Letter to the Editor Molecular characterization of an extensively drug-resistant Acinetobacter baumannii isolated from a corn culture soil
2021-12-22
Dear Editor,
Soil is the most biodiverse habitat on Earth and it has been estimated that one gram of soil can contain up to one billion bacterial cells(Fierer, 2017). The microorganisms of soil produce a wide variety of secondary metabolites,including antibiotics. However, soil is also a reservoir of antimicrobial resistance genes(ARGs)(Zhuet al.,2019).The natural ARGs in soil form an intrinsic resistome,which means that they have arisen independently of human activity.In addition, many soil functions from chemical, physical,and biological perspectives have a direct impact on human health(Galánet al.,2013).
Acinetobacter baumanniihas emerged as the major opportunistic pathogen and has been reported in different types of infections, primarily in immunocompromised patients.Acinetobacter baumanniihas intrinsic resistance to several antimicrobials.Furthermore,it also has a high capacity to acquire new resistance mechanisms,and multidrug-resistant(MDR)bacteria have been spreading through several niches(Michalopoulos and Falagas,2010;McConnellet al.,2013).Antimicrobial resistance is considered to be a global threat to public health, and carbapenem-resistantA. baumanniiis classified as a critical pathogen by the World Health Organization(WHO,2017).
Special attention has been given to antimicrobialresistant bacteria isolated from humans,and a few studies have evaluated the occurrence of resistant bacteria and ARGs in bacteria found in environments such as soil.Recently,an antimicrobial-resistant bacterium carrying plasmid-mediated resistance genes was described in a soil sample(Morettoet al.,2020).Furthermore,the One Health concept has now recognized that this is a truly inter-domain problem because the environment,humans,and animals are interrelated components(Tiedjeet al., 2019). Therefore, this study aimed to characterize an environmental extensively drug-resistant(XDR)A.baumanniiisolated from soil.
Soil sample(1 g)was initially added to 5 mL of Brain Heart Infusion(BHI)broth(Oxoid,UK)and after 18 h at 37°C,100 μL was seeded onto MacConkey agar(Oxoid,UK).A negative lactose colony was selected and stored at-80°C in BHI plus 15%glycerol.Genomic DNA was obtained using the QIAamp DNA Mini Kit(Qiagen,Germany)according to the manufacturer’s instructions.The bacterial identification was performed by sequencing the 16S rDNA and 23S rDNA genes, and through the detection of theblaOXA-51-likegene by polymerase chain reaction(PCR)(Weisburget al.,1991;Huntet al., 2006; Woodfordet al., 2006). The BLAST algorithm(http://blast.ncbi.nlm.nih.gov/Blast.cgi)was used to analyze the sequences deposited in GenBank.
Antimicrobial susceptibility was ascertained by determining the minimum inhibitory concentration(MIC)using the broth dilution method and Etest(bioMérieux,USA)according to Clinical and Laboratory Standards Institute(CLSI)(2018).The antimicrobials tested were ampicillin-sulbactam,piperacillin-tazobactam, ceftazidime, cefotaxime, ceftriaxone,cefepime,imipenem,meropenem,tetracycline,gentamicin,tobramycin,levofloxacin,ciprofloxacin,polymyxin B,colistin,and trimethoprim-sulfamethoxazole.The XDR classification was determined according to Magiorakoset al.(2012).
The PCR method was used to detect ARGs in the following groups:β-lactams(blaCMY,blaSHV,blaCTX-M-Gp1,blaCTX-M-Gp2,blaCTX-M-Gp8,blaCTX-M-Gp9,blaVEB,blaPER,blaGES,blaIMP,blaVIM,blaKPC,blaOXA-1-like,blaOXA-23-like,blaOXA-40-like,blaOXA-48-like,andblaOXA-58-like),tetracyclines(tetA,tetB,tetC,tetD,andtetM),aminoglycosides(aac(3’)-Ia,aac(3’)-IIa,aac(6’)-Ib,aac(6’)-Ih,ant(2”)-Ia,aph(3’)-VI,andaph(3’)-Ia),and sulfonamides(sul1,sul2,andsul3)(Noppe-Leclercqet al.,1999;Nget al.,2001;Kerrnet al.,2002; Perreten and Boerlin,2003; Woodfordet al.,2006;Dallenneet al., 2010). For polymyxins, mutations in thepmrAandpmrBgenes were screened by sequencing usingA.baumanniiATCC 17978 as the wild type strain(GenBank accession no.CP000521.1)(Arroyoet al.,2011).
Polymerase chain reaction-based replicon typing(ABPBRT) was performed to detect the plasmids belonging to 19 homology groups(GRs)(Bertiniet al.,2010).Plasmid extraction was performed using the Plasmid Mega Kit (Qiagen, Germany) according to the manufacturer’s instructions.The molecular weight of the plasmid was determined using BioNumerics software 5.1(Applied Maths,Belgium)after standard 0.8%agarose gel electrophoresis.Multilocus sequence typing(MLST)analysis was performed using the primers and conditions described in theA.baumanniiMLST database of the University of Oxford, UK(http://pubmlst.org/abaumannii). The new alleles and sequence type(ST)were submitted to the MLST website to be assigned.The phylogenetic relationships among closely related STs were determined by the eBURST V3 algorithm(http://eburst.mlst.net/).
TheA. baumanniiS402 isolate (GenBank accession nos.MG100205(16S rDNA),MG100206(23S rDNA),and MK506783 (blaOXA-51-like)) was obtained from a soil that had previously been used to cultivate corn in Ribeirão Preto City,São Paulo State,Brazil.This isolate was resistant to almost all CLSI recommended antimicrobials and had high MICs,except for fluoroquinolones(MIC:1 μg mL-1),which meant that it was classified as an XDR(Table I).The S402 isolate presented the following ARGs:β-lactams(blaSHV,blaCTX-M-Gp9,blaGES),tetracyclines(tetB),aminoglycosides(aac(6’)-Ib,aph(3’)-Ia),and sulfonamides(sul1)(GenBank accession Nos. MG188752-MG188754 and MK506779-MK506782).This isolate also harbored one plasmid,which wasca.63 Kb in size and classified as GR6.All the ARGs found were confirmed by PCR using the plasmid DNA recovered after electroelution.
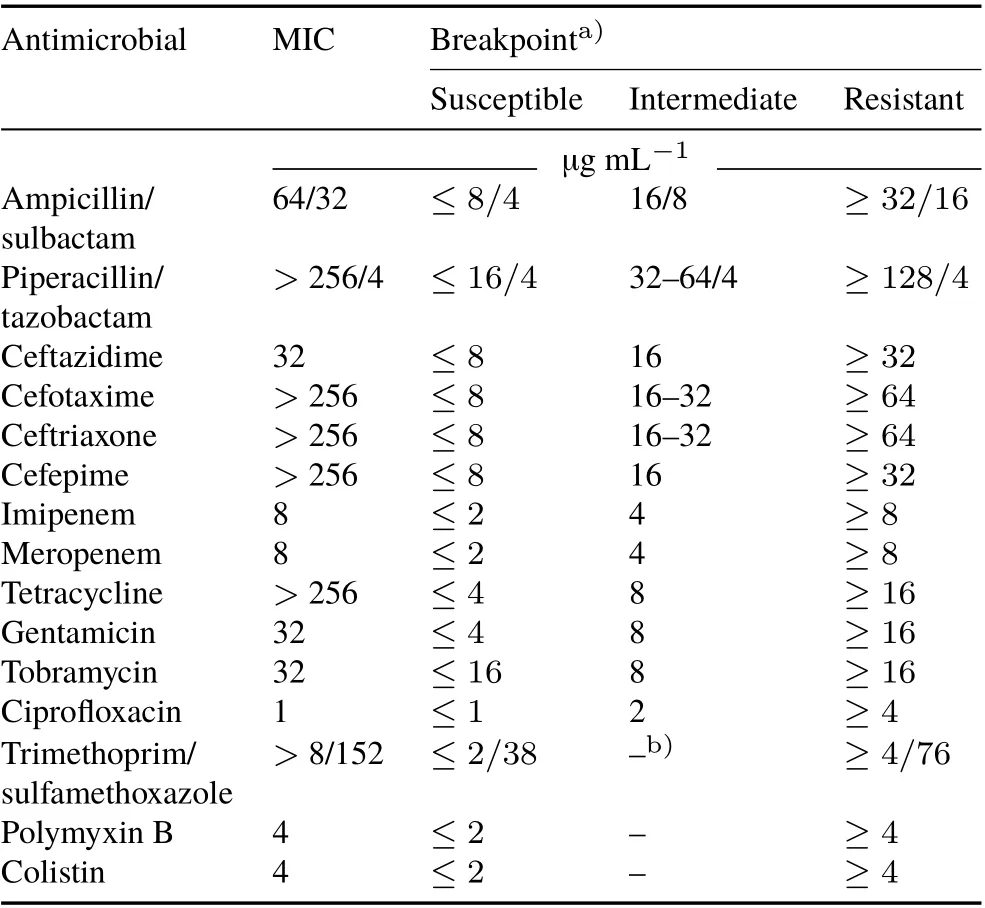
TABLE I Minimum inhibitory concentrations (MICs) and breakpoints of the antimicrobials tested against the extensively drug-resistant Acinetobacter baumannii S402 isolated from a soil in Brazil
Mutations in PmrA (Ile18Thr, Thr44Asn, Ser119Thr,and Thr168Ser)and PmrB(Lys45Gln,His54Tyr,Thr58Ala,Phe65Leu, Lys67Arg, Ile76Leu, His86Asn, Gln110Glu,Ile112Val, Arg165Ser, Ile171Val, Lys179Met, Asp193Asn,Glu210Asp,Gln255His,Glu320Asp,Ile322Asn,His362Asn,Thr417Ser, and Leu439Val) were also detected (GenBank accession no.MK660500-MK660501).The MLST analysis showed five new alleles,which weregltA(allele 111),gyrB(176),gdhB(183),recA(118), andcpn60(113), and two alleles that had already been described,gpi(132)andrpoD(129),which were responsible for generating the new ST1606(singleton).
Several studies have shown increased resistance profiles inA.baumanniifrom different sources.However,there have only been a few studies that used environmental isolates(Ciceket al.,2014;Zekaet al.,2014;Furlanet al.,2018).The occurrence of XDRA.baumanniiin clinical settings greatly limits treatment options for infections caused by this bacterium.Besides,XDRA.baumanniistrains are epidemiologically significant due to their extensive resistance to multiple antimicrobials and their high probability of being resistant to all approved antimicrobials(Magiorakoset al.,2012).The presence of XDRA.baumanniiin the environment confirms the silent dissemination of this pathogen and the ARGs.There are two reports of XDRA.baumanniiin soil samples,both of which came from Croatia(Hrenovicet al.,2017;Dekicet al.,2020).
The S402 isolate showedβ-lactamase-encoding genes that are not commonly reported inA. baumannii, such asblaSHVandblaCTX-M-Gp9.In addition,it carriedblaGES,which is commonly reported in clinical isolates ofA.baumannii(Hujeret al.,2006;Naaset al.,2007;Bonninet al.,2011;Potronet al.,2011).In addition,the S402 isolate harbored other ARGs for aminoglycosides,tetracyclines,and sulfonamides, which have already been reported in clinical and environmental isolates ofA.baumannii(Coyneet al.,2011;Bonninet al.,2013;Atasoyet al.,2015;Wanget al.,2017).The presence oftetBandsul1in isolates ofA.baumannii,including those resistant to carbapenems,has been reported extensively,and the results from this study also showed that they were present(Khorsiet al.,2015;Wanget al.,2017).
Mutations in the PmrAB two-component regulatory system have been closely related to polymyxins resistance and have already been described inA.baumannii(Beceiroet al.,2011;Thi Khanh Nhuet al.,2016).These bacteria have their own plasmid types because the replicons are different from those described for enterobacteria andPseudomonas aeruginosa. Among the GRs, GR6 contains the replicase Aci6,which belongs to the Rep superfamily pfam03090 and has no iterons(Bertiniet al.,2010).Plasmids belonging to the GR6 group have been detected in MDRA.baumannii,which contains several ARGs,including those detected in this study (Poviloniset al., 2013; Leungtongkamet al.,2018).
Soil is a complex environment where it is difficult to fully understand the occurrence and dissemination of the resistome. However, several factors can select ARGs and consequently, MDR bacteria (D’Costaet al., 2011). The presence of XDRA.baumanniiin soil is closely related to the illegal disposal of hospital waste(Hrenovicet al.,2017).In addition,soil pH is an important environmental factor that contributes to the survival ofA.baumanniiin soil(Dekicet al.,2020).Human activities are increasing the resistome,which will promote the dissemination of ARGs across the globe(Zhuet al.,2019).
In conclusion,this study reports an XDRA.baumannii(S402)that was obtained from soil taken from a field cultivated with corn,which is widely used as a human and animal feed and,consequently,can spread to several niches.To the best of our knowledge,this is the third report in the world and the first report in Brazil of an XDRA.baumanniiobtained from soil that is resistant to carbapenems and polymyxins,two of the main antimicrobials used in the treatment of different infections.
ACKNOWLEDGEMENTS
This study was financially supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo(FAPESP),Brazil(No.2018/19539-0).The authors thank the FAPESP(No. 2018/01890-3)and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior(CAPES),Brazil(No.88882.180855/2018-01)(Finance code 001)for fellowships.The authors also thank the team of curators from theAcinetobacter baumanniimultilocus sequence typing (MLST)website(https://pubmlst.org/abaumannii/)sited at the University of Oxford,UK for curating the data and making them publicly available.
杂志排行
Pedosphere的其它文章
- Distribution characteristics and diversities of cbb and coxL genes in paddy soil profiles from southern China
- Assessment of compost and three biochars associated with Ailanthus altissima(Miller)Swingle for lead and arsenic stabilization in a post-mining Technosol
- Synthesis of an eco-friendly nanocomposite fertilizer for common bean based on carbon nanoparticles from agricultural waste biochar
- Letter to the Editor Soil carbon availability affects nitrogen transformation under irrigated lucerne
- Short-term microbial responses to soluble inorganic P input in a tropical lowland rain forest in Amazonia
- Impacts of silver nanoparticles on enzymatic activities,nitrifying bacteria,and nitrogen transformation in soil amended with ammonium and nitrate
