Distribution characteristics and diversities of cbb and coxL genes in paddy soil profiles from southern China
2021-12-22WentaoPENGYanWANGXiuxiuZHULiufengXUJuanZHAOZhongliCUIandHuiCAO
Wentao PENGYan WANGXiuxiu ZHULiufeng XUJuan ZHAOZhongli CUI and Hui CAO
1College of Life Sciences/Key Laboratory of Agricultural Environmental Microbiology,Ministry of Agriculture and Rural Affairs,Nanjing Agricultural University,Nanjing 210095(China)
2Rubber Research Institute,Chinese Academy of Tropical Agricultural Sciences,Haikou 571101(China)
(Received October 21,2018;revised March 19,2019)
ABSTRACT The Calvin-Benson-Bassham cycle and Wood-Ljungdahl pathway are two well-known ones in the six carbon sequestration pathways,but the current knowledge of their occurrence in different layers of agricultural soil profiles is poor.In this study, the diversities of three genes encoding ribulose-1,5-bisphosphate carboxylase/oxygenase(RubisCO),i.e.,genes encoding the green-like(cbbLG)and red-like(cbbLR)forms of RubisCO I and encoding RubisCO II(cbbM),and the gene encoding carbon monoxide dehydrogenase large subunit(coxL)from five paddy soils in southern China were investigated by real-time quantitative polymerase chain reaction,restriction fragment length polymorphism(RFLP)analysis,and clone library.The abundances of the four genes ranged from 107 to 109 copies g-1 soil,and the cbbLR gene outnumbered the other three genes in all soil samples,suggesting important roles they play in carbon dioxide(CO2)fixation.In addition,it was found that the copy numbers of cbbLR and cbbLG decreased with increasing soil depth,while the copy numbers of cbbM and coxL decreased in the shallow depths but increased with increasing soil depth.The results of RFLP showed a larger Shannon index(H)in the deeper soil layers among the four gene clone libraries,indicating that the community diversity in these soil layers was greater.The cbbLG gene had relatively low diversity(at genus level),and most of the sequences were classified as Sideroxydans and Thiobacillus.In contrast,the highly diverse groups were found in the other three gene clone libraries(cbbLR,cbbM,and coxL),most of which were distantly related to known sequences,even forming separate clusters.In summary,this study provides a new insight into CO2 fixers along agricultural soil profiles by comparing four bacterial genes.
Key Words: Calvin-Benson-Bassham cycle,carbon dioxide fixation,carbon monoxide dehydrogenase,ribulose-1,5-bisphosphate carboxylase/oxygenase
INTRODUCTION
The carbon content in terrestrial ecosystems is thrice as large as that in the atmospheric environment(Falkowskiet al.,2000).Hence,terrestrial ecosystems are recognized as the major sinks of global carbon dioxide(CO2)emissions.Finding sustainable ways to reduce the“greenhouse effect”caused by CO2has been the focus of global attention(Gibson,1996).Consequently,exploring effective methods of CO2-fixing is important,with microorganisms found to play an important role in carbon (C) sequestration, estimated to possess a potential global sequestration of 0.6 to 4.9 Pg C year-1across the Earth’s terrestrial area(Yuanet al.,2012a).
A total of six C fixation pathways are known to exist among the processes of microbe metabolism(Bar-Evenet al., 2010). The Calvin-Benson-Bassham (CBB) cycle is deemed to be the most important and widely distributed pathway(Tabita,1999).In the CBB cycle,ribulose 1,5-bisphosphate carboxylase/oxygenase(RubisCO)participates in CO2fixation and catalyzes a rate-limiting step;therefore,RubisCO is thought to be a marker of C-fixing autotrophs.Although four isoforms of RubisCO(I,II,III,and IV)have been identified (Tabita, 1999), the isoform RubisCO III exists only in some thermophilic archaea(Katoet al.,2012),while isoform RubisCO IV is not involved in the CBB cycle(Ashidaet al.,2003).These forms are less relevant because of their negligible C-fixing abilities. The major isoform RubisCO I is widely distributed among both eukaryotes and prokaryotes and may be subdivided into four subtypes as IA, IB, IC, and ID (Tabita, 1988). The four subtypes are further classified into red-like (IA and IB) and green-like forms (IC and ID) based on their phylogenetic distances(Watson and Tabita,1997).Primers for thecbbLR(encoding the red-like form)andcbbLG(encoding the green-like form)genes have been designed to explore the functional microbial groups (Selesiet al., 2005). The red-like group usually includes photosynthetic and obligate autotrophic bacteria,whereas the green-like group includes facultative autotrophic microbes(Yuanet al.,2012a).RubisCO II is hypothesized to be similar to the ancestor of RubisCO enzymes, owing to its adaptability to high CO2concentrations prior to the increase in atmospheric O2levels(Tabita,1988;Elsaied and Naganuma,2001).ThecbbMgene encoding RubisCO II is widespread among anaerobicα-Proteobacteria and closely related to sulfur metabolism(Giriet al.,2004).
The Wood-Ljungdahl pathway,also known as the acetylcoenzyme A(CoA)pathway,is a C fixation pathway found in anaerobic microorganisms.For example,some bacteria such as acetogens, sulfate-reducing bacteria, and methanogens do not have a CBB cycle,so they use the acetyl-CoA pathway for CO2fixation. For the pathway, the key enzyme is carbon monoxide(CO)dehydrogenase(CODH),which catalyzes the reduction of CO2to CO(Hügler and Sievert,2011).The enzyme exists widely in anaerobic C sequestering microorganisms,and it is a dimer composed of heterotrimers including small (coxS), medium (coxM) and large (coxL)subunits (Dobbeket al., 2002). ThecoxLgene is often used as a molecular marker to assess the diversity of COoxidizing bacteria in environmental samples(Dunfield and King,2004).In addition,thecoxLgene is divided into two genetic subtypes;however,there is evidence that type II is not functional, while type I has genetic and biochemical characteristics(Cunliffe,2011).
In recent years,there have been many studies on CO2fixation in different ecosystems,including forest soils(Hardy and King, 2001; Kristensenet al., 2011), marine systems(Yanget al., 2001; Tolli and Taylor, 2005), wetland peats(Rich and King,1999),and even extreme environments like volcanic deposits(Huberet al.,2000;Weber and King,2010)and salt marshes(King,2007).However,few studies have simultaneously focused on the distribution characteristics of the four C sequestration-related genes in paddy soils,especially considering different soil profiles.
Therefore, the aim of our study was to investigate the community abundances and phylogenetic diversities of four genes(cbbLG,cbbLR,cbbM,andcoxL)in paddy soils(taking soil type,crop type,and soil profile into consideration)by real-time quantitative polymerase chain reaction (qPCR),restriction fragment length polymorphism(RFLP)analysis,and clone library. Furthermore, the soil physicochemical properties were determined to identify the key factors driving the distribution of these microbes.
MATERIALS AND METHODS
Soil sampling and physicochemical analyses
Samples of five paddy soils(including two types of soils)were collected from five sites in March 2012 in a non-tilled agricultural field in the Guangde region,Xuancheng City,Anhui Province of China.The area has a subtropical monsoon climate with an average annual temperature of 15.4°C and an average annual precipitation of about 1 300 mm.The soils were identified as Fe-leachic Gleyi-Stagnic Anthrosols and Typic Fe-leachi-Stagnic Anthrosols based on the Chinese Soil Taxonomy Classification System.Two sites numbered“1”were planted with oil rape, and the other sites were planted with wheat.For each site,three master horizons were identified and separately sampled.These included the plough layer(PL,15—20 cm),plow pan(PP,20—30 cm),and percogenic horizon (PH, 50—60 cm). The sample classification information is shown in Table I.
After removing 1—2 cm of topsoil,the samples in three different layers were collected and placed in a sterile paper bag.Some soil samples were air-dried at room temperature and sieved with a 2-mm grid for physicochemical analyses including pH, SOC, and total nitrogen (TN), phosphorus(TP), and potassium (TK) as well as available N (AN), P(AP),and K(AK)according to the methods described by Bao(2000). The rest of the fresh soil was stored at 4 and-20°C for further analysis.The detailed physicochemical properties of soils are listed in Table SI(see Supplementary Material for Table SI).
DNA extraction and PCR amplification
Soil DNA was extracted by E.Z.N.A.®soil DNA kit and the extracted DNA was assessed for integrity by 0.75%(weight/volume)agarose gels,followed by the amplification of the fragment carryingcbbLR,cbbLG,cbbM,andcoxL.The primers are described in Table II,and the PCR reaction mixtures and protocols were used as previously described,with a minor modification(King,2003;Katoet al.,2012).Each 50 μL reaction mixture contained 5.0 μL 10×buffer,5.0 μL MgCl2(2.5 mmol L-1),5.0 μL deoxy-ribonucleoside triphosphate (dNTP) mixture (250 μmol L-1), 1.0 μL of each primer(0.5 pmol μL-1),0.5 μLTaqDNA polymerase(2.5 U per 50 μL), 1.0 μL template DNA, and 31.5 μLsterilized water.The PCR conditions were as follows:initial denaturation at 94°C for 2 min,30 cycles of denaturation at 94°C for 30 s,annealing at 65°C for 30 s,and extension at 72°C for 1 min,and a final extension at 72°C for 10 min.Three copies of each DNA sample were amplified,and the PCR product was gel-purified using a DNA recovery kit and then stored at 4°C.
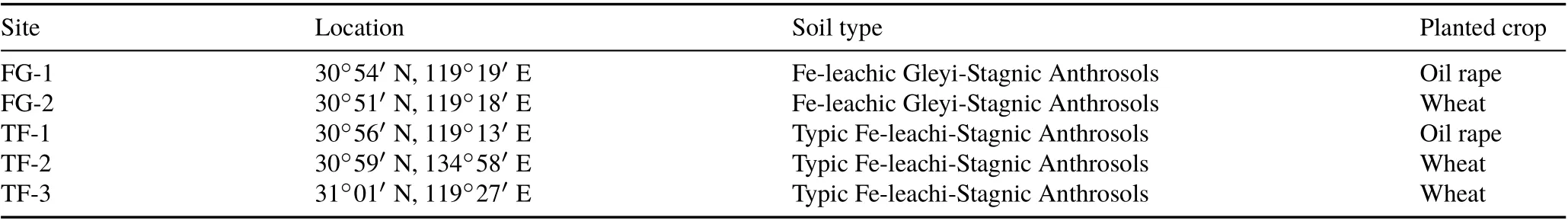
TABLE I Geographic location and soil classification parameters for the five sampling sites

TABLE II Selected primer sets for amplification
Real-time qPCR of four genes
For qPCR analysis,the amplified gene fragments were used in the preparation of standard substances.During this process,purified fragments were cloned into TA vectors,and the positive clones with the correct fragments were used for the extraction of the plasmid.The plasmid DNA concentration was determined on a NanoDrop 2000 spectrophotometer(Thermo Scientific,USA),and the copy number of the corresponding gene was calculated according to a previously described formula(Leeet al.,2008).Ten-fold serial dilutions of the plasmid DNA were prepared from 103to 108and subjected to a qPCR assay in triplicate to generate an external standard curve.At the same time,DNAs extracted from different soil samples were added into identical reaction mixtures in triplicate and amplified using the same program.In particular,10 μL of reaction mix contained 5 μL of SYBR Premix ExTaq(Takara Bio Inc., Japan), 0.2 mol L-1of each primer(Invitrogen,China),0.2 μL of ROX reference dye (provided with SYBR Premix ExTaq), 1 μL of soil DNA template(approximately 5 ng μL-1),and 3.4 μL of sterilized water.An ABI Prism 9700 real-time PCR system(Applied Biosystems,USA)was used and data analysis was performed automatically using 7500 software(version 2.0).
Construction of clone libraries and RFLP analysis
For the construction of clone library,the purified PCR products were ligated into the pMD-19T vector.The resulting ligation products were used to transform intoEscherichia colicompetent cells and cultured at 37°C.Then the positive monoclonal colony was selected and transferred to the new antibiotic plate. The clones grown at this time were the constructed clone libraries.
The diversities ofcbbL-,cbbM-,andcoxL-containing(C-fixing)microbial community were estimated by RFLP analysis.Reamplification was performed with the positive clones as the template, and then the tandem tetrameric restriction endonucleasesHhaI andRsaI were used for double digestion analysis of amplified inserts. The enzymatic cleavage product was subjected to 12%polyacrylamide gel electrophoresis to obtain an RFLP fingerprint,and the operational taxonomic units (OTUs) were defined by the Tannon digital image analysis software based on the fingerprint. Representative clones from RFLP patterns were chosen for sequencing(GenScript Biotechnology Co.Ltd.,China)before performing multiple alignments in ClustalW(Thompsonet al.,1997).
Statistical analysis
Microbial community diversity was quantified using the Shannon index (H) and Chao1 index based on the results from RFLP analysis.The obtained sequences were aligned with known gene sequences from the GenBank database using Basic Local Alignment Search Tool(BLAST),and sequences with identity>97% were identified as an OTU.The number of OTUs and diversity indices for four clone libraries were estimated using PAST (version 3.25)(Hammeret al., 2001). The Bray-Curtis algorithm was applied to calculate the similarity among clone libraries,and phylogenetic relations were determined by a neighborjoining analysis with 1 000 bootstrap replicates in MEGA 5.0.Redundancy analysis(RDA)was implemented by Canoco(version 5.0),and the rest of the figures were constructed by OriginPro(version 2018).
Nucleotide sequence accession numbers
The sequences described in this paper were deposited in the GenBank database.Accession numbers are as follows:cbbLR, KJ731002—KJ731290;cbbLG, KJ730844—KJ73-1001;cbbM,KJ731291—KJ731525;andcoxL,KJ468241—KJ468390 and KJ731526—KJ731797.
RESULTS
Abundances of four genes in three profiles
The abundances of bacterial RubisCO- and CODHrelated genes detected by the quantitative PCR assay varied among different layers of paddy soils. The bacterial gene abundance levels were measured with a high amplification efficiency(>95%)and accurate standard curve(R2>0.99).The abundance of the four genes ranged between 107and 109copies g-1soil and generally followed the order ofcbbLR >cbbM >coxL >cbbLG(Fig.1).Consistent with the results of physicochemical properties, gene abundance in the PL was significantly higher than that in the PP and PH,up to 10.5×109copies g-1soil. However, two patterns of the lowest gene abundance were observed as follows:thecbbLgenes(includingcbbLRandcbbLG)were the lowest in the PH andcbbMandcoxLin the PP. When considering the influence of soil type,it was found that the highest and lowest copy numbers for the four genes were all detected in TF-1 and TF-3,respectively.
Diversity analysis based on RFLP results
A total of 4 580 positive clones were selected, and thecbbLR,cbbLG, andcbbMgenes had no less than 80 clones in each library,while thecoxLgene had around 30 clones.Diversity indices,includingHand Chao1 index,were calculated based on RFLP results.Results(Tables III and SII,see Supplementary Material for Table SII)revealed that the largerHvalue among the four gene clone libraries was observed in the deeper soils,which meant that the community diversity of deeper soils was greater.There were few common trends in the effects of soil type and aboveground vegetation on the diversity of the four gene carriers;however,some were observed in certain gene libraries. The microbial species richness(characterized by Chao1 estimator)seemed to be significantly higher in the FG soil(remarkably in the PL)forcbbMgene and in the oil rape-planted soil(in the PL)forcoxLgene.
Phylogenetic analyses of cbbLR clone library
For thecbbLRgene, 289 representative clones were sequenced and 47 defined OTUs were used to build a phylogenetic tree,together with reference sequences downloaded from the GenBank database. As shown in Figs. 2a and S1 (see Supplementary Material for Fig. S1), all the sequences could be grouped intoα-Proteobacteria(25.6%),β-Proteobacteria(51.7%),and three unknown clusters(22.7%).Obvious differences were observed in the distribution ratio between two types of soils (pie charts in Fig. 2a). In the FG soil,α-Proteobacteria predominated in the PL and gradually declined with an increase in the soil depth,whileβ-Proteobacteria were distributed in the opposite manner.The distribution with soil layer observed in the TF soil was similar to that in the FG soil,but with a lower abundance ofα-Proteobacteria(relative abundance ranging from 5.3%to 39.3%)and a higher abundance ofβ-Proteobacteria,especially in the PH(ranging from 67.9%to 84.2%).At the genus level,six known genera were identified andRubrivivax,Bradyrhizobium,and unknown cluster 2 dominated the clone libraries(42.8%to 84.0%).The relative abundance ofBradyrhizobiumandAlcaligenesdecreased as the soil depth increased(59.2%to 5.8%and 32.5%to 3.8%,respectively),butRubrivivaxabundance increased from 3.8%to 62.4%.In addition,unknown cluster 2,one of the abundant groups in our study,had the highest abundance in the PH of the FG soil,but exhibited the lowest abundance in the TF soil.
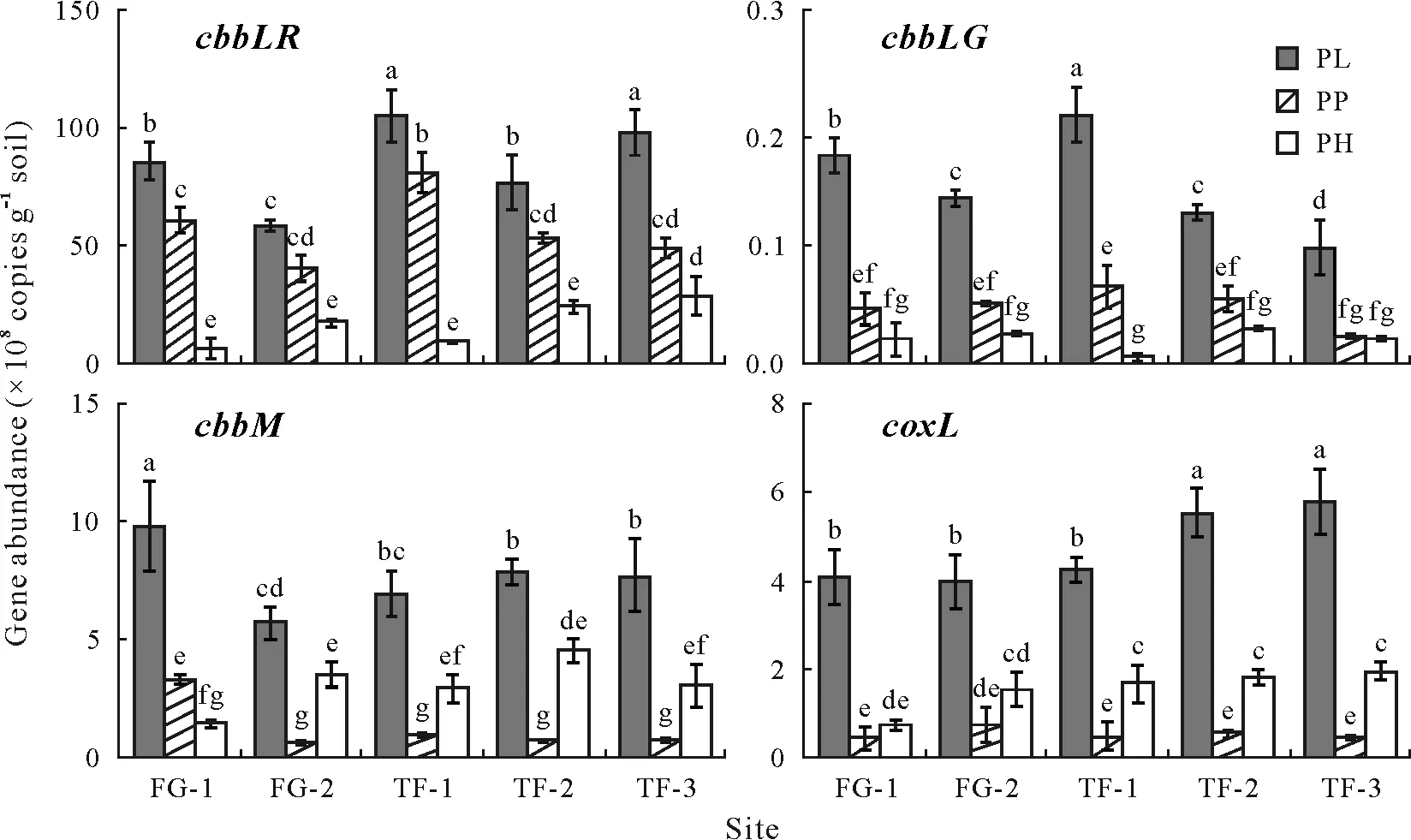
Fig.1 Abundances of the three genes encoding ribulose-1,5-bisphosphate carboxylase/oxygenase(RubisCO),i.e.,encoding the green-like(cbbLG)and red-like(cbbLR)forms of RubisCO I and encoding RubisCO II(cbbM),and the gene encoding carbon monoxide dehydrogenase large subunit(coxL)detected in different layers of paddy soils collected from five sites.Vertical bars represent the standard errors of means(n=3).Bars with the same letter(s)are not significantly different at P <0.05.See Table I for the detailed descriptions of the sampling sites.PL=plough layer;PP=plow pan;PH=percogenic horizon.
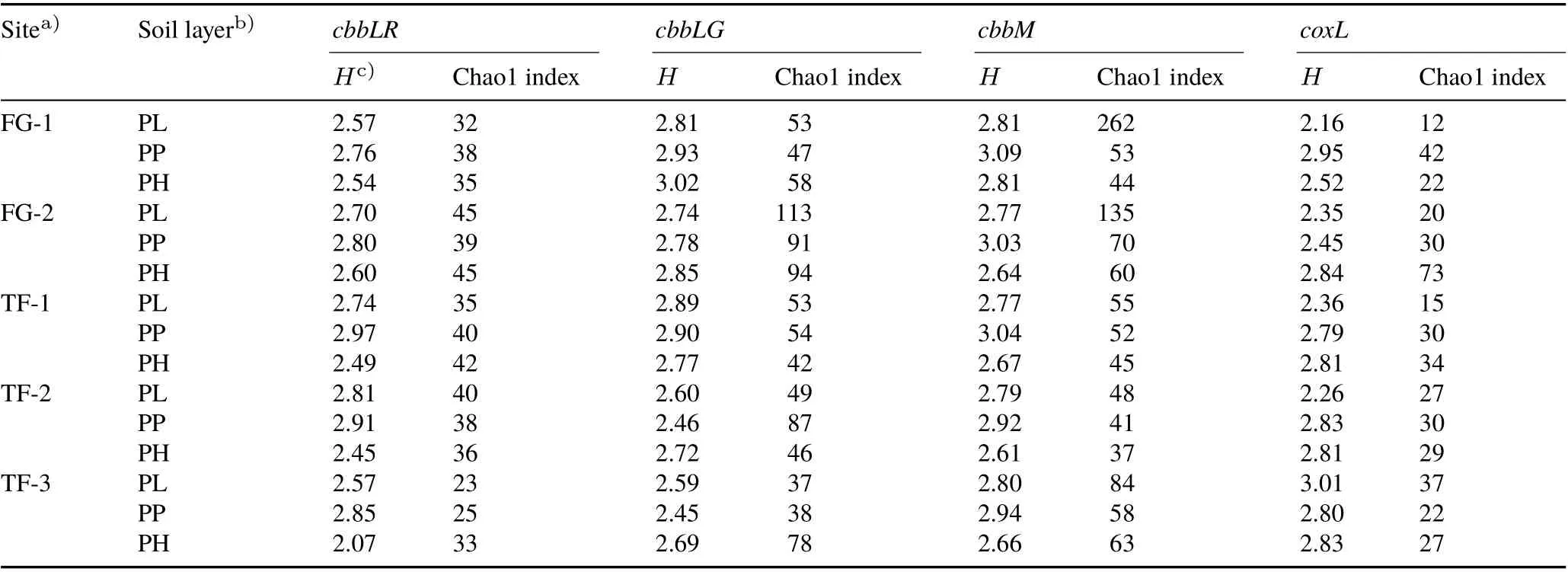
TABLE III Diversity analysis,based on restriction fragment length polymorphism results,for the three genes encoding ribulose-1,5-bisphosphate carboxylase/oxygenase(RubisCO),i.e.,genes encoding the green-like(cbbLG)and red-like(cbbLR)forms of RubisCO I and encoding RubisCO II(cbbM),and the gene encoding carbon monoxide dehydrogenase large subunit(coxL)detected in different layers of paddy soils collected from five sites
Phylogenetic analyses of cbbLG clone library
For thecbbLGgene,158 clones were sequenced based on the results of PCR-RFLP,and 28 OTUs were identified.Most sequences were closely related to three groups(79.2%to 91.5%)and group 1 showed an absolute predominance among the clone libraries(71.8%to 81.6%)(Fig. S2, see Supplementary Material for Fig.S2).Groups 1 and 2 were phylogenetically linked toβ-Proteobacteria,leading to the dominant position ofβ-Proteobacteria.In addition,a small proportion ofα-Proteobacteria in the PL and PP(0.8%to 3.5%)and Cyanobacteria(0.9%to 1.7%)were detected in certain gene libraries (Fig. 2b). In contrast to thecbbLRlibrary,β-Proteobacteria were less affected by different soil layers. Further analyses at the genus level revealed a high similarity between groups 1 and 2 andSideroxydans lithotrophicusES-1(CP001965)(similarity of amino acid sequences ranging from 93.6%to 100%in group 1 and 91.8%to 95.9% in group 2). The uncultured bacteria, clustered with group 3, were found to be widely distributed in the underground aquifer and farmland soils. Aside from the genus mentioned above,a small number of chemoautotrophs and phototrophs such asThiobacillus,Nitrobacter, andProchlorothrixwere observed.
Phylogenetic analyses of cbbM clone library
A total of 289 clones were sequenced incbbMclone libraries,and 40 OTUs were identified at 97%similarity.The phylogenetic tree shows that all the sequences were grouped into five clusters,including four classes of Proteobacteria and one group without reference sequences(Fig.S3,see Supplementary Material for Fig.S3).Pie charts with the relative abundance of microbes at the phylum level in the five soil samples are illustrated in Fig.2c.Of note,the variation trend and relative abundance of microbes in soil profiles were alike among the five soil samples,such thatcbbMcarriers may be less affected by either soil type or aboveground vegetation.The analysis of distribution variation among soil profiles revealed an increase in the abundance ofα-Proteobacteria(21.8%—28.7%to 36.6%—44.2%)andγ-Proteobacteria(0%—3.2%to 7%—11.8%)and a decline in that ofβ-Proteobacteria(42.1%—48.3%to 22.1%—29.1%)with an increase in the soil depth.A total of eight known genera were detected,as per the results of BLAST and phylogenetic analysis,withSulfuricellaandMagnetospirillumaccounting for approximately 50%of clones.Among the identified microbes,six genera followed a similar trend in soil profiles.The highest relative abundance ofSulfuricellaandRhodobacterwas observed in the PL, which decreased with an increase in the soil depth.On the other hand,Magnetospirillum,Thiodictyon,andDechloromonasshowed exactly opposite trend.In addition,Acidithiobacilliaexhibited an arch-shaped distribution,owing to their high abundance in the PP.
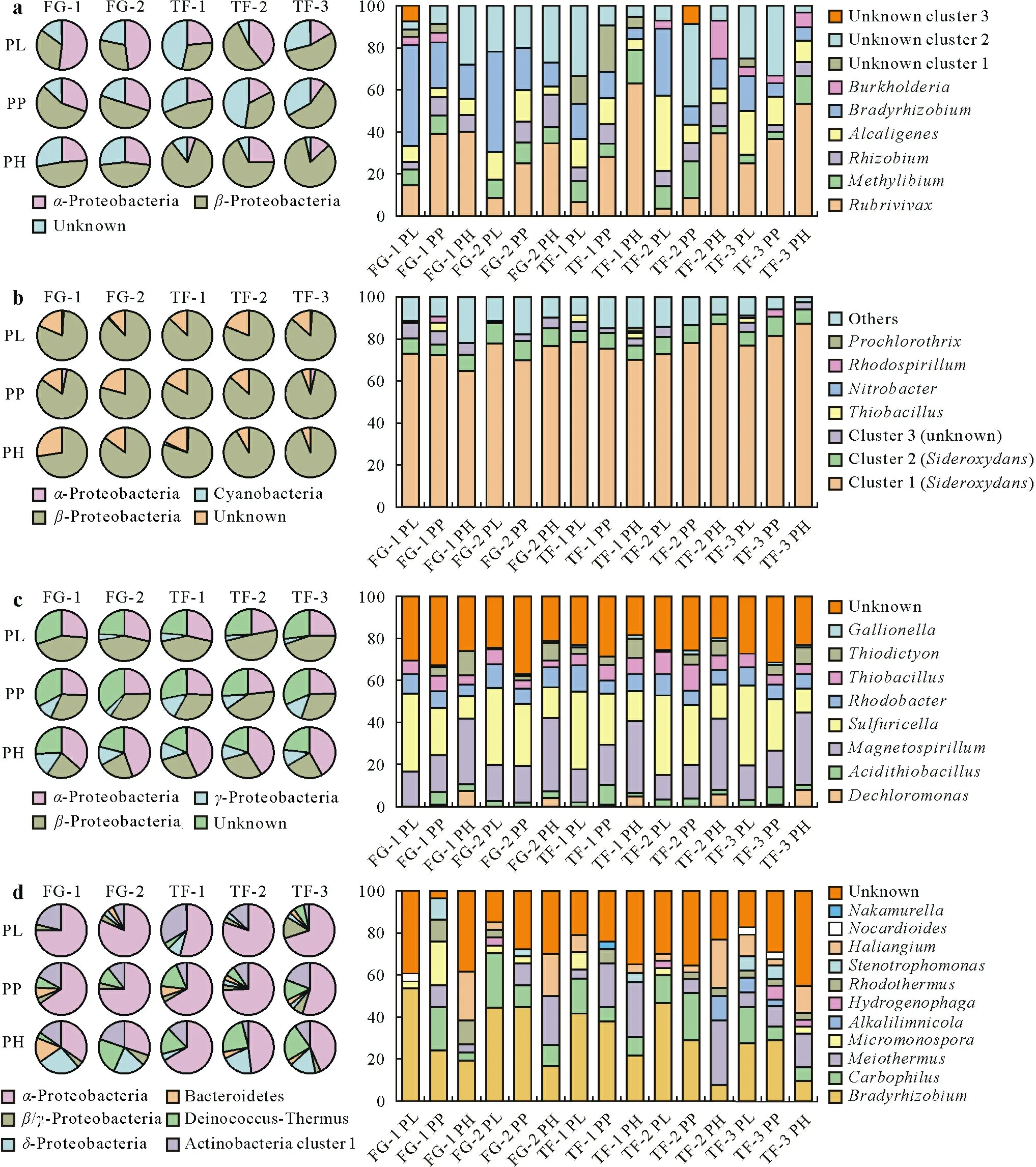
Fig.2 Percentages of various taxa contained in four gene clone libraries:three encoding ribulose-1,5-bisphosphate carboxylase/oxygenase(RubisCO),i.e.,genes encoding the red-like(cbbLR)(a)and green-like(cbbLG)(b)forms of RubisCO I and encoding RubisCO II(cbbM)(c),and one encoding carbon monoxide dehydrogenase large subunit(coxL)(d),detected in different layers of paddy soils collected from five sites.The pie chart represents the relative abundance of each phylum,and the histogram represents the relative abundance of each genus.See Table I for the detailed descriptions of the sampling sites FG-1,FG-2,TF-1,TF-2,and TF-3.PL=plough layer;PP=plow pan;PH=percogenic horizon.
Phylogenetic analyses of coxL clone library
For CODH carriers, 422coxLgene clones were sequenced and these were subdivided into 99 OTUs.A large proportion of clones (an average of 32.5%) lacked reference sequences, owing to the shortage of studies on this gene.Nonetheless,the most diverse microbes included Proteobacteria,Bacteroidetes,Deinococcus-Thermus,and Actinobacteria(Figs.2d and S4,see Supplementary Material for Fig.S4).α-Proteobacteria dominated in 15 clone libraries at a proportion of 30.0%to 81.5%and were at the lowest level in the PH.Deinococcus-Thermus,another abundant phylum in clone libraries,generally followed the trend of increasing with deepening of soil profiles.δ-Proteobacteria seldom appeared in the PL and PP,but their abundance dramatically increased in the PH.Among the 11 defined genera,Bradyrhizobium,Carbophilus,andMicromonosporaoutweighed the other eight genera at an average proportion of 53.5%.The three genera regularly distributed in soil profiles includedBradyrhizobium,which followed the trend of decrease with increasing soil depth;Meiothermus,which showed exactly opposite situation by increase with increasing soil depth;andHaliangium, which showed no remarkable difference between the PL and PP but exhibited a remarkable increase in its abundance in the PH.
Relationships between bacterial community structure and soil characteristics
The RDA results of the RFLPs showed that the soil samples can be clearly divided into three regions according to the soil profile(Fig.3).This indicates that the composition of the C-fixing microbial community exhibited significant differences in different soil profiles.In addition,TN(P=0.002), SOC (P= 0.002), AN (P= 0.002), TP (P=0.002),TK(P=0.004),and AK(P=0.006)significantly explained the diversity of community structure ofcbbLRcontaining bacteria in all paddy soils (Fig. 3a). For thecbbLGclone library,SOC(P=0.002),TN(P=0.002),AN(P=0.002),TP(P=0.002),AP(P=0.006),TK(P=0.004),and pH(P=0.006)correlated well with community variations incbbLG-containing bacteria(Fig.3b).For thecbbMclone library,SOC(P=0.002),TN(P=0.002),AN(P=0.002),TP(P=0.002),TK(P=0.002),AP(P=0.006),and AK(P=0.006)all contributed to the variations incbbM-containing microbial communities(Fig.3c).For thecoxLclone library,SOC(P=0.002),TN(P=0.002),AN(P=0.006),TP(P=0.002),and TK(P=0.004)were significantly correlated with the changes incoxL-containing microbial communities.

Fig.3 Redundancy analysis(RDA)of restriction fragment length polymorphisms of the three genes encoding ribulose-1,5-bisphosphate carboxylase/oxygenase(RubisCO),i.e.,genes encoding the red-like(cbbLR)and green-like(cbbLG)forms of RubisCO I and encoding RubisCO II(cbbM),and the gene encoding carbon monoxide dehydrogenase large subunit (coxL) detected in different layers of paddy soils collected from five sites. See Table I for the detailed descriptions of the sampling sites FG-1,FG-2,TF-1,TF-2,and TF-3.SOC=soil organic C;TN=total N;TP=total P;TK=total K;AN=available N;AP=available P;AK=available K;PL=plough layer;PP=plow pan;PH=percogenic horizon.
DISCUSSION
This study compared the abundances and diversities of four genes(cbbLG,cbbLR,cbbM,andcoxL)in five paddy soils.Results from qPCR(Fig.1)showed high abundance of RubisCO-and CODH-related genes in all soils ranging from 107to 109copies g-1soil,indicating a formerly underestimated role that CO2-fixers play in agricultural systems.The copy numbers of the four genes were higher than those of marker genes of many important processes in paddy soils,such asamoAgene of ammonium oxidation(105to 107copies g-1soil)(Jia and Conrad,2009)andnifHgene of N fixation(105to 106copies g-1soil)(Wanget al.,2012).Moreover,a research on14CO2labeling indicates that autotrophic microbes account for 4%of the total fixed CO2in terrestrial ecosystems each year (Yuanet al., 2012a). Therefore, it can be inferred that the bacteria carrying RubisCO-related genes contribute significantly to C sequestration and nutrient turnover in paddy soils. In addition, the order of the four gene copy numbers was similar to that previously presented(Xiaoet al.,2014),where thecbbgene abundance was in the order ofcbbLR >cbbM >cbbLGin five paddy soils from South China.On the basis of the significant positive correlation between RubisCO activity andcbbgene copy numbers(Yuanet al.,2012a;Xiaoet al.,2014),it may be speculated thatcbbLRcarriers possess a greater C-fixing potential thancbbLGandcbbMcarriers.
Another notable phenomenon is that the copy numbers of all genes gradually decreased with increasing soil depth.The same pattern was also found when exploring the abundance ofcbbLgene in different depth soils,which may be related to the organic matter content in the soil layer (Wuet al.,2014).Although all the genes exhibited the most abundant copy numbers in the top horizon,the locations of the lowest copy numbers were different.ThecbbLgenes(cbbLRandcbbLG)were least abundant in the PH,whereas genescbbMandcoxLwere sparse in the PP. This observation may be attributed to different adaptations to the anaerobic and oligotrophic environment.cbbMandcoxLare thought to have existed in ancient times and could be more adaptive to the early Earth conditions(lack of oxygen and organic matter)(Ferry and House,2006;Tabitaet al.,2007).Therefore,an unpredictable rise in their abundance was observed in the PH, suggesting that certain microbes are more active and adaptive in the reducing environments that are abundant in reducing substances.On the contrary,thecbbLgene,which is widely distributed among evolved C-fixing microbes,plants,and algae(Tabita,1999),was less adaptive to anaerobic and innutritious environments.
Based on the results of PCR-RFLP and phylogenetic analyses,the microbial groups carryingcbbLGandcbbLRgenes in the soil were mainly autotrophic microorganisms.Selesiet al.(2005)cloned and sequenced 24cbbLgreen-like genes from a long-term experimental farmland in Germany and found that the main group wasNitrobacter.In this study,Thiobacillus denitrificansandNitrobacter hamburgensiswere also detected in various soil layers,but the dominant group wasSideroxydans lithotrophicus.This may be related to the soil type. The soil collected in this experiment is in a state of long-term flooding with low redox potential and rich in Fe2+ions. Theoretically, this environmental condition is suitable for the growth of iron-oxidizing bacteria.The main groups of microorganisms carryingcbbLRgene areRubrivivax,Variovorax,Methylibium,Alcaligenes,andPseudonocardia. A study exploring the diversity ofcbbLRgenes in the soil of no-tillage paddy field,through Cstabilized isotope labeling techniques,found such autotrophic C sequestering microorganisms asBradyrhizobium,Rubrivivax,Rhodopseudomonas,Rhodospirillum,Methylibium,andVariovorax(Qianet al.,2015),consistent in part with the results of our study.
At present, there are few reports on the diversity ofcbbMgenes in farmland ecosystems,with the majority focused on the aquatic environment,volcanic vents,and other habitats(Wanget al.,2009;Konget al.,2012).A previous study detected the microbial community structure ofcbbMgenotypes in long-term snow-covered lake water by clone library and quantitative fluorescence technique,and found that some special obligatory autotrophic microorganisms inhabited lake water, which are mainly related to ammonia and reduced iron and sulfur (Konget al., 2012). The ecological environment of this study is similar,as there is also no sunlight and lack of photoautotrophic producers,and here we found that chemical auxotrophic bacteria such asThiobacillusandSulfuritaleawere the main components of the microbial community carrying thecbbMgene. In thecoxLclone libraries,both heterotrophic(Meiothermus)and mixotrophic genera(all those remaining)were present.AlthoughcoxLcarriers usually exhibit CO-oxidation characteristics(King and Weber,2007),their potential for CO2fixation is non-negligible as CO2was confirmed to be used by two dominant genera,includingBradyrhizobium(Badger and Bek, 2008)andCarbophilus(Meyer, 2015). AscoxLcarriers have been more thoroughly studied,the bacteria carrying CO oxidase may preferentially use organic substrates(Weber and King,2012;Cunliffe,2013).This may serve as the primary reason for the enrichment ofBradyrhizobiumandCarbophilusin the upper layers.
The RDA results showed that the diversity of the four genes in different soils was not identical and mainly related to soil physicochemical properties. Among the soil properties,TN,SOC,AN,TP,and TK were significant factors affecting the diversity of the four genes(Fig.3).The SOC is an important environmental factor, which can provide all kinds of inorganic elements or even organic matter for microorganisms,and is conducive for the growth of facultative autotrophic bacteria(Yuanet al.,2012b).Research has found that ammonia oxidation is coupled to CO2fixation by highly diverse archaea and bacteria(Jenniferet al.,2011),and our results also suggest the possible link between N and microbial CO2fixation.
CONCLUSIONS
This work highlights the potential of CO2fixation and reveals the variations in C-fixing microbial communities in different profiles of paddy soils.Of course,it is necessary to use other techniques such as isotope labeling and highthroughput sequencing in future research.Such techniques will eventually provide a better understanding of the important role of autotrophy in C cycle.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China(Nos.42077026 and 41371262).The authors thank the anonymous referees for their focused review and constructive comments/suggestions that greatly improved the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material for this article can be found in the online version.
杂志排行
Pedosphere的其它文章
- Letter to the Editor Molecular characterization of an extensively drug-resistant Acinetobacter baumannii isolated from a corn culture soil
- Assessment of compost and three biochars associated with Ailanthus altissima(Miller)Swingle for lead and arsenic stabilization in a post-mining Technosol
- Synthesis of an eco-friendly nanocomposite fertilizer for common bean based on carbon nanoparticles from agricultural waste biochar
- Letter to the Editor Soil carbon availability affects nitrogen transformation under irrigated lucerne
- Short-term microbial responses to soluble inorganic P input in a tropical lowland rain forest in Amazonia
- Impacts of silver nanoparticles on enzymatic activities,nitrifying bacteria,and nitrogen transformation in soil amended with ammonium and nitrate
