Functional analysis of the orphan genes Tssor-3 and Tssor-4 in male Plutella xylostella
2021-06-02LITianpuZHANGLiwenLIYaqingYOUMinshengZHAOQian
LI Tian-pu ,ZHANG Li-wen ,LI Ya-qing ,YOU Min-sheng ,ZHAO Qian
1 State Key Laboratory of Ecological Pest Control for Fujian-Taiwan Crops,Institute of Applied Ecology,Fujian Agriculture and Forestry University,Fuzhou 350002,P.R.China
2 Joint International Research Laboratory of Ecological Pest Control,Ministry of Education,Fuzhou 350002,P.R.China
3 Key Laboratory of Integrated Pest Management for Fujian-Taiwan Crops,Ministry of Agriculture and Rural Affairs,Fuzhou 350002,P.R.China
Abstract Orphan genes are genes with no sequence homologues in other species.Here,we identifi ed two orphan genes,namely,Tssor-3 and Tssor-4,in Plutella xylostella.Both genes contained a signal peptide sequence,suggesting their functions as secreted proteins.Expression pattern analysis based on real-time quantitative PCR (qPCR) showed that both orphan genes were specifically expressed in all male gonads except the testes.The expression of both the orphan genes peaked at the male adult stage.Immunofluorescence assays suggested that the two proteins were seminal proteins,indicating their potential roles in male reproductive regulation.To further explain their functions,we knocked down the expression of these two genes by RNA interference (RNAi).The results showed that the expression of Tssor-3 and Tssor-4 was significantly downregulated at 24 h after injection compared to that of the controls.Biological assays showed that the number of laid eggs and the hatching rate of offspring eggs were significantly reduced when the expression of Tssor-3 and Tssor-4 was reduced,suggesting that the two orphan genes played a role in male fertility in P.xylostella.Our results provide evidence that orphan genes are involved in male reproductive regulation,which is important for male fitness during evolution.
Keywords:Plutella xylostella,orphan genes,male fertility,RNA interference
1.Introduction
Orphan genes are genes that have no homologous sequences in other species and are also known as species-specific genes (Domazet-Loso and Tautz 2003).Previous studies have proven that orphan genes are closely related to growth,development,morphological diversity,and environmental adaptability of species(Domazet-Loso and Tautz 2003;Liet al.2015;Thumeckeet al.2017).For example,the orphan gene Qua-Quine Starch (QQS) inArabidopsisthalianaregulated protein composition by affecting the separation of carbon and nitrogen between proteins and carbohydrates (Liet al.2015).In salamander,destruction of the orphan geneProd1significantly affected the development of the two preaxial digits (Kumaret al.2015).Two orphan genes discovered inTriboliumcastaneum,namely,Tc-flipflop1andTc-flipflop2,played an important role in limb bud eversion in embryos (Thumeckeet al.2017).Another orphan gene fromDrosophilamelanogaster,flightin,enhanced the flight strength of wings and improved survival adaptability (Domazet-Loso and Tautz 2003).
Recent studies have found that some orphan genes specifically regulate male reproduction and are related to male adaptive fitness.Therefore,studies have proposed the“out of testis”hypothesis for the origination of some orphan genes (Kaessmann 2010).For example,in wheat,researchers mapped the orphan geneMs2by map-based cloning,proving that this gene was related to male sterility in barley andBrachypodium(Niet al.2017).In rice,researchers identified 1 921 orphan genes,most of which were expressed after sexual maturity and had a stronger ability to withstand environmental stress than non-orphan genes (Guoet al.2007).In grape,seven orphan genes were organ specific and were specifically expressed in reproductive organs (Liet al.2013).
The diamondback moth (DBM),Plutellaxylostella,belonging to Lepidoptera,is a worldwide migratory pest and harms mainly cruciferous plants (Furlonget al.2013).Because of its wide host range (Fathiet al.2010)and strong reproductive capacity (Liet al.2016),DBM is difficult to control.According to statistics,the annual loss and control cost due toP.xylostellaworldwide has reached US$4–5 billion (Zaluckiet al.2012;Furlonget al.2013).Therefore,studying the potential mechanisms related to reproductive fitness will provide new insights for better control of this pest.Multiple studies have shown that orphan genes are involved in the regulation of male reproduction (Kaessmann 2010).Therefore,targeting orphan genes that are related to male reproduction is expected to provide new clues to understand the male reproductive fitness of DBM.In this study,two orphan genes inP.xylostella,namedTssor-3andTssor-4,were identified.Further comparative analysis validated their identification as orphan genes by showing that they had no homologous sequences in other species based on a BLAST search against the NR database (https://www.ncbi.nlm.nih.gov/).Real-time quantitative PCR(qPCR) analysis revealed thatTssor-3andTssor-4were specifically expressed at the male adult stage and in all the gonads except the testes.Immunofluorescence assays confirmed that the Tssor-3 and Tssor-4 proteins were located in male gonads,i ncluding the testes,seminal vesicle and accessory gland.Further,functional analysis based on RNA interference (RNAi) showed thatTssor-3andTssor-4were related to male fertility.Our work provides evidence for the hypothesis that orphan genes are associated with male reproductive fitness and provides new clues to understand the mechanism of male reproductive success inP.xylostella.
2.Materials and methods
2.1.Insects
The wide tape strain,Geneva 88 (G88),was collected from the New York State Agricultural Experiment Station,USA in 1988.This strain was fed an artificial diet and reared in a growth chamber at (26±2)°C and (65±5)%relative humidity (RH) with a photoperiod of 16 h light/8 h dark.The adults were fed a 10% honey solution as the food supply.
2.2.RNA extraction and cDNA synthesis
Total RNA was isolated from the first-day male adults ofP.xylostellausing an Animal Tissue RNA Extraction Kit (Promega,Madison,WI,USA) according to the manufacturer’s instructions.The concentration of RNA was assessed by NanoDrop 2000 spectrophotometry(NanoDrop,Wilmington,DE,USA) by measuring the OD at 260 nm.The integrity of the RNA samples was quantified by 1% agarose electrophoresis.The cDNAs were synthesized using FastKing gDNA Dispelling RT SuperMix (Tiangen,Beijing,China) under the following conditions:42°C for 15 min and 95°C for 3 min.
2.3.Cloning and sequencing
Gene-specific primers (Appendix A) for the coding sequences (CDSs) ofTssor-3andTssor-4were designed by a primer design tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).The CDSs of the two orphan genes were amplified using PCR in a 25-μL reaction system (12.5 μL of 2×Phanta Max Buffer,1 μL of cDNA template,1 μL of primers,0.5 μL of dNTP Mix,0.5 μL of Phanta Max Super-Fidelity DNA Polymerase,8.5 μL of nuclease-free water).The PCR program was set as follows:95°C for 3 min;34 cycles of 95°C for 15 s,60°C for 30 s and 72°C for 30 s;and a final extension step at 72°C for 5 min.The PCR products were purified with a gel extraction kit (Omega,Norcross,GA,USA).The purified PCR products were subcloned into the pJET1.2 vector (Thermo,Waltham,MA,USA) and sent to Biosune Biotech Company (Fuzhou,China) for final sequencing.
2.4.Sequence analysis
The cloned sequences were translated by Translate Server (https://web.expasy.org/translate/).The signal peptides were predicted by SignalP ver.5.0 (http://www.cbs.dtu.dk/services/SignalP/).The functional domains were predicted with SMART (http://smart.embl-heidelberg.de/).
2.5.Expression profiles of Tssor-3 and Tssor-4
Samples were collected at different developmental stages ofP.xylostella,including eggs,first to third instar larvae,fourth instar female larvae,fourth instar male larvae,female pupae (first-to fourth-day female pupae were mixed together),male pupae (first-to fourth-day male pupae were mixed together),female adults (first-to fifthday female adults were mixed together) and male adults(first-to fifth-day male adults were mixed together).Different tissues,including the head,thorax,gonads except testes,and testes,were collected from the firstday male adults.In addition,male pupae at each stage from the first-to fourth-day were collected separately.Moreover,male adults at each stage from the first-to fifth-day were collected separately.All the samples were immediately frozen in liquid nitrogen and stored at–80°C until use.We prepared four biological replicates for each sample.Total RNA was extracted for each sample following the procedure described in Section 2.2.qPCR analysis was performed to validate the expression patterns of the two orphan genes (primers are listed in Appendix B).qPCR was performed on an ABI Prism 7500 Fast Detection System (Applied Biosystems,Carlsbad,CA,USA).The qPCRs were performed in 20 μL reaction mixtures (Promega,Madison,WI,USA) containing 7 μL of nuclease-free water,10 μL of 2×GoTaqqPCR Master Mix,0.4 μL of each primer (10 μmol L–1),0.2 μL of CXR Reference Dye and 2 μL of sample cDNA (500 ng μL–1).The qPCR cycling conditions were set as follows:95°C for 10 min;40 cycles of 95°C for 15 s and 60°C for 30 s;melt curve step at 95°C for 15 s;60°C for 30 min;and 95°C for 15 s.Relative quantification was performed using the comparative 2–ΔΔCTmethod (Livak and Schmittgen 2001).The RIBP (Rlk/Itk-binding protein) gene was used as a control to normalize target gene expression and correct for sample-to-sample variation.
2.6.lmmunofluorescence assay
To locate the Tssor-3 and Tssor-4 proteins,an immunofluorescence assay was performed.The gonads of male adults were used as the experimental sample,while the midgut and silk glands of fourth instar male larvae were used as the control.All the samples were fixed in paraformaldehyde (4%) at 4°C overnight.After washing three times with 1×PBS (pH 7.2–7.4),the tissues were incubated with 0.1% Triton X-100 at room temperature for 4 h.After washing three times with 1×TBST,the tissues were incubated with 3%blocking buffer at room temperature for 1 h.Then,the samples were washed three times with 1×TBST and incubated with purified rabbit anti-Tssor-3 antiserum and rabbit anti-Tssor-4 antiserum (dilution 1:200) at 4°C overnight.After washing three times with 1×TBST,the tissues were incubated with HRP-goat anti-rabbit IgG(Boster,Wuhan,China) at a dilution of 1:200 at room temperature for 2 h in the dark.After washing three times with 1×TBST,the tissues were incubated with one droplet of DAPI (Yeasen,Shanghai,China) at room temperature for 30 min in the dark.After washing three times with 1×TBST,the tissues were placed on glass slides,with one drop of glycerin added,and a coverslip was gently placed on the sample.The results were observed using a laser scanning confocal microscopy (Leica,Wetzlar,Germany).
2.7.RNAi assay
Primers (Appendix C) for the template of the transcription reactions were designed by a primer design tool(https://www.ncbi.nlm.nih.gov/tools/primer-blast/).The amplification program was set as follows:95°C for 3 min;35 cycles of 95°C for 30 s,55°C for 30 s and 72°C for 30 s;and a final extension step of 72°C for 5 min.The control gene (enhanced green fluorescent protein (EGFP))was synthesized by the same procedure.After purifying the PCR products,dsRNAs (dsTssor-3,dsTssor-4,and dsEGFP) were synthesized using a T7 RiboMAXTM Express RNAi System (Promega,Madison,WI,USA)following the manufacturer’s instructions.The dsRNAs were diluted to 4 μg μL–1using nuclease-free water.The fourth-day male pupae were injected with 207 nL (828 ng)of dsRNAs in the abdomen using a Nanoliter 2010 injector(WPI,Sarasota,FL,USA).Surviving male adults were collected at 12,24,36,and 48 h after dsRNA injection.Four replicates were set for each treatment.
2.8.Bioassays after RNAi
To assess the effect of RNAi,we determined the relative expression of the orphan genesTssor-3andTssor-4after injection.We collected experimental males (injected with dsTssor-3and dsTssor-4) and control males (injected with dsEGFP) every 12 h after injection to assess the relative expression ofTssor-3andTssor-4.
To determine the effect of the orphan genesTssor-3andTssor-4on the male fertility ofP.xylostella,we assessed the reproductive ability of the males,including the number of laid eggs and the hatching rate of offspring eggs.The experimental males were mated with virgin females 12 h after dsRNA injection,and the eggs were collected every 12 h thereafter.The number of laid eggs and the hatching rate of offspring eggs were calculated accordingly.Ten independent replicates were set.The independentt-test was used for statistical analysis.
3.Results
3.1.Characterization of Tssor-3 and Tssor-4
The orphan genesTssor-3andTssor-4were cloned from male adultP.xylostella.The full-length sequence ofTssor-3was 303 bp long without introns (accession number:MW436694),encoding a protein of 100 amino acid residues with a signal peptide containing 21 amino acid residues (Fig.1-A).
The full-length sequence ofTssor-4was 6 306 bp long with one intron (accession number:MW436695).The CDS ofTssor-4was 384 bp long with one intron,encoding a protein of 127 amino acid residues with a signal peptide containing 17 amino acid residues (Fig.1-B).No functional domain was identified in the two genes based on the domain prediction analysis,which is consistent with the characteristics of orphan genes.
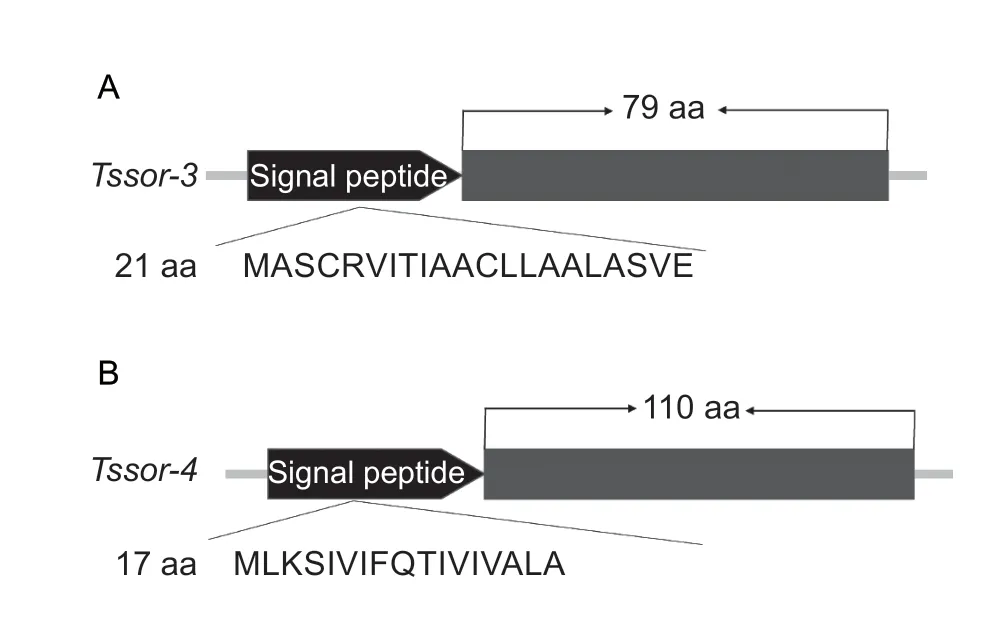
Fig.1 Gene structures of Tssor-3 (A) and Tssor-4 (B).
3.2.Expression patterns of Tssor-3 and Tssor-4
The expression patterns ofTssor-3andTssor-4were analysed at different developmental stages and in different tissues.The results showed that the orphan genesTssor-3andTssor-4were specifically highly expressed at the male adult stage,and the relative expression level ofTssor-3andTssor-4was 2.36 and 5.27,respectively (Fig.2-A and B).To further resolve the stage-specific expression patterns ofTssor-3andTssor-4,the male pupal stage (first-to fourth-day male pupae)and male adult stage (first-to fifth-day male adults) were further examined.The results showed that the expression of bothTssor-3andTssor-4began on the fourth day of the male pupal stage.However,Tssor-3reached the highest relative expression on the first and second day of the male adult stage,and the relative expression on the second day of the male adult stage was 7.71-fold higher than that on the fourth day of the male pupal stage(F5,12=51.395,P<0.001) (Fig.2-C).TheTssor-4gene reached the highest relative expression on the first day of the male adult stage,which was 4.92-fold higher than that on the fourth day of the male pupal stage (F5,12=79.554,P<0.001) (Fig.2-D).
We further analysed the tissue expression patterns for the two orphan genes.We investigated the tissues of male adults,including the head,thorax,all gonads except testes,and testes.All tissues were dissected from male adults within one day post eclosion.The results showed thatTssor-3andTssor-4were specifically expressed in the gonads (except the testes),and the relative expression levels ofTssor-3andTssor-4were 72.35 and 45.63,respectively.(Fig.2-E and F).
3.3.lmmunofluorescence assay validation
We then analysed the expression patterns ofTssor-3andTssor-4at the protein level.The gonads of male adults,including the testes,seminal vesicle and accessory gland,were used as the experimental group (Fig.3-A),while the midgut and silk glands of fourth instar male larvae were used as the control group.The results of the immunofluorescence assay confirmed that the Tssor-3 and Tssor-4 proteins were located in male gonads,including the testes,seminal vesicle and accessory gland,whereas no signals were detected in the midgut or silk glands (Fig.3-B and C).This finding,combined with the finding that these two genes encoded signal peptides,showed thatTssor-3andTssor-4encoded the seminal proteins in males ofP.xylostellaand may be involved in the regulation of male reproduction.
3.4.RNAi of Tssor-3 and Tssor-4 influenced male fertility
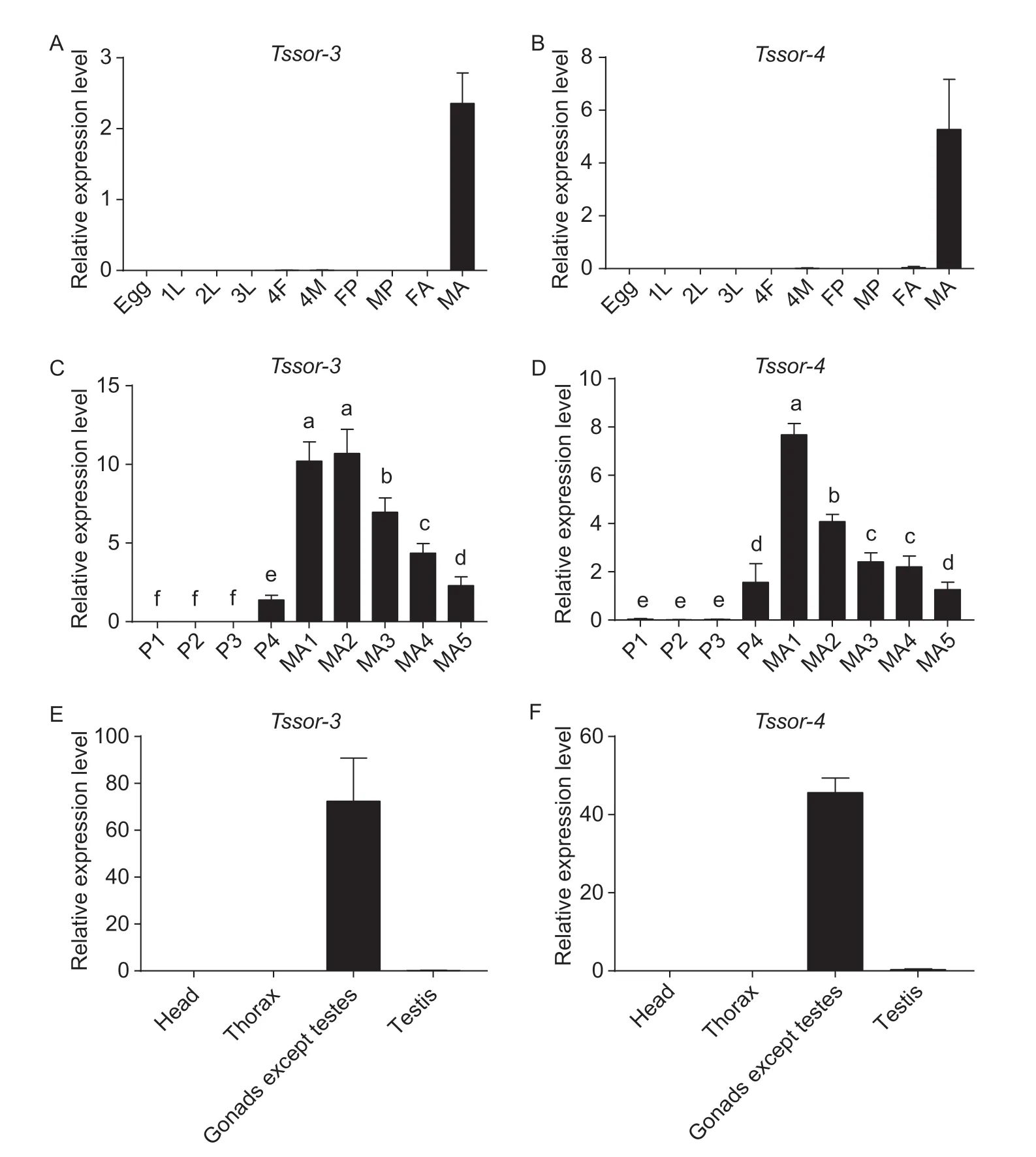
Fig.2 Expression patterns of Tssor-3 and Tssor-4 at the mRNA level.A,C and E,the relative expression of Tssor-3 at different developmental stages,male pupae and adults,and different tissues,respectively.B,D,and F,the relative expression of Tssor-4 at different developmental stages,male pupae and adults,and different tissues,respectively.1L,2L and 3L,indicate first,second and third instar larvae,respectively;4F,fourth instar female larvae;4M,fourth instar male larvae;FP,female pupa;MP,male pupa;FA,female adult;MA,male adult.P1,P2,P3 and P4 indicate first-,second-,third-and fourth-day male pupa,respectively;MA1,MA2,MA3,MA4 and MA5 indicate first-,second-,third-,fourth-and fifth-day male adult,respectively.The relative expression level is represented as the mean±SD (n=4).Different letters above the bars represent significant differences (P<0.05).
After injecting dsRNAs into the abdomen of the fourthday male pupae,qPCR was performed to examine the effects.The results showed that the relative expression ofTssor-3andTssor-4was significantly decreased at 24 h (by 29 and 31%,respectively) (t6=3.048,P=0.023;t6=3.374,P=0.015) (Fig.4-A and B).We then tested male fertility in the treated samples.Male fertility was assessed by determining the number of laid eggs and the hatching rate of offspring eggs.As shown in Fig.4-C and E,suppression ofTssor-3significantly reduced the number of laid eggs at 24 h (by 35%) (t16=2.816,P=0.012)and the hatching rate of offspring eggs at 24 and 36 h(by 14 and 20%,respectively) (t16=5.025,P=0. 000124;t10.614=3.892,P=0.003).Similarly,downregulation ofTssor-4significantly reduced the number of laid eggs at 12 h (by 65%) (t6.549=2.743,P=0.031) (Fig.4-D) and the hatching rate of offspring eggs at 12,24 and 36 h (by 12,22 and 8%,respectively) (t6=4.509,P=0.004;t10=4.494,P=0.001;t10=3.260,P=0.009) (Fig.4-F).Our results proved that downregulation of the orphan genesTssor-3andTssor-4could significantly influence male fertility inP.xylostella.
4.Discussion
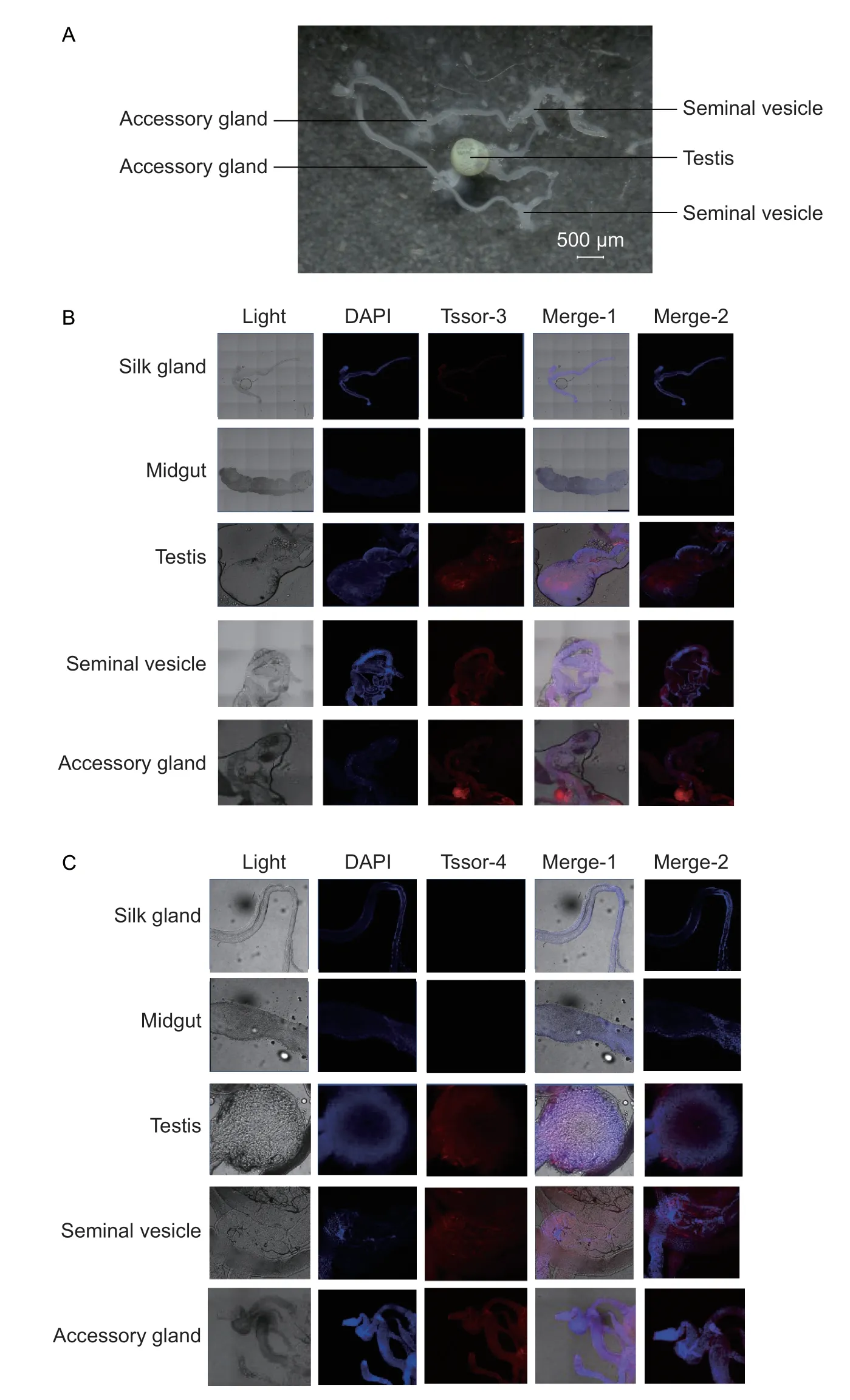
Fig.3 Expression patterns of Tssor-3 and Tssor-4 at the protein level.A,the gonad of male adult.B,immunofluorescence result for the Tssor-3 protein.Merge-1 is a composite of light DAPI and Tssor-3;Merge-2 is a composite of DAPI and Tssor-3.C,immunofluorescence result for the Tssor-4 protein.Merge-1 is a composite of light,DAPI and Tssor-4;Merge-2 is a composite of DAPI and Tssor-4.
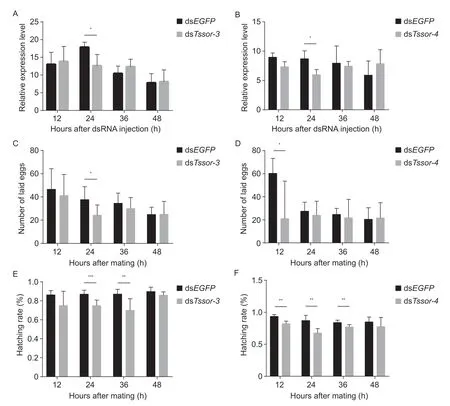
Fig.4 Functional analysis of Tssor-3 and Tssor-4.A and B,the relative expression of Tssor-3 and Tssor-4 after RNAi.C and D,the number of laid eggs after injecting dsTssor-3 and Tssor-4.E and F,the hatching rate of offspring eggs after injecting dsTssor-3 and Tssor-4.The relative expression level is represented as the mean±SD (n=4).*,** and *** above the bars represent significant differences at P<0.05,P<0.01 and P<0.001,respectively.
Multiple studies have proven that orphan genes can explain species-specific evolutionary fitness,especially male reproductive success.For example,the orphan protein encoded by the orphan geneMs2can lead to male sterility in wheat,barley andBrachypodium(Niet al.2017).However,few such genes have been reported in pest insects,especially inP.xylostellathat possesses a high reproductive capability.This study identified two orphan genes,namely,Tssor-3andTssor-4,that were specifically expressed at the male adult stage and in all gonads except the testes ofP.xylostella,encoding seminal proteins.The full-length sequence ofTssor-3was 303 bp long without introns and encoded a protein of 100 amino acid residues.Tssor-4contained one intron and had a CDS length of 384 bp,encoding a protein of 127 amino acid residues.Both orphan genes had a signal peptide and had no known functional domains.These findings were consistent with previous studies of orphan genes with short gene sequences,short peptide sequences and a small number of introns (Yanget al.2013;Xuet al.2015).For example,compared with genes with a wider phylogenetic distribution,primate orphan genes show unique characteristics:higher tissue specificity,faster evolution,and shorter peptide length(Chenet al.2020).
In addition to the sequence characteristics,previous studies have proven that orphan genes are more specifically expressed in certain tissues or at certain developmental stages.For example,in a study of rice orphan genes,the expression of most orphan genes began after sexual maturity (Guoet al.2007).In our study,we also found thatTssor-3andTssor-4were specifically expressed at the male adult stage,and the relative expression levels ofTssor-3andTssor-4were 2.36 and 5.27,respectively,which were >3-fold higher than that at other stages.Notably,these two orphan genes were expressed in all the gonads except the testes,and the relative expression levels ofTssor-3andTssor-4were 72.35 and 45.63,respectively.Male accessory glandular tissue is an important reproductive tissue in insects that synthesizes and secretes accessory glandular fluid.Accessory glandular fluid is the most important component in insect semen besides spermatozoa (Gaoet al.2008).Accessory glandular tissue contains amino acids,vitamins and other nutrients,which protect,store and activate sperm and enhance the competitiveness of sperm (Kinget al.2011).Male accessory glandular secretions had a strong effect on the reproductive activity of females;for example,the mating inhibition of females was regulated by accessory glandular fluid,as previously reported inCeratitiscapitata(Miyatakeet al.1999) andD.melanogaster(Chapmanet al.2003).Based on the sequence characteristics (signal peptides) and expression patterns,we propose that these two orphan genes encode seminal proteins inP.xylostella.This hypothesis was further validated by the results of an immunofluorescence assay,which showed that the Tssor-3 and Tssor-4 proteins were located in the gonads of male adults,including the testes,seminal vesicles and accessory glands.Seminal fluid proteins are mainly produced in the accessory gland of male insects and cause many physiological and post-mating behavioural changes (Yuet al.2016).InDrosophila,seminal fluid proteins affect female receptivity,sperm storage,sperm competition and so on (Chapman 2001).In honeybee,seminal fluid is very effective in keeping sperm alive,which is directly related to reproduction (Kinget al.2011).Thus,we propose that these two genes,which may encode seminal proteins,are closely related to male reproduction and evolutionary male fitness inP.xylostella.
RNAi has become a commonly used technique in the study of gene functions and has been successfully applied in many insects,including Diptera (Mohr 2014),Lepidoptera (Gonget al.2013;Liuet al.2014;Yanget al.2014),Coleoptera (Yoonet al.2016),Hemiptera(Christiaenset al.2014),and Orthoptera (Wynantet al.2012) insects.In this study,after injecting dsTssor-3and dsTssor-4,the relative expression of bothTssor-3andTssor-4was significantly downregulated at 24 h(by 29 and 31%,respectively).Bioassays were performed after RNAi to investigate the changes in male fertility after downregulation ofTssor-3andTssor-4.After injecting dsTssor-3,the number of laid eggs was reduced significantly at 24 h (by 35%),whereas after injecting dsTssor-4,the number of laid eggs was reduced significantly at 12 h (by 65%).Furthermore,downregulation ofTssor-3led to a significant decrease in the hatching rates of offspring eggs at 24 h and 36 h (by 14 and 20%,respectively),and downregulation ofTssor-4significantly reduced the hatching rates of offspring eggs at 12,24 and 36 h (by 12,22 and 8%,respectively).Our results proved that these two genes were involved in male fertility regulation inP.xylostella,which further supported the assumption that orphan genes were related to male reproductive and evolutionary fitness.Previous studies also found that orphan genes were closely related to male fertility.For example,the orphan protein encoded by the orphan geneMs2can lead to male sterility in wheat,barley andBrachypodium(Niet al.2017).
Because the actual effect of RNAi is relatively shortlived,further functional research is limited.Given the fact that these two orphan genes are relatively short,it is difficult to select the target site for CRISPR;therefore,it remains a challenge to study the mechanisms associated with these two orphan genes.
5.Conclusion
The orphan genesTssor-3andTssor-4were cloned fromP.xylostella.The qPCR results showed thatTssor-3andTssor-4were specifically expressed at the male adult stage and in all the gonads except the testes.The immunofluorescence results revealed that proteins encoded byTssor-3andTssor-4were located in male adult gonads.After RNAi,the egg production of females and the hatching rate of offspring eggs were significantly reduced.All the results demonstrated that the orphan genesTssor-3andTssor-4indeed play an important role in the control ofP.xylostella.
Acknowledgements
This work was supported by the Natural Science Foundation of Fujian Province,China (2020J01525),the National Natural Science Foundation of China(31320103922 and 31230061) and the Major Science and Technology Projects in Fujian Province,China(2018NZ010100130).We thank American Journal Experts for the language editing.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Receptor-like kinase OsASLRK regulates methylglyoxal response and content in rice
- Heredity and gene mapping of a novel white stripe leaf mutant in wheat
- Construction of a high-density adzuki bean genetic map and evaluation of its utility based on a QTL analysis of seed size
- Effects of temperature and solar radiation on yield of good eatingquality rice in the lower reaches of the Huai River Basin,China
- Difference in corn kernel moisture content between pre-and postharvest
- The effect of elevating temperature on the growth and development of reproductive organs and yield of summer maize
