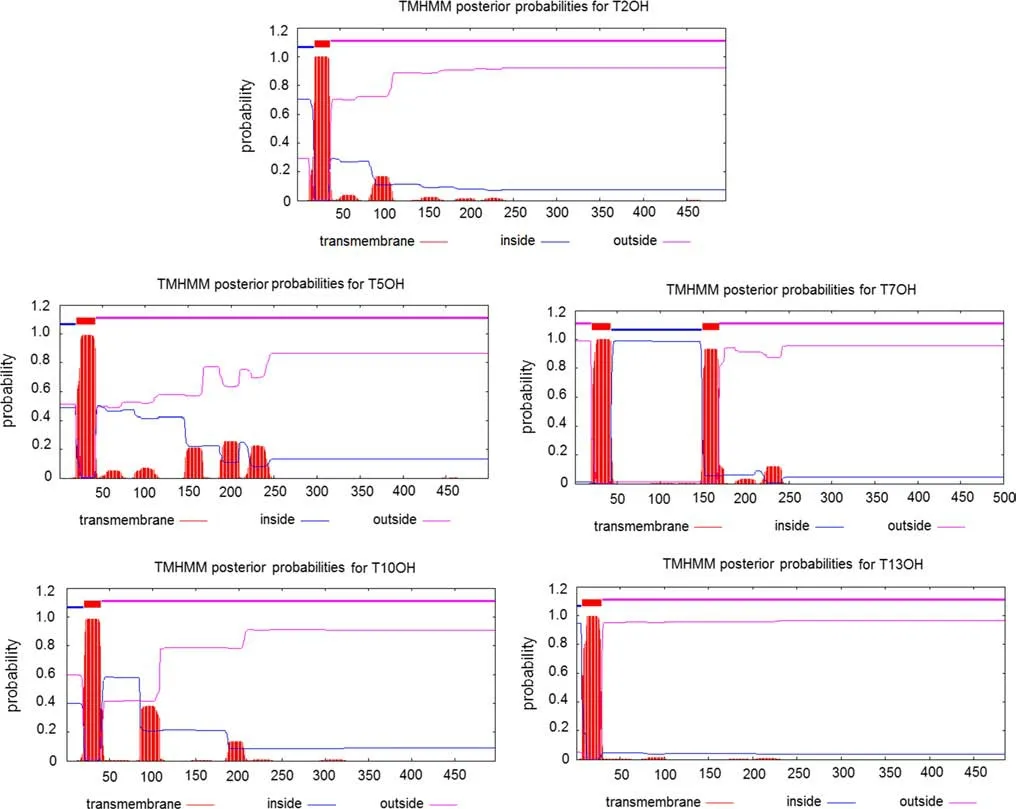
Fig. 1 TMHMM-2.0 results for the transmembrane analysis of f ive of the 13 enzymes involved in Taxol biosynthesis in yews. See Table 1 for protein names

Fig. 2 Netphos 3.1 results for the phosphorylation analysis of the 13 enzymes involved in Taxol biosynthesis in yews. See Table 1 for protein names
Expression patterns in dif ferent organs and after dif ferent treatments
The transcriptome data of the roots, stems, and leaves (needles) ofTaxus yunnanensis(SRP127697) were reassembled and used to analyze the expression patterns of the 13 genes.DBAT,T10OH,TAT,BAPT,TS, andT5OHhad higher expression in roots than leaves;T13OH,T2OH,GGPPS,TBT,DBTNBT,T7OH, andPAMhad more transcripts in stems than in roots (Fig. 3 a).
RNA-seq data (SRP096539) showed that the expression patterns of the 13 genes dif fered under low temperature conditions (Fig. 3 b).TS,GGPPS,DBAT,T10OH,T5OH, andTATwere up-regulated after cold treatment, butT2OH,TBT,T7OH,T13OH, andPAMwere down-regulated.
Transcriptome data after GA3and MeJA treatment, were also analyzed (SRP133888 and SRP083793 for MeJA; RNAseq data for GA3were obtained by our group and are unpublished). After GA3treatment, 11 of the 13 target genes were up-regulated;T10OHandT7OHwere not (Fig. 3 c). All 13 target genes were up-regulated at dif ferent times after MeJA exposure (Fig. 3 d).T10OH,T7OH, andT5OHwere rapidly up-regulated after 0.5 h, but down-regulated at 1 h and 2 h.At 3 h and 24 h, expression levels ofT10OHandT7OHwere higher, but were lower at 18 h. The transcript levels of other genes were relatively higher at 1, 2, and 18 h.
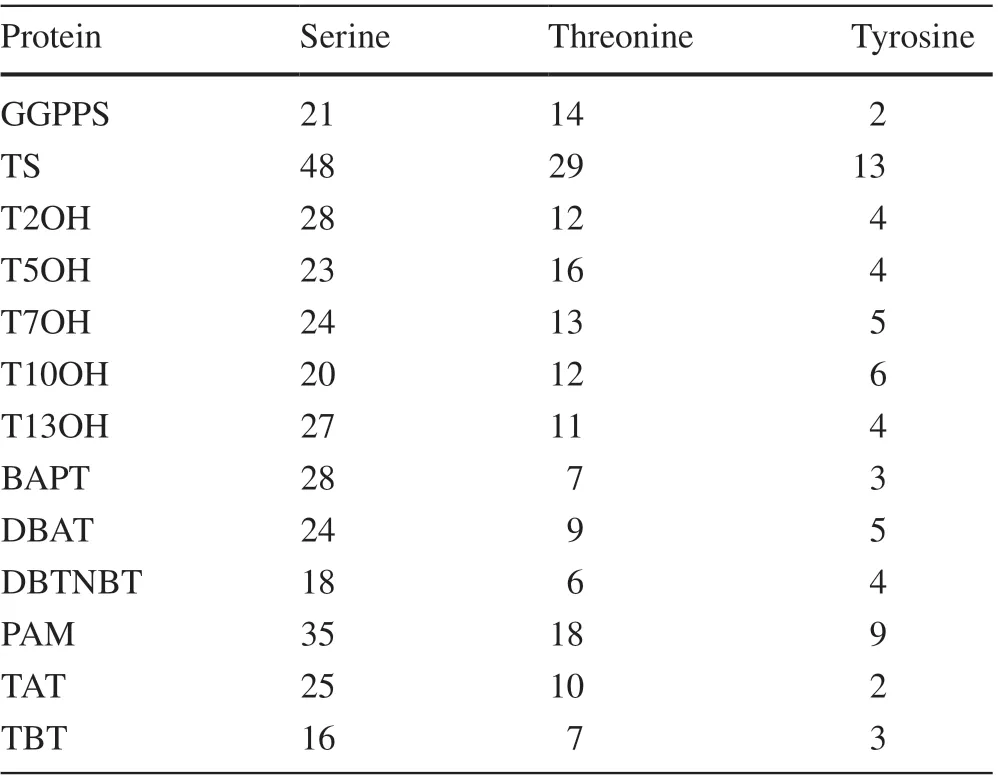
Table 2 Summary of Netphos 3.1 results for phosphorylation sites of the 13 target enzymes involved in Taxol biosynthesis in yews. See Table 1 for protein names
qRT-PCR
The 9 of the 13 target genes selected for expression analysis after drought, salinity, and GA3exposure had similar expression patterns; they were up-regulated after drought,salinity, and COR treatments, and up-regulated 3- to 6-fold under PEG and NaCl stresses (Fig. 4). However,T2OHandT13OHhad signif icantly higher expression,and were up-regulated more than 15-fold by COR, while other genes had about 5-fold lower expression.T13OHhad lower expression after PEG and NaCl treatment, and expression ofDBATwas similar to that ofT13OHafter PEG exposure.
Discussion
Toward enhancing Taxol production, previous studies showed that elicitors and environmental stresses can stimulate the synthesis of Taxol and other secondary metabolites(Chen et al. 2013; Wang et al. 2016; Sarmadi et al. 2018).Plants respond defensively to environmental changes by altering their gene expression to increase stress tolerance,and concurrently promote the production of secondary metabolic substances (Martin et al. 2002; Nejat and Mantri 2017).
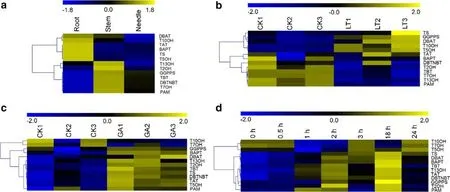
Fig. 3 Heat maps of transcriptome analysis of the expression patterns of the 13 genes in the Taxol biosynthetic pathway in yews in a various organs and after treatment with b low temperature, c GA 3 and d MeJA. See Table 1 for protein names

Fig. 4 Expression of 9 target genes after PEG 6000 (20%), NaCl(1 mol/L NaCl), and COR (1 μmol/L) treatments of yews as determined by quantitative real-time PCR using TBC41 as the reference gene. See Table 1 for protein names
ABA regulates a range of developmental and growth processes, especially to confer tolerance to environmental conditions, such as drought, salinity, low temperature, heat, and pathogens (Mauch-Mani and Mauch 2005). Here, promoter component analysis indicated that, except forBAPT, the promoter sequences ofTS,T5OH,T10OH,T13OH,DBBT,DBTNBT, andPAMcontained the ABRE element involved in ABA responsiveness. Moreover,T10OH,T13OH,BAPT,andDBTNBTalso contained MBScis-acting elements involved in drought-inducibility (Table S2). Furthermore,the promoter sequences ofBAPTandDBTNBThadcis-acting elements (TC-rich repeats) involved in defense and stress responsiveness (Table S2). PEG can enhance the production of taxanes inTaxuscell and tissue cultures (Sarmadi et al.2019). Treatment with 2% or 3% PEG increase 10-DAB and Taxol contents signif icantly; 3% PEG increases production of Taxol by 2.5 times compared with the control (Sarmadi et al. 2019). Here, 9 out of 13 genes related to Taxol synthesis were up-regulated about f ive times compared to that of the control after PEG and salinity treatment (Fig. 4), indicating that drought and salinity could enhance the production of Taxol. In addition, the LTRcis-elements were found in the promoter region ofT5OHandDBTNBT, indicating that these genes can be induced by cold treatment (Table S2).The RNA-seq data also indicated thatT5OH,DBAT,TS,GGPPS, andT10OHwere up-regulated after cold treatment,whileBAPT,DBTNBT,T2OH,TBT,T7OH,T13OH, andPAMwere down-regulated (Fig. 3 b). These genes may have been af fected by many factors that could potentially interact with each other. Moreover, abiotic stresses might af fect the levels of taxanes because the genes are regulated by these stresses. The molecular interactions among ABA, abiotic stresses, and Taxol synthesis and their regulatory mechanisms are very complex and require further research.
Hormones, especially MeJA, SA, and GA, are known to enhance the expression of genes involved in Taxol biosynthesis and improve the synthesis and accumulation of Taxol in yews or cell cultures of yews (Yukimune et al. 1996; Chen et al. 2008; Li et al. 2012; Onrubia et al. 2013 ; Sun et al.2013; Kashani et al. 2018; Sarmadi et al. 2018). COR has been used as an elicitor to improve the production of Taxol by inducing JA biosynthesis (Cusido et al. 2014). Here, thecis-acting elements involved in MeJA and GA responsiveness were found in the promoter regions of genes such asTS,DBAT,DBBT,T13OH,DBTNBT, andPAM(Table S2).G-box and GCC-box, involved in response to MeJA were found in the promoter sequences ofTSandT5OH(Dai 2008;Zhang et al. 2015; Yang et al. 2018). In this study, a G-box was found in the promoter sequences of seven genes but notDBAT(Table S2). Moreover, the expression of all 13 genes were up-regulated after MeJA inducement at dif ferent time (Fig. 3 d), and 9 of the 13 genes were up-regulated after COR inducement (Fig. 4). These results were consistent with those of previous studies (Li et al. 2012; Onrubia et al. 2013;Sun et al. 2013; Kashani et al. 2018; Sarmadi et al. 2018).In addition, because the biosynthetic pathway for GA3(a diterpenoid) shares a common substrate (GGPP) with that of Taxol, there might have been competition between the two pathways for GGPP. Although a key regulator (MYC2 TF) involved in JA signal transfer has been identif ied and the mechanisms of the interactions between ERF TF and downstream genes have been studied (Lenka et al. 2015;Zhang et al. 2015; Yang et al. 2018), the molecular mechanisms underlying of hormone-induced paclitaxel synthesis are still unknown and need further study.
Factors such as theTaxusspecies, growing conditions(e.g. altitude, temperature, light, and soil), age and sex of plants, and season af fect the abundance of Taxol or taxanes in the bark, shoots, stems, needles, and roots (Wheeler et al.1992; Liu et al. 2006; Cameron and Smith 2008; Onrubia et al. 2011; Liu et al. 2011; Nasiri et al. 2016). The Taxol content in the roots and bark of the trunk and branches is usually higher than in other plant parts, while the precursors, 10-DAB and BAC III are higher in the needles and stems (Liu et al. 2006; Onrubia et al. 2011). For example, inT. chinensis, the quantity of Taxol is higher in trunk, root,and branch bark, and BAC III is higher in both old and fresh needles (Liu et al. 2006). Moreover,GGPPSandT10OH,which function before 10-DAB formation have higher expression levels in fresh needles; however, the expression ofDBAT, which functions after 10-DAB formation is higher in the bark of branches and roots (Liu et al. 2006). Here, the expression of Taxol biosynthetic genes, includingGGPPSandT10OH, was higher in roots and stems, with lower transcripts in needles ofT. yunnanensis(Fig. 3 a). Transcriptome data showed that expression ofGGPPSandT10OHwas lower in needles, which may be due to dif ferences in age, sampling season, growing conditions, in particular, theTaxusspecies; needles ofT. chinensisandT. yunnanensisare known to dif fer in the abundance of Taxol. The expression pattern of these 13 genes was partly consistent with the conclusion that Taxol mainly accumulates in the roots or bark and that precursors, such as 10-DAB and BAC III are mainly synthesized in the needles and branches of yews (Onrubia et al. 2011). Although the mechanism of Taxol synthesis and transport and the precursors in the roots and aerial parts are still not fully understood, in the future, species and new varieties with high Taxol, 10-DAB, and/or BAC III levels should be screened, and cultivation practices that promote Taxol biosynthesis should be developed for sustainably producing these important natural compounds from yews.
Terpenoid synthases are usually localized in the cytosol(sesquiterpene synthases) or plastids (monoterpene synthases and diterpene synthases) (Bohlmann et al. 1998).Here, we also found that the 13 Taxol biosynthesis-related enzymes were located in dif ferent organelles (Table 1). For example, TS was predicted to be located in the chloroplast,which is consistent with a previous report that it is translated as a preprotein with an N-terminal targeting sequence for localization and processing in the plastids (Williams et al.2000). The f ive hydroxylases were located in the endoplasmic reticulum with the putative N-terminal membrane insertion region, which is similar to other cytochrome P450s(Kaspera and Croteau 2006; Brignac-Huber et al. 2016).However, a new hydroxylase gene (TcCYP725A22) that has been cloned fromTaxus wallichianavar. chinensisand predicted to have no signal peptide, and was conf irmed to localize at the cell membrane in a sub-cellular localization experiment using onion epidermal cells (Liao et al. 2019a,b). Moreover, signal peptide analysis showed that none of the 13 target proteins contained a signal peptide. Proteins enter organelles or sub-organelles through some mechanism to complete their specif ic functions. Although determining the subcellular localization of a protein using bioinformatics is convenient and rapid, it is still necessary to verify the subcellular localization of important proteins, such as the enzymes in Taxol biosynthetic pathway, to obtain important clues on the function of the proteins.
Additionally, phosphorylation site analysis showed that the 13 enzymes had many phosphorylation sites (Fig. 2,Table 2). In animals, the expression of P450 depends on the regulation of phosphorylation and ubiquitin (Brignac-Huber et al. 2016). Although the gene expression is mainly regulated at the transcriptional level, post-transcriptional regulations, such as splicing and stabilization of RNA and synthesis and degradation of proteins, also have an important ef fect on gene expression. Limited research is available on Phosphorylation modif ications of Taxol biosynthetic enzymes and the related regulatory mechanisms need further investigation.
Conclusions
The promoter sequences containedcis-elements involved in ABA, GA, MeJA, and abiotic stress responses (including drought, low temperature, and heat), which may explain why these factors could stimulate taxane biosynthesis in yews. Moreover, the expression results from the RNA-seq and qRT-PCR analyses supported this idea, because the related genes involved in Taxol biosynthesis were upregulated under these conditions. Furthermore, according to RNA-seq results, the 13 Taxol biosynthesis-related genes were highly expressed either in roots or stems; needles had fewer transcripts. These 13 target enzymes are located in dif ferent organelles, suggested that the precursors of Taxol may be transported within cells. In addition, there may be regulation after transcription, such as phosphorylation modif ication, which af fects the expression of genes for Taxol synthesis. Further studies are need to elucidate the complicated mechanisms involved in the biosynthesis of Taxol and its regulation and interpret the complex network of taxane biosynthesis in yews.
Authors’ contribution YYF and QDY conceived and designed this study. YYF, JLY, ZKK, LX and YLY performed the experiments and analyzed the data, JLY and WS performed data collection and statistical analysis. CDF and QDY contributed suggestions. YYF, JLY, and ZKK wrote the manuscript. All authors approved the f inal version of the paper.
Compliance with ethical standards
Conf lict of interestThe authors declare that they have no conf lict of interest.
杂志排行
Journal of Forestry Research的其它文章
- Genome-wide identif ication and cold stress-induced expression analysis of the CBF gene family in Liriodendron chinense
- Decay rate of Larix gmelinii coarse woody debris on burned patches in the Greater Khingan Mountains
- Characterizing conservative and protective needs of the aridland forests of Sudan
- Point-cloud segmentation of individual trees in complex natural forest scenes based on a trunk-growth method
- Accuracy of common stem volume formulae using terrestrial photogrammetric point clouds: a case study with savanna trees in Benin
- Appropriate search techniques to estimate Weibull function parameters in a Pinus spp. plantation
