ldentifcation of blast-resistance loci through genome-wide association analysis in foxtail millet (Setaria italica (L.) Beauv.)
2021-06-24LlZhijiangJlAGuanqingLlXiangyuLlYichuZHlHuiTANGShaMAJinfengZHANGShuoLlYandongSHANGZhonglinDlAOXianmin
Ll Zhi-jiang,JlA Guan-qing,Ll Xiang-yu,Ll Yi-chu,ZHl Hui,TANG Sha,MA Jin-feng,ZHANG Shuo,Ll Yan-dong,SHANG Zhong-linDlAO Xian-min
1 College of Life Sciences,Hebei Normal University,Shijiazhuang 050024,P.R.China
2 Institute of Crop Breeding,Heilongjiang Academy of Agricultural Sciences,Harbin 150086,P.R.China
3 Institute of Crop Sciences,Chinese Academy of Agricultural Sciences,Beijing 100081,P.R.China
4 Institute of Plant Protection,Heilongjiang Academy of Agricultural Sciences,Harbin 150086,P.R.China
Abstract Blast disease caused by the fungus Magnaporthe grisea results in significant yield losses of cereal crops across the world. To date,very few regulatory genes contributing to blast resistance in grass species have been identified and the genetic basis of blast resistance in cereals remains elusive. Here,a core collection of foxtail millet (Setaria italica) containing 888 accessions was evaluated through inoculation with the blast strain HN-1 and a genome-wide association study (GWAS) was performed to detect regulators responsible for blast disease resistance in foxtail millet. The phenotypic variation of foxtail millet accessions inoculated with the blast strain HN-1 indicated that less than 1.60% of the samples were highly resistant,35.25% were moderately resistant,57.09% were moderately susceptible,and 6.08% were highly susceptible. The geographical pattern of blast-resistant samples revealed that a high proportion of resistant accessions were located in lower latitude regions where the foxtail millet growing season has higher rain precipitation. Using 720 000 SNP markers covering the Setaria genome,GWAS showed that two genomic loci from chromosomes 2 and 9 were significantly associated with blast disease resistance in foxtail millet. Finally,eight putative genes were identified using rice blast-related transcriptomic data. The results of this work lay a foundation for the foxtail millet blast resistance biology and provide guidance for breeding practices in this promising crop species and other cereals.
Keywords:foxtail millet,blast resistance,core collection,GWAS
1.lntroduction
Blast caused by the fungusMagnaporthe griseais considered as an important fungal disease in cereal crops worldwide due to its widespread destructiveness and occurrence under conducive environments,and has been reported to occur on more than 50 grass species (Ou 1987).Hybridization tests among blast pathogen isolates from rice (Oryza sativa),wheat (Triticum aestivum),and foxtail millet (Setaria italica) have indicated that they are close relatives of the same pathogen species (Katoet al.2000),suggesting similar pathogenic responses of cereal crops to blast strain inoculation (Couchet al.2005).
Blast-related studies in rice have been extensively reported including identification of different isolate genotypes,screening of resistant varieties,genetic analysis of resistance characteristics,mining of resistance QTLs,and cloning of resistance genes (Valent and Khang 2010;Wanget al.2016;Xiaoet al.2016). Genome-wide association study (GWAS) has been quickly developed as an effective approach for QTL mining and functional gene identification from natural populations,and has been widely used in rice (Huanget al.2012;Hoanget al.2019;Zhanget al.2019),maize (Zea mays) (Wenget al.2011;Liet al.2013;Farfanet al.2015),soybean (Glycine max) (Hwanget al.2014),and other crops (Liuet al.2019) to research agronomic characteristics. GWAS has also shown its power for studying crop disease and pest resistance characters in maize (Samayoaet al.2015),rice (Wanget al.2014;Liet al.2017,2019;Fenget al.2019),and other crops (Yeet al.2019;Sheret al.2020). With 805 158 SNPs identified from 517 accessions of rice,126 significant QTL loci for blast resistance were mined out,among which seven known blast-resistance genes were included,indicating GWAS is a powerful tool for disease-resistance gene mining.
Foxtail millet was domesticated in China more than 10 500 years ago and has contributed greatly to the development of oriental human civilization (Yanget al.2012). It is still a staple cereal in dry land agriculture in northern China,India and other Asian countries,and is considered as a strategic crop to deal with the drier and warmer climate in the future (Diaoet al.2014). With a total annual growing area of approximately two million hectares,mostly in the North China Plain,Northeast China and the Northwest Loess Plateau,foxtail millet grain is cooked into diverse staple daily foods in the countryside and the straw is used as a high-quality feed for animals like sheep,goats and cows (Austin 2006;Doust and Diao 2017). Blast is one of the most severe diseases in foxtail millet and usually causes yield losses greater than 30%,although grain losses can be more than 80% for the most sensitive cultivars in certain environments. Blast used to be a major disease mostly occurring in the North China Plain before 2010,where high level of precipitation and temperature creates a humid environment suitable for blast occurrence during the foxtail millet growing season. However,blast has spread into the foxtail millet growing area in Northeast China and the Northwest Loess Plateau in recent years,where the low level of rainfall and cool temperature previously made it difficult for the pathogenic fungus to develop,and has become a major disease in these regions,which might be related with climate changes (Li Zet al.2016).
Blast fungi can infect all organs of foxtail millet in different growing stages,resulting in seedling blast,leaf blast,stem blast,and panicle blast,among which panicle blast is the most harmful for grain yield (Lianget al.1959;Yanet al.1985,1988). The infection capability of differentM.griseaisolates varies greatly. Seven hundred and 11 foxtail millet blast isolates collected from 10 provinces in the 1980s were classified into 32 strains belonging to seven groups using six genotypes of foxtail millet (Yanet al.1985). By evaluating the pathogenicity ranges of 60 foxtail millet varieties to 10 blast isolates collected from northern China,a pathogenicity testing system for blast isolates consisting of seven foxtail millet varieties was established and the 10 isolates were classified into four groups (Li Zet al.2016). Field estimation of blast resistance values using 1 637 accessions of foxtail millet by Wu (1985) showed that 15% of the tested accessions were resistant to blast. A similar experiment was carried out by Liuet al.(1990). However,because of the difficulty in controlling conditions in the field,the results might be not accurate. Measurement of blast resistance values was carried out by Xieet al.(1984) and Wanget al.(1985) in artificial greenhouse conditions and many foxtail millet landraces with blast resistance in the seedling stage were identified. Further study indicated that the blast resistance capability of foxtail millet landraces in the seedling stage was positively correlated with that in the late growing stage,although there might be some differences. However,there have been no reports on QTL mining and gene cloning for blast resistance in foxtail millet.
A primary core collection of foxtail millet germplasm from the Crop Gene Bank of China,Institute of Crop Sciences (ICS),Chinese Academy of Agricultural Sciences (CAAS),was previously constructed and used as a GWAS panel. Resequencing of the panel accessions with the Illumina methodology generated a foxtail millet haplotype map with 720 000 common SNPs (Jiaet al.2013). One strongly infectious blast strain in foxtail millet,namely HN-1,was isolated (Li Zet al.2016),with which this study aimed to test the resistance of the foxtail millet primary core collections. QTL mining was also carried out using the susceptibility rankings of each accession to the HN-1 isolate. The QTLs identified in this trial will promote blast resistance biology and breeding in foxtail millet and other cereal crops,such as rice.
2.Materials and methods
2.1.Foxtail millet germplasm sampling and planting
A core collection of foxtail millet previously reported by Jiaet al.(2013) was sampled for blast resistance evaluation (Appendix A). A total of 888 accessions from the core collection were planted during the spring of 2014 in Harbin,Heilongjiang Province,China,in a greenhouse environment using a nutrient pot nursery. Twenty seeds were planted in each pot and seedling thinning was conducted when three leaves had expanded. Finally,10 healthy,uniform individuals were retained in each pot. For each accession of foxtail millet,three repeats comprising a total of 30 individuals were used for evaluations.
2.2.Blast strain propagation and inoculation
The HN-1 strain was isolated from infected leaves of the variety T58,which was planted in Nanbin Experimental Station in Sanya City,Hainan Province,China,in late November of 2012 (Li Zet al.2016),and its pathogenicity was verified in 2013 by inoculation. HN-1 was then preserved at 4°C and was transferred to sorghum grain medium for propagation. After culturing at 25°C for 15-20 days,hyphae were eluted and then laid in an enamel tray,which was covered with 1-2 damp filter papers. The enamel tray was then covered using etamine and cultured at 25°C for 4-5 days. Spores of HN-1 were collected and suspended using sterile water at a concentration of 1×105cfu mL-1.After spraying the spores during the 4-to 5-leaf stages,all inoculated accessions were kept under high temperature (28°C) and humidity (>80%) conditions for 13 h while continuously covered with a plastic membrane. The reaction types were investigated at 7 days after inoculation.The scab density and distribution pattern across the seedling leaves were used as grading standards for foxtail millet blast. A total of nine grades of susceptibility were recorded for further analysis:immune (grade 0),highly resistant (grades 1-3),moderately resistant (grades 4-5),moderately susceptible (grades 6-7),and highly susceptible (grades 8-9) (Lu 2006),which was quite similar to the criteria for rice blast evaluation.
2.3.Genome-wide association analysis of blast resistance-related loci
A previously reported hapmap consisting of 720 000 SNPs of the foxtail millet primary core collection was used for GWAS of blast resistance (Jiaet al.2013) to the HN-1 isolate following the TASSEL 5.0 pipeline using the mixed linear model (PCA+K). The whole-genome significance cutoff was set at 10-7based on permutation tests described previously (Churchill and Doerge 1994). A Manhattan plot was created using the R package qqman (Turner 2014). Annotated genes (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Sitalica) surrounding (±100 kb) relevant significant loci were selected as candidates for further analysis of putative blast resistanceregulating genes.
2.4.Analysis of candidate foxtail millet genes using rice transcriptomic data
All candidate genes were subjected to BlastP analysis to identify corresponding rice homologs from Phytozome 12.0 (https://phytozome.jgi.doe.gov/pz/portal.html).Transcriptomic data reported by Li Wet al.(2016) were retrieved for evaluation of differentially expressed genes responsive to blast strain inoculation in rice,and all the foxtail millet candidate genes were assessed according to the list of differentially expressed rice genes. Finally,the expressional data of eight foxtail millet candidate genes were determined through SIFGD (http://structuralbiology.cau.edu.cn/SIFGD/).
3.Results
3.1.Phenotypic variation of foxtail millet to blast strain HN-1 inoculation
A total of 888 foxtail millet accessions were inoculated using the blast strain HN-1 in artificial greenhouse conditions and dramatic variation in blast scab density and distribution patterns among all accessions was observed at the seedling stage. To determine the degree of seedling resistance to the blast pathogen in foxtail millet,a scale of nine grades of disease susceptibility was used to classify all accessions based on scab size and density on fully expanded leaves 7 days after inoculation (Fig.1-A). Among all 888 accessions tested,no immune accessions were identified,implying all foxtail millet accessions were susceptible to the HN-1 isolate. Only 14 samples were identified as highly resistant (grades 1-3),representing less than 1.60% of the samples tested,including Damaomaogu,Huangbangtou,Danpinian,Jinpingguzi,and Qitouhuang. Fifty-four samples were identified as highly susceptible (grades 8-9) varieties,representing 6.08% of all samples. Several well-known varieties such as Dajinmiao,Jinxiangyu,Baishagu,Hongguzi,Yangu 13,Changgu 4,Daqingmiaoyuci,Xiaohonggu,and Yazizui were classified into this cluster. The number of accessions belonging to the moderately resistant (grade 4-5) and moderately susceptible (grade 6-7) groups was 313 and 507,respectively,comprising 35.25 and 57.09% of the samples tested. A nearly normal distribution of accession numbers was observed for the graded subgroups (Fig.1-B),indicating the evaluation was reliable.
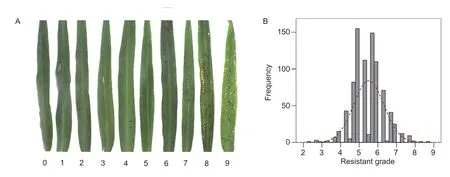
Fig.1 Blast strain inoculation in a foxtail millet (Setaria italica (L.) Beauv.) core collection. A,grade standards for resistance evaluation. B,frequency distribution of foxtail millet accessions.
3.2.Geographical distribution of blast-resistance accessions across the world
To clarify the relationship between blast resistance and the geographical origins of the foxtail millet germplasm,the geographical pattern of blast resistance according to the cultivation regions of relevant samples was determined.Accessions sourced from Canada,Kenya,South Africa,and Bengal showed moderate and high levels of blast resistance,while varieties derived from East Asia,Europe and North Africa displayed moderate and high levels of susceptibility to blast disease (Appendix A). Given that China is the main country of foxtail millet cultivation across the world,the spread of blast resistance of Chinese foxtail millet accessions was analyzed. Based on the rainfall distribution of China,collections sourced from southern regions with higher levels of rainfall (over 400 mm yr-1) showed higher frequency of resistance to blast disease,while varieties derived from northern regions with lower levels of rainfall (less than 400 mm yr-1) displayed relatively lower frequency of resistance to blast disease. For landraces collected in China,the resistance frequencies in accessions sampled from different locations were:Guangxi (100%),Hainan (100%),Tianjin (85.71%),Jiangsu (66.67%),Heilongjiang (64.29%),Jilin (61.54%),Liaoning (53.33%),Shandong (44.44%),Beijing (44.12%),Henan (44.12%),Hubei (40%),Sichuan (40%),Hebei (38.89%),Guizhou (33.33%),Tibet (33.33%),Xinjiang (28.57%),Inner Mongolia (26.32%),Qinghai (25%),Shaanxi (20.83%),Shanxi (20.00%),Gansu (15.00%),and Ningxia (12.50%). For improved cultivars developed in China,the resistance frequencies of samples collected from different breeding programs were:Jinan of Shandong (100%),ICS of CAAS (100%),Jinzhou of Liaoning (100%),Heze of Shandong (100%),Xi’an of Shaanxi (75%),Zhengzhou of Henan (60%),Cangzhou of Hebei (57.14%),Zhangjiakou of Hebei (50%),Tieling of Liaoning (50%),Anyang of Henan (45.83%),Lüliang of Shanxi (45%),Gongzhuling of Jilin (42.86%),Harbin of Heilongjiang (40%),Taiyuan of Shanxi (40%),Changzhi of Shanxi (33.33%),Yan’an of Shaanxi (33.33%),Chifeng of Inner Mongolia (33.33%),Hohhot of Inner Mongolia (31.25%),Nenjiang of Heilongjiang (28.57%),Changchun of Jilin (28%),Chengde of Hebei (27.27%),Shijiazhuang of Hebei (26.67%),Lanzhou of Gansu (20%),Chaoyang of Liaoning (20%),Datong of Shanxi (0%),Baoding of Hebei (0%),and Baicheng of Jilin (0%). These results clearly indicated that both landraces and improved cultivars possess similar patterns of blast disease resistance according to the rainfall distribution in China.
3.3.Candidate loci associated with blast resistance identifed through GWAS
To detect genomic regions responsible for blast resistance in foxtail millet,a GWAS was conducted using a previously released haplotype-map of Setaria. A total of 154 significant association loci were identified on chromosomes 2 and 9 (Fig.2-A and B). Because the average distance of linkage disequilibrium decay is 100 kb in the foxtail millet genome,a total of 57 annotated genes within ±100 kb intervals of the significant loci were inferred as candidates correlated with blast disease resistance,among which six annotated genes (Seita.9G361500,Seita.9G361200,Seita.9G361900,Seita.9G361800,Seita.2G097500,andSeita.2G097400) were predicted as kinase-related proteins and eight (Seita.2G096800,Seita.2G097400,Seita.2G097500,Seita.9G361900,Seita.9G361800,Seita.9G359100,Seita.9G359600,andSeita.9G361000) possessed similar protein domains to the reported disease-resistance genes identified in other cereal crop species (Appendix B).
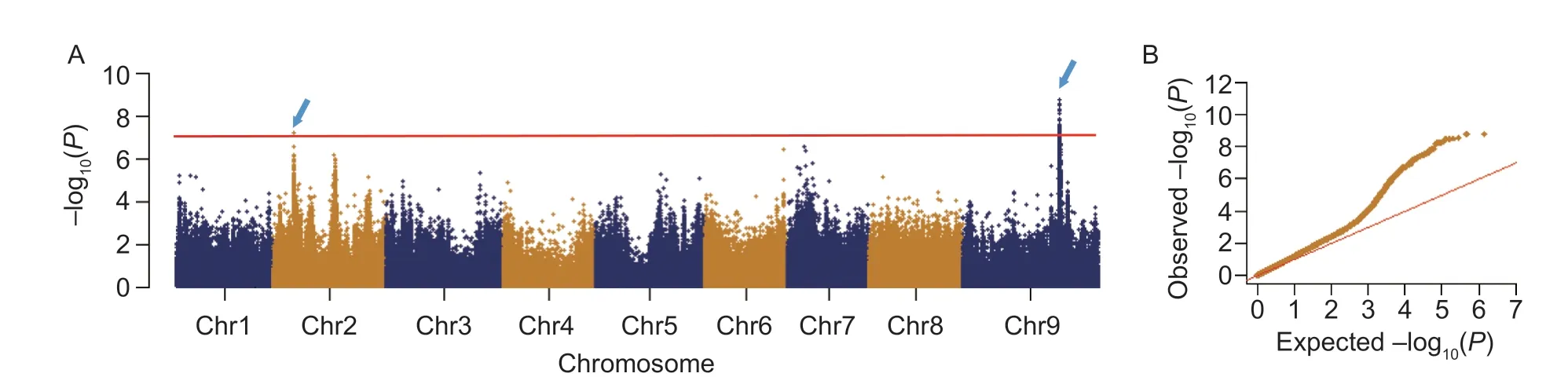
Fig.2 Genome wide association study of blast disease resistance in foxtail millet. A,Manhattan plot of genome-wide association study (GWAS) results. The broken line represents the significance cutoff value and the blue arrows indicate two significant loci on chromosomes 2 and 9. B,quantile-quantile plot for the GWAS results.
3.4.Prediction of putative blast-resistance genes in foxtail millet
To narrow down the scale of candidate genes detected by GWAS in foxtail millet,transcriptome results obtained in rice (Li Wet al.2016) were used for further comparison and investigation (Table 1). Eight rice genes homologous to foxtail millet candidates were detected as differentially expressed genes after blast strain inoculation between resistant and susceptible rice accessions (Li Wet al.2016),among which one SHR5-receptor-like kinase encoding gene (LOC_Os08g10300;homologous toSeita.2G097500andSeita.2G097400) was a vital blast resistance regulator that has been confirmed in rice through a transgenic overexpression approach,implying thatSeita.2G097500andSeita.2G097400are putative blast-resistance genes in foxtail millet.
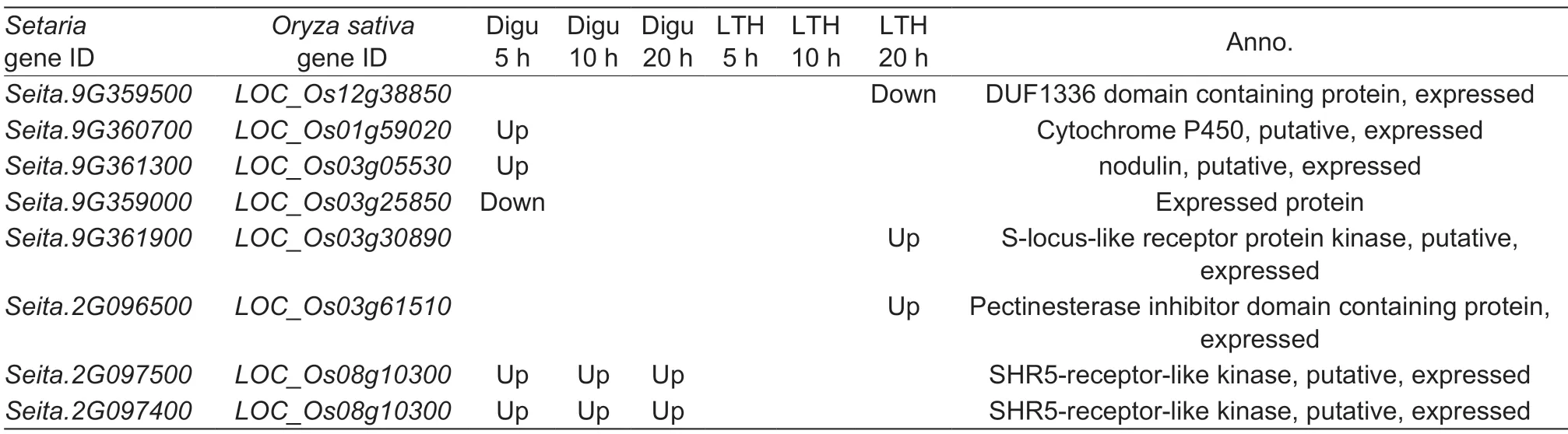
Table 1 Candidate foxtail millet genes inferred from differentially expressed homologs identified in rice for blast disease resistance1)
To clarify the spatio-temporal expression patterns of the candidate genes correlated to blast resistance,RNA-seq data released previously (Youet al.2015) were retrieved for analysis (Fig.3). Among the eight candidate genes,Seita.9G361900andSeita.9G359500showed relatively high expression levels in leaves,implying these two genes might be important contributors to leaf blast resistance in foxtail millet. Furthermore,three candidates includingSeita.9G361300,Seita.2G096500andSeita.9G360700exhibited nearly zero expression in leaves,which was consistent with the triggered expression patterns of many disease response genes. Further functional verification using knock-out,complementation,overexpression and haplotype analyses of these putative genes is currently underway.
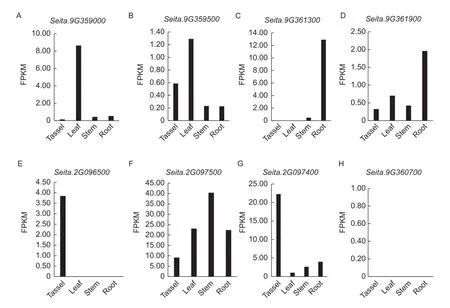
Fig.3 Spatio-temporal expression of candidate genes correlated with blast resistance in foxtail millet. A-H,fragments per kilobase million (FPKM) values of the candidate genes Seita.9G359000 (A),Seita.9G359500 (B),Seita.9G361300 (C),Seita.9G361900 (D),Seita.2G096500 (E),Seita.2G097500 (F),Seita.2G097400 (G),and Seita.9G360700 (H) across four different organs of foxtail millet.
4.Discussion
4.1.Response of foxtail millet germplasm to blast strain inoculation
It has been reported that the blast fungi isolated from finger millet,rice,foxtail millet,common millet,and wheat are closely related and can hybridize with each other (Katoet al.2000),implying similar pathogenic ability of cereal crop blast isolates. Several large-scale characterizations of blast resistance in rice (Liet al.2017) and foxtail millet (Yanet al.1985) have been conducted to screen for blast-resistance genotypes. In this trial,a worldwide collection of foxtail millet accessions was evaluated through inoculation under artificial greenhouse conditions with the blast strain HN-1 isolated from southern China,where blast disease frequently occurs both in rice and foxtail millet. Less than 1.60% of samples were identified as highly resistant lines,suggesting significant improvement of blast resistance in foxtail millet varieties is essential for future breeding programs. The geographical distribution of blast-resistant lines revealed that collections sourced from higher longitudes were more sensitive to blast disease,while samples cultivated at lower longitudes showed greater resistance to blast strain inoculation. The reason might be that the rainfall in lower longitude regions is higher than that at higher longitudes,which creates a humid environment for the fungal pathogen to grow in the foxtail millet growing season;thus,human selection in lower longitude regions has developed accessions that have better blast-resistance genotypes.Especially in China,the blast resistance of both landraces and elite cultivars was correlated with the rainfall level in the corresponding ecological regions. These results imply that environmental adaptation of foxtail millet to specific regions of blast disease occurrence might be the result of natural long-term selection pressure from the blast fungus. Similar results were also observed in other studies on foxtail millet germplasm (Wuet al.1985;Li Zet al.2016). It can be inferred that high intensity selection might be an effective approach to achieve clear improvement in blast resistance in foxtail millet varieties.
The nearly normal distribution of the numbers of foxtail millet germplasm accessions in response to inoculation with the blast strain HN-1 suggested that modern breeding selection pressure on blast resistance is limited in foxtail millet and there are diverse genotypes that can be used for future breeding. Collections sorted as landraces possessed a much higher frequency of samples resistant to blast disease compared with improved elite varieties,suggesting that several blast-resistance genes might have been lost in conjunction with yield and quality improvements in modern foxtail millet breeding due to their small side-effect on agronomic characters under field conditions (Zuoet al.2015). Furthermore,fewer highly sensitive samples were identified in elite cultivars compared with landraces,implying that blast disease-susceptible accessions were negatively selected during improvement of modern cultivars.
In this trial,only 36.83% of foxtail millet germplasm accessions were identified as highly or moderately resistant to blast fungi;a similar result was also reported by Liuet al.(1990). It has been suggested that fewer highly resistant accessions can be identified under greenhouse conditions compared with field conditions (Xieet al.1984),and positive correlations among resistance at the seedling,jointing and seed filling stages have been observed (Wanget al.1985),which implies that the results of this study could also be valuable for guiding future breeding programs in foxtail millet.
4.2.Candidate regulators’ contribution to blast resistance in foxtail millet
Blast isolates from foxtail millet and rice are closely related (Tosaet al.2006;Tosa and Chuma 2014) and can hybridize with each other (Couchet al.2005),which implies that foxtail millet and rice share similar mechanisms in their respective blast disease resistance process. Therefore,genetic results obtained from rice were used for comparative analysis.Molecular dissection of the regulatory basis of blast resistance in crop species has been previously conducted in rice using both GWAS (Wanget al.2014;Liet al.2019) and transcriptomic approaches (Li Wet al.2016) to identify vital genes contributing to blast disease resistance. To date,more than 100 genomic loci have been detected and 28 functional genes have been isolated from rice (Denget al.2017;Xiaoet al.2017;Zhaoet al.2018),among which NBS-LRR domain-containing proteins are considered vital regulators responsible for blast disease resistance. To elucidate the genetic basis of blast disease resistance in rice,Li Wet al.(2016) analyzed the transcriptomic difference between blastresistant and -susceptible varieties after inoculation with a blast isolate,and a leucine-rich repeat kinase from the LRR VIII-2 subfamily (LOC_Os08g10300) was confirmed as a key regulator that enhanced resistance toM.oryzae. In this trial,many candidate genes detected through GWAS in foxtail millet were kinases or leucine-rich domain-containing proteins,especiallySeita.2G097500andSeita.2G097400,which were homologs ofLOC_Os08g10300in rice,and might play important roles in blast disease resistance in foxtail millet. Further verification of the exact functions of these genes is necessary.
In this trial,eight candidate genes were identified using rice transcriptome data (Li Wet al.2016) and two of them (Seita.9G361900andSeita.9G359500) showed high expression levels in leaves,suggesting they are potentially important regulators controlling leaf-related blast characters. Moreover,many candidate genes identified through GWAS were considered potential functional genes because of their homologs in other cereal crops.For instance,Seita.9G361800encodes a wall-associated kinase (WAK). WAKs have been reported as important regulators responsible for disease resistance in wheat and rice (Yanget al.2014),and expression ofOsWAK1could be induced by blast fungi inoculation (de Oliveiraet al.2014).Seita.9G361900encodes an S-locus-like receptor protein kinase (CBRLK),whose homolog inArabidopsishas been confirmed as a negative-regulator of disease resistance signaling towards a virulent bacterial pathogen. Functional confirmation of these genes is currently underway.
After whole-genome comparison between blastresistance genes reported in rice and the candidates identified in this study,little overlap (exceptSeita.2G097500andSeita.2G097400) was detected between the two cereal species. This may be due to the different blast strains used for inoculation,which could lead to divergent results in the genetic response (Wanget al.2014). Moreover,the limited overlap of resistance genes identified between different crop species implies that genetic dissection of functional genes from more cereal crops is essential for deciphering the genetic basis of blast disease resistance in the future to promote pyramiding of superior resistance alleles through marker-assisted selection in future breeding programs of cereal crop species.
5.Conclusion
Evaluation of blast resistance of the foxtail millet core collections revealed less than 2% of the samples were highly resistant to blast fungi inoculation during the seedling stage. Geographical distribution pattern suggested more highly resistant accessions were located in lower latitude regions with higher rain precipitation during the foxtail millet growing season. Genome-wide association study detected two genomic loci from Chr.2 and Chr.9 in Setaria which were significantly associated with blast resistance in foxtail millet. Candidate genes inferred through comparative genetic approaches will lay the foundation for future foxtail millet blast-resistance breeding across the world.
Acknowledgements
This research was supported by the National Key R&D Program of China (2018YFD1000706 and 2018YFD1000700),the National Natural Science Foundation of China (31871630 and 31771807),the earmarked fund for China Agriculture Research System of MOF and MARA (CARS-06-13.5),the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences,and the Heilongjiang Academy of Agricultural Sciences Research Program,China (2018JJPY005). We thank Robbie Lewis,MSc,from Liwen Bianji,Edanz Group China (www.liwenbianji.cn/ac),for editing a draft of this manuscript.
Declaration of competing interest
The authors declare that they have no confilct of interest.
Appendices associated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Effects of plant density and nitrogen rate on cotton yield and nitrogen use in cotton stubble retaining fields
- Adoption of small-scale irrigation technologies and its impact on land productivity:Evidence from Rwanda
- Lignin metabolism regulates lodging resistance of maize hybrids under varying planting density
- Natural nematicidal active compounds:Recent research progress and outlook
- lmproving grain appearance of erect-panicle japonica rice cultivars by introgression of the null gs9 allele
- Comparative transcriptome analysis of different nitrogen responses in low-nitrogen sensitive and tolerant maize genotypes
