Heredity and gene mapping of a novel white stripe leaf mutant in wheat
2021-06-02LlHuijuanJlAOZhixinNlYongjingJlANGYumeiLlJunchangPANChaoZHANGJingSUNYulongANJunhangLlUHongjieLlQiaoyunNlUJishan
Ll Hui-juan,JlAO Zhi-xin,Nl Yong-jing,JlANG Yu-mei,Ll Jun-chang,PAN Chao,ZHANG Jing,SUN Yu-long,AN Jun-hang,LlU Hong-jie,Ll Qiao-yun,NlU Ji-shan
1 National Centre of Engineering and Technological Research for Wheat/National Key Laboratory of Wheat and Maize Crop Science/Henan Agricultural University,Zhengzhou 450046,P.R.China
2 Shangqiu Academy of Agricultural and Forestry Sciences,Shangqiu 476000,P.R.China
Abstract Spotted leaf (spl) mutant is a type of leaf lesion mimic mutants in plants.We obtained some lesion mimic mutants from ethyl methane sulfonate (EMS)-mutagenized wheat (Triticum aestivum L.) cultivar Guomai 301 (wild type,WT),and one of them was named as white stripe leaf (wsl) mutant because of the white stripes on its leaves.Here we report the heredity and gene mapping of this novel wheat mutant wsl.There are many small scattered white stripes on the leaves of wsl throughout its whole growth period.As the plants grew,the white stripes became more severe and the necrotic area expanded.The mutant wsl grew only weakly before the jointing stage and gradually recovered after jointing.The length and width of the flag leaf,spike number per plant and thousand-grain weight of wsl were significantly lower than those of the WT.Genetic analysis indicated that the trait of white stripe leaf was controlled by a recessive gene locus,named as wsl,which was mapped on the short arm of chromosome 6B by SSR marker assay.Four SSR markers in the F2 population of wsl×CS were linked to wsl in the order of Xgpw1079–Xwmc104–Xgwm508-wsl–Xgpw7651 at 7.1,5.2,8.7,and 4.4 cM,respectively and three SSR markers in the F2 population of wsl×Jimai 22 were linked to wsl in the order of Xgwm508–Xwmc494–Xgwm518-wsl at 3.5,1.6 and 8.2 cM,respectively.In comparison to the reference genome sequence of Chinese Spring (CS),wsl is located in a 91-Mb region from 88 Mb (Xgwm518) to 179 Mb (Xgpw7651) on chromosome 6BS. Mutant wsl is a novel germplasm for studying the molecular mechanism of wheat leaf development.
Keywords:wheat (Triticum aestivum L.),mutant,white stripe leaf (wsl),heredity,gene mapping
1.lntroduction
Plant disease lesion mimic mutants refer to those mutants which spontaneously form necrosis symptoms similar to the pathogen infected plants in the absence of pathogen infection or obvious adversity (Johalet al.1995).The chloroplast degradation,and the decreases of photosynthetic pigment content and the photosynthetic area result in the decrease of photosynthesis,which in turn causes the yield loss of lesion mimic mutants.Lesion mimic mutants are generally more resistant to pathogens,especially fungal pathogens,than their wild types (WT) (Dietrichet al.1994;Liet al.2009).The typical phenotypes of lesion mimic mutants are similar to hypersensitive response (HR),and most of them are related to either programmed cell death (PCD) and reactive oxygen accumulation caused by allergic reactions (Jianget al.2003;Séverineet al.2003;Wang 2005),or to reactive oxygen intermediates (ROIs),salicylic acid (SA),jasmonic acid(JA),and ethylene (ET) (Wang 2005).The environmental conditions during plant growth,such as light (Fusetet al.1993),humidity (Jambunathanet al.2001),temperature (Huet al.1996),and nutritional status,also impact the formation of disease-like spots.
Lesion mimic mutants have been found from a variety of plant species,such as maize (Zea mays)(Johalet al.1995;Huet al.1998),barley (Hordeum vulgareL.) (Rostokset al.2003;Perssonet al.2009),Arabidopsis (Arabidopsis thaliana)(Brodersenet al.2002),rice (Oryza sativaL.) (Moriet al.2007;Shanget al.2009;Wanget al.2015),peanut(Arachis hypogaeaLinn.) (Badigannavaret al.2002),and soybean (Glycine maxL.)(Alet al.2019).In rice,many disease-like spot mutants have been developed,and most of the mutated genes have been successfully mapped and cloned,such asSPL28(Qiaoet al.2010),spl5(Jinet al.2015),OsLSD1(Wanget al.2005),NOE1(Linet al.2012),Spl7(Yamanouchiet al.2002),spl11(Zenget al.2004),lmr(Fekihet al.2015),andOsACDR1(Kimet al.2009).
In recent years,a large number of wheat lesion mimic mutant genes have been reported.Wheat line Ning7840 has adult plant resistance to leaf rust and shows lesion mimic symptoms during heading,which are controlled by a recessive genelmon chromosome 1BL (Li and Bai 2009).A lesion mimic plant was found in a segregating population of common wheat cross Yanzhan 1/Zaosui 30.The yield per plant and thousand-grain weight of this mutant are lower than those of its WT,and they are controlled by two pairs of recessive geneslm1andlm2(Yaoet al.2009).A white spot mutantlm3has small and discrete white spots on its leaves,and it is highly resistant to powdery mildew.The trait is controlled by a single partially dominant gene on 3BL (Wanget al.2016).Mutant HLP (hypersensitive-like phenotype) was obtained by EMS mutagenesis,and the mutant exhibits tiny,discrete,white lesions once the fifth or sixth leaf has emerged,and its adult plant resistance to leaf rust is enhanced (Kamlofskiet al.2007).Spot leaf mutant C591 (M8) is a dominant mutant.The chlorophyll content of this mutant is significantly lower than the WT parent when spots spread to the whole leaf and leaf sheath,but the yield is not significantly different from its WT parent(Nair and Tomar 2001).Although the yield obtained for a speckled mutant M66 was low,it is highly resistant to pathogens (Kinane and Jones 2001).The mutant I30 has white disease-like spots and long streaks on its leaves from the trifoliate stage,and cell death and H2O2accumulation occur in the white disease like spots.The white diseaselike spot of I30 is controlled by a recessive nuclear gene on chromosome 6DS (Liet al.2017).
Phenotypes of lesion mimic mutants range from similar allergic reactions to large amounts of chloroplast loss or death(Liet al.2009).AIM9 is a recessive gene controlling lesion mimic mutant with leaf chlorosis.The chloroplast shapes of AIM9 change from long elliptic to circular,and the chloroplasts separates from the cell wall and moves to the center of the cell (Luo and Ren 2006).The wheat speckle mutant LF2010,which is controlled by a recessive nuclear gene,shows yellow non-necrotic spots on its leaves at the three-leaf stage,and the spots are induced by light and temperature (up 8°C) (Duet al.2014).Although there are some reports about wheat lesion mimic mutants,the molecular genetic mechanisms are still unknown.Gene mapping and cloning of lesion mimic mutant genes will assist in further elucidating the molecular mechanisms of the occurrence and formation of plant disease-like spots,understanding the related genes of plant cell development and apoptosis,as well as the signaling pathways of plant disease-resistance and defense genes (Wuet al.2008;Matinet al.2010).
Mutantwslwas obtained from wheat cultivar Guomai 301 mutagenized with EMS.The typical trait ofwslis the white stripe leaves appearing from the seedling stage onward.The heredity of the white stripe leaf is stable.This study analyzes the morphological and genetic characteristics ofwsland provides a new germplasm for the study of the molecular mechanisms of lesion mimic mutants in wheat.It will lay a foundation for the further utilization of mutations of thewslgene and its properties.
2.Materials and methods
2.1.EMS treatment and selection of the mutants
The wheat cultivar Guomai 301 was bred in our laboratory,the National Centre of Engineering and Technological Research for Wheat,Henan Agricultural University.In October 2012,about 1 kg grains of Guomai 301 were soaked with cool water for 4 h,treated with 0.4% EMS solution at 0°C for 2 h,at 20°C for 14 h,washed with fresh water for 4 h,then the EMS treated grains were sown in the experimental field of Shangqiu Experimental Farm,Henan Province,China (34°25´N,115°39´E,49 m a.s.l.).The progeny of the mutagenized Guomai 301 were planted in Shangqiu from 2012–2015.After the selection,a stable-heredity mutant ofwslwas obtained and planted in our experimental field at Houwang Village,Xingyang City,Henan,China (34°25´N,115°39´E,49 m a.s.l.) from 2016 onward.The field experiments were carried out in a completely randomized design as described by Duanet al.(2015).
2.2.Morphological analysis
The plants ofwslwere observed from seedling to maturity stages.Photos were taken at typical developmental stages with a camera (Nikon Coolpix 4500,Nikon Corporation,Tokyo,Japan).The agronomic traits,including plant height,spike length,spike number per plant,thousand-grain weight,and others,were investigated at the maturity stage.The data were analyzed using SPSS17.0 Software.
2.3.Genetic analysis
The reciprocal crosses of the homozygous mutantwslwith Chinese Spring (CS) and Jimai 22 were made in 2017.The phenotypes of the hybrid plants at F1were observed in 2018.The phenotypes of the segregating populations at F2and F2:3were observed and recorded in 2019 and 2020,respectively.The segregation ratios of the segregating populations at F2and F2:3were calculated.The goodness of fit test for a Mendelian ratio was performed with a Chisquare (χ2) andt-test.
2.4.SSR assay
Two F2populations derived from the crosses ofwsl×CS andwsl×Jimai 22 were used to map the mutated genewsl,and verify the results between the two.The genomic DNAs ofwsl,CS,Jimai 22,and the segregating plants in the F2populations were extracted from leaf tissues as described by Cota-Sánchezet al.(2006).Bulked segregant analysis (BSA) (Michelmoreet al.1991) was used to identify microsatellite markers linked to thewslgene.Two DNA bulks were assembled by using equal amounts of DNA from 10 normal and 10 white stripe leaf plants for each segregating population.Markers showing polymorphisms between both parental DNA templates and between the two DNA bulks were further used to assay the individuals comprising the mapping populations.A microsatellite assay was utilized to locatewslon wheat chromosomes and to find closely linked simple sequence repeat (SSR) markers.A total of 886 wheat SSR markers evenly distributed on the 21 pairs of wheat chromosomes were used to detect the polymorphism loci linked towsl.The SSR markers employed in this study included GWM (Röderet al.1998),GDM (Pestsovaet al.2000),CFD (Guyomarc’het al.2002),GPW (Guptaet al.2002),CFA (Sourdilleet al.2003),WMC (Somerset al.2004),BARC (Songet al.2005),and some were selected from GrainGenes 2.0 (http://www.wheat.pw.usa.gov/).
2.5.PCR
PCR amplification and product analysis were carried out as described by Niuet al.(2008) and Duanet al.(2015).PCR amplification reactions were performed with a Thermal Cycler (Thermo Scientific,Massachusetts,USA).The amplification reactions were carried out using the following profile:94°C for 5 min;followed by 35 cycles of 94°C for 30 s,50–60°C (based on primer annealing temperature)for 30 s and 72°C for 30 s;and a final elongation at 72°C for 10 min before cooling to 4°C.PCR products were separated in 8% non-denatured polyacrylamide gels(acrylamide:bisacrylamide=19:1) at room temperature.A total of 2 μL of each sample was loaded and electrophoresis was performed in 1×TBE (90 mmol L–1Tris-borate,2 mmol L–1of EDTA,pH 8.3) buffer at U=500 V,I=200 mA,P=80 W for 1 h,then visualized by silver staining (Bassamet al.1991).
2.6.Linkage analysis and genetic mapping
The F2mapping population derived from crosswsl×CS comprised 200 individuals,and the F2mapping population derived from crosswsl×Jimai 22 comprised 192 individuals.We randomly selected 20 individuals from the F2population to amplify and verify the initially screened SSR markers,which showed polymorphisms both between parental DNA templates and between the two DNA bulks,and to calculate the crossing-over percentage.An SSR marker with a crossing-over percentage between 0 and 50 was identified as a linkage marker.The linkage maps of linkage markers and the mutant genewslwere drawn using IciMapping 4.1 Software (http://www.isbreeding.net),with a LOD threshold>3.0.The software calculated genetic distance (cM) with the Kosambi function (Kosambi 1944).
2.7.Chromosomal assignment of the SSR markers
Chromosomal locations of thewsl-linked microsatellite markers were carried out using a set of CS nulli-tetrasomic lines.Genomic DNAs extracted from these CS nullitetrasomic lines were used to perform PCR reactions with the primers of thewsllinkage markers.
3.Results
3.1.Characterization of the mutant wsl
We have obtained a lesion mimic mutant from EMS mutagenized Guomai 301,namedwsl.The phenotype ofwslwas obviously different from its parent Guomai 301 at the seedling stage in the field.Small scattered white stripes appeared on the leaves ofwslfrom the seedling stage,and they accompanied the plant during the whole growth period.The mutantwslgrew only weakly before jointing stage and gradually recovered after jointing stage (Fig.1).As the plants grew,the white stripes became more severe and the necrotic area expanded (Fig.2).The plant height,spike length,internode number,and the spikelet number per spike ofwslwere the same as those of Guomai 301,however,the flag leaf length,flag leaf width,spike number per plant,and thousand-grain weight ofwslwere significantly lower than those of Guomai 301 (Table 1).
3.2.lnheritance of the trait wsl
A plant was considered as awslindividual if it had the typical white stripes on its leaves.The plants at F1derived from the reciprocal crosses ofwslwith CS and Jimai 22 had the normal phenotypes,which indicated that the mutated gene was a nuclear recessive gene.The segregation ratios of individuals with white stripe leaf and normal phenotypes in the F2populations fitted the Mendelian ratio of 1:3 (χ2<χ20.05=3.84) (Table 2).The segregation ratios of homozygouswsllines,heterozygous lines and homozygous normal leaf plant lines in the F2:3family lines fitted the Mendelian ratio of 1:2:1 (χ2<χ20.05=5.99) (Table 3),and the segregation ratios ofwslto normal leaf plants in the F2:3heterozygous lines also fitted the Mendelian ratio of 1:3 (χ2<χ20.05=3.84) (Table 4).These data indicated that the white stripe leaf trait was controlled by a pair of nuclear recessive genes.Thus,the gene controlling the white stripe leaf trait ofwslmutant was named aswsl.

Fig.1 The individual plants of Guomai 301 and mutant wsl at different developmental stages.A,Guomai 301 (left) and wsl(right) plants at the seedling stage.B,Guomai 301 (left) and wsl(right) plants at the jointing stage.C and D,Guomai 301 and wsl plants at the booting stage,respectively.Scale bars=5 cm.
3.3.Linkage analysis of the mutated gene wsl
In the F2population ofwsl×CS,four (Xwmc104,Xgwm508,Xgpw7651,and Xgpw1079) of the 886 SSR primer pairs could amplify polymorphic products both between parental DNA templates and between the two DNA bulks,which indicated that SSR markersXwmc104,Xgwm508,Xgpw7651,andXgpw1079were linked towsl(Fig.3).Similarly,we found three linkage markersXgwm518,Xwmc494andXgwm508in the F2population ofwsl×Jimai 22(Fig.4).The primers of the SSR markers linked withwslare listed in Table 5.
3.4.All the wsl linked markers were on 6B

Fig.2 The leaves of Guomai 301 and mutant wsl at different developmental stages.A,leaves of Guomai 301 (left) and mutant wsl (right) at the seedling stage.B and C,top fully opened leaves and flag leaves of Guomai 301 (left) and mutant wsl (right) at the jointing and booting stages,respectively.
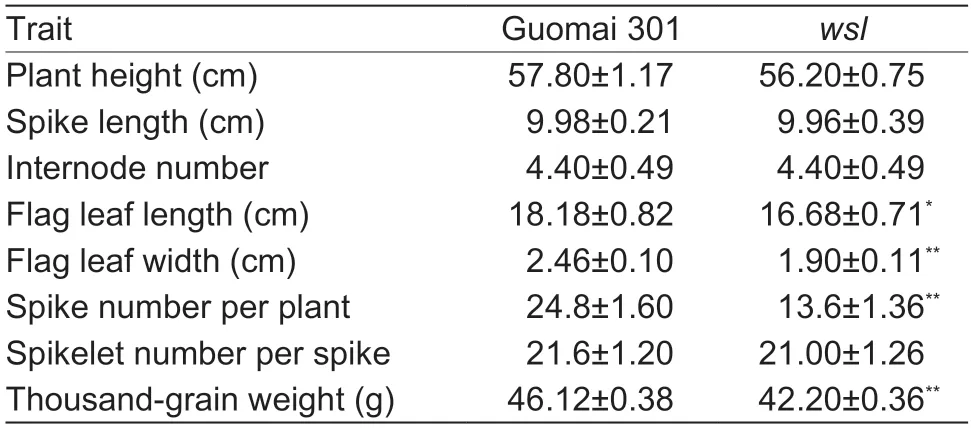
Table 1 Comparison of the main agronomic traits between Guomai 301 and wsl

Table 2 Genetic analysis of the white stripe leaf trait in the four segregating populations at F2
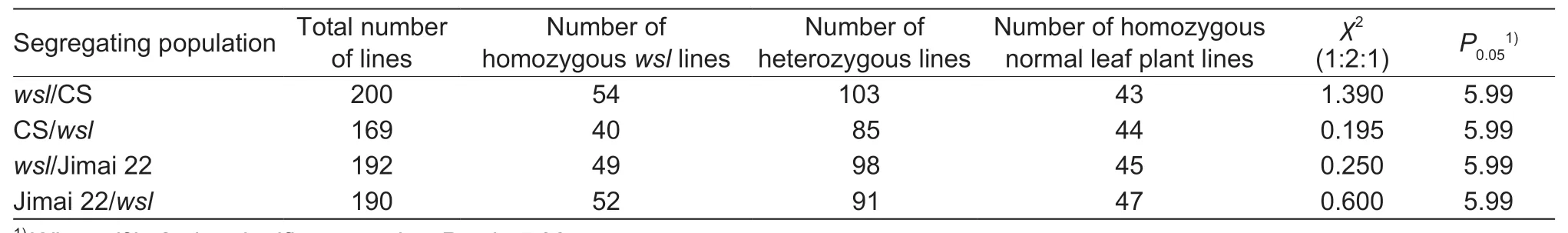
Table 3 Genetic analysis of the white stripe leaf trait in the four segregating populations at F2:3
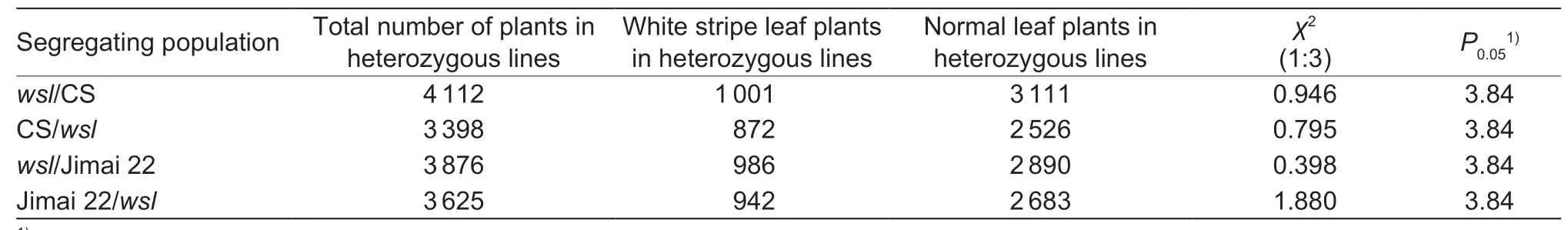
Table 4 The segregation ratios of heterozygous lines in F2:3
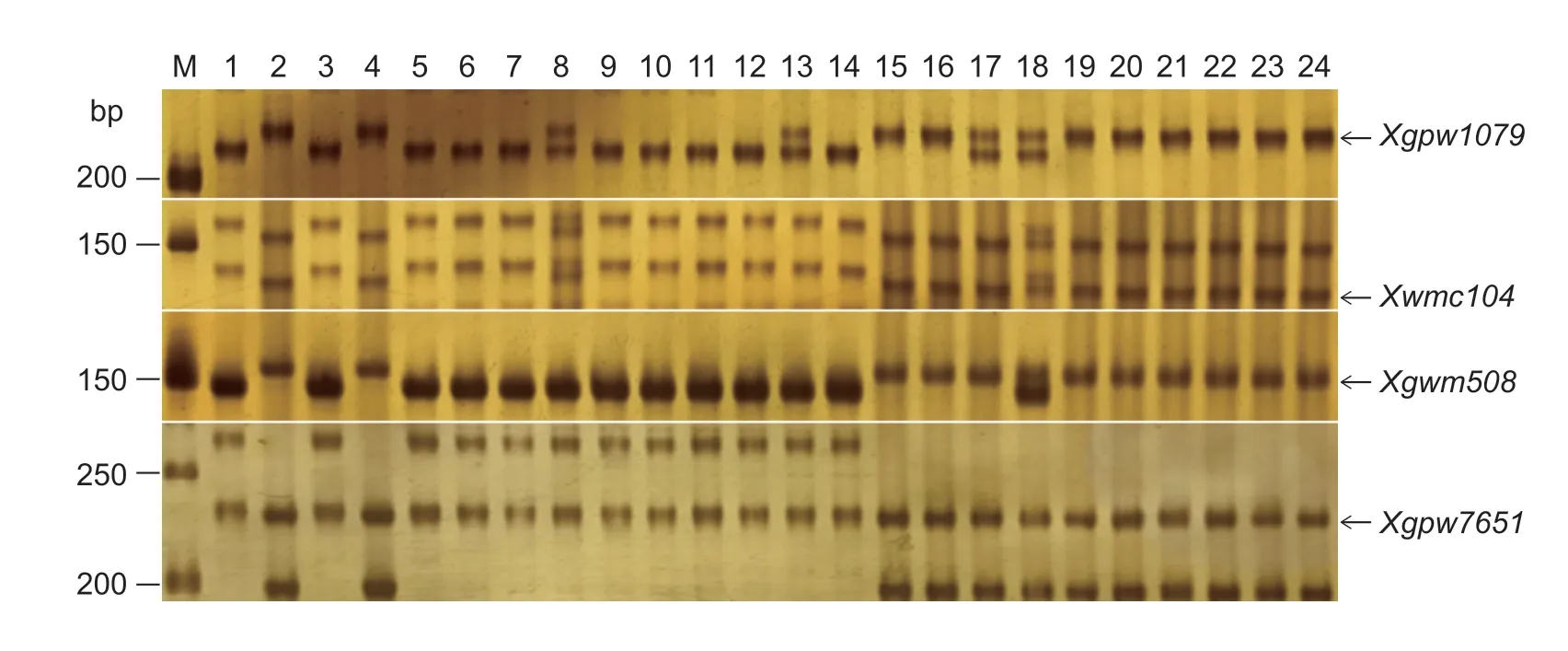
Fig.3 PCR products amplified with the polymorphic SSR primers in the F2 population of wsl×Chinese Spring (CS).M,DNA molecular weight marker;lane 1,wsl;lane 2,CS;lane 3,white stripe leaf plant DNA pool;lane 4,normal plant DNA pool;lanes 5–14,the homozygous white stripe leaf individuals in the F2 population;lanes 15–24,the homozygous normal individuals in the F2 population.Arrows indicate the polymorphic bands.
A set of CS nulli-tetrasomic lines was used to verify the chromosomal locations ofwsllinked SSR markersXgwm518andXgpw7651.The target bands ofXgwm518andXgpw7651were absent in CS 6B nullisomic/6D tetrasomic line,which demonstrated that these two markers andwslwere on chromosome 6B (Fig.5).Similarly,CS 6A nullisomic/6D tetrasomic,6B nullisomic/6D tetrasomic and 6D nullisomic/6A tetrasomic lines were used to verify the chromosomal locations of the SSR markersXgwm508,Xwmc104,Xwmc494,andXgpw1079.The target bands of these four SSR markers were also absent in CS 6B nullisomic/6D tetrasomic line,which demonstrated that these four markers andwslwere on chromosome 6B (Fig.6).These sixwsllinked markers in both populations were all located on 6BS.Therefore,the SSR assay located the mutated genewslon wheat chromosome 6BS.
3.5.Genetic mapping of the mutated gene wsl
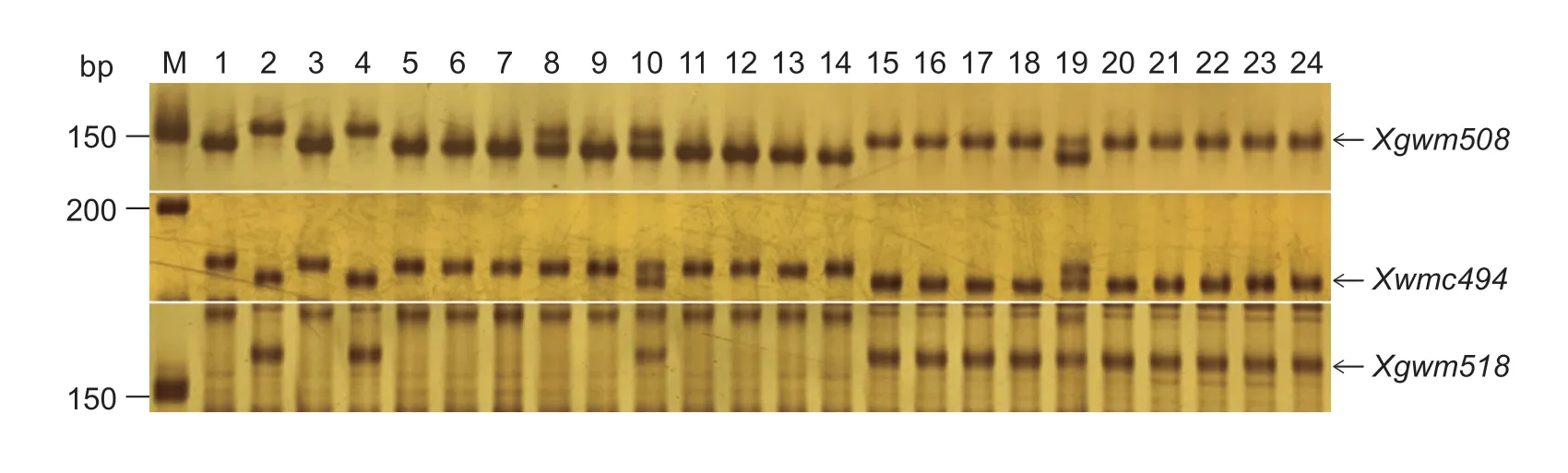
Fig.4 PCR products amplified with the polymorphic SSR primers in the F2 population of wsl×Jimai 22.M,DNA molecular weight marker;lane 1,wsl;lane 2,Jimai 22;lane 3,white stripe leaf plant DNA pool;lane 4,normal plant DNA pool;lanes 5–14,the homozygous white stripe leaf individuals in the F2 population;lanes 15–24,the homozygous normal individuals in the F2 population.Arrows indicate the polymorphic bands.
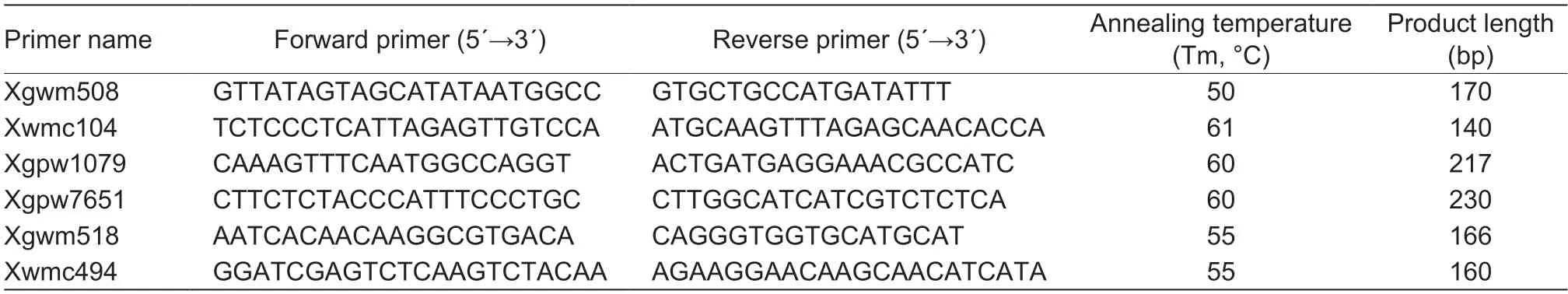
Table 5 DNA sequences of the SSR primers amplified polymorphic markers linked to wsl
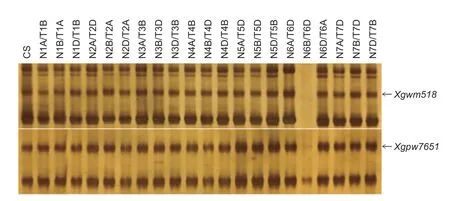
Fig.5 Chromosomal location of the PCR products amplified with the SSR primers Xgwm518 and Xgpw7651in a set of Chinese Spring (CS) nulli-tetrasomic lines.Arrows indicate the target bands.
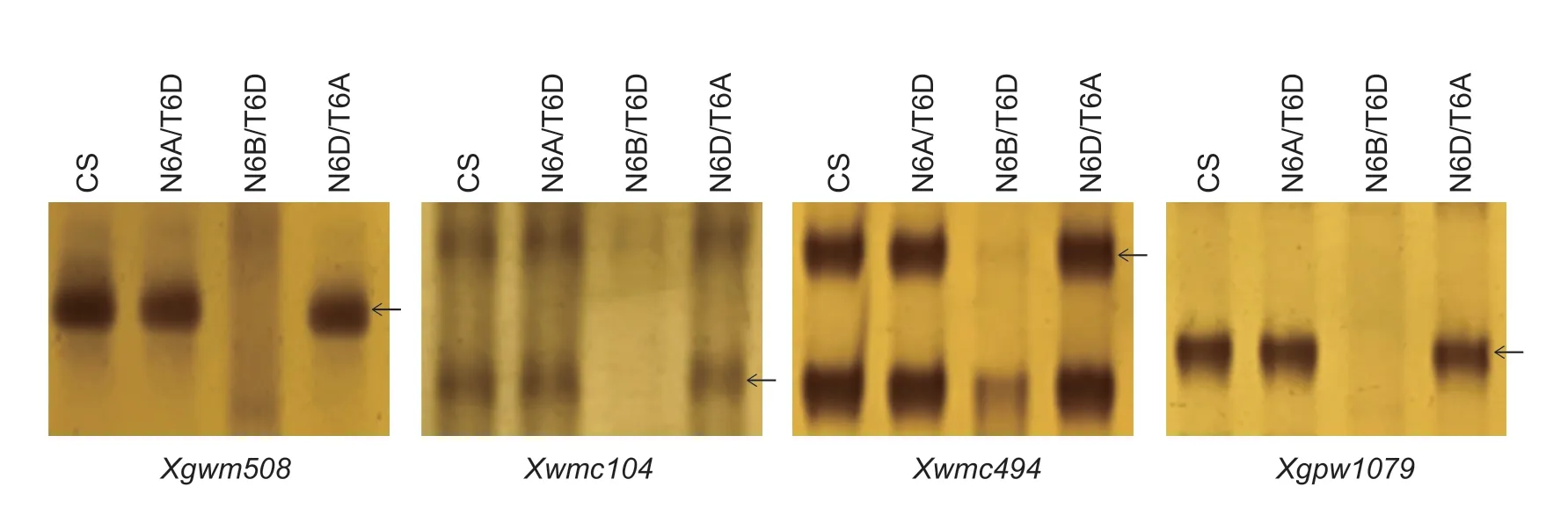
Fig.6 Chromosomal location of the PCR products amplified with the SSR primers Xgwm508,Xwmc104,Xwmc494,and Xgpw1079 in Chinese Spring (CS) N6A/T6D,N6B/T6D and N6D/T6A lines.Arrows indicate the target bands.
The amplification data of these markers in the F2populations were analyzed (Appendix A),and the linkage maps were drawn.In the F2population ofwsl×Jimai 22,the linkage order and genetic distances wereXgwm508–Xwmc494–Xgwm518-wslat 3.5,1.6 and 8.2 cM,respectively (Fig.7-A).In the F2population ofwsl×CS,the linkage order and genetic distances wereXgpw1079–Xwmc104–Xgwm508-wsl–Xgpw7651at 7.1,5.2,8.7,and 4.4 cM,respectively(Fig.7-B).The genewslwas located in the same region in the populations derived from crosseswsl×Jimai 22 andwsl×CS,and they were mutually verified (Fig.7-A and B).Thewslgene was located in the region between SSR markersXgwm518andXgpw7651.According to the reference genome sequence of CS (http://plants.ensembl.org),Xgwm518andXgpw7651are located at physical locations of 88 and 179 Mb on chromosome 6BS,respectively,thus thewslgene was physically mapped to a region from 88–179 Mb on chromosome 6BS (Fig.7-C).The gene collinearity map comparison showed that the mutated genewsland the mutated geneI30(Liet al.2017) were in the homoeologous regions on 6BS and 6DS,respectively(Fig.7-C and D).
4.Discussion
4.1.wsl is a novel leaf lesion mimic mutant
In this study,we obtained a wheat lesion mimic mutantwsl.The phenotype of mutantwslis almost the same as wheat lesion mimic mutant I30 (Liet al.2017),but it is significantly different from other lesion mimic mutants such as Ning7840 (lm;Li and Bai 2009),lm3 (Wanget al.2016),HLP (Kamlofskiet al.2007),and C591 (Nair and Tomar 2001).The wheat lesion mimic genes are on chromosome 3BS (lm1),4BL (lm2),3BL (lm3),1BL (lmin Ning7840),and 6DS (I30),respectively.In this study,genetic analysis and gene mapping showed that the mutated genewslwas a recessive gene on chromosome 6BS.Therefore,thewslis a novel lesion mimic gene in wheat.
A number of genes associated with lesion mimic phenotypes in rice have been cloned.Four lesion mimic mutants of rice were similar to wheat mutantwslin phenotype,however,the genes associated with lesion mimic phenotypes in rice,zn(Liet al.2010),z3(Kimet al.2018),z15(Fenget al.2019),andzl16(Liuet al.2018),were on the collinear regions of wheat chromosomes 4,7,1,and 2 respectively.Since none of them are in the 6BS collinear region where wheatwslis mapped,these data suggest thatwslis different from these rice genes.
4.2.Genes wsl and I30 are in the homoeologous regions of 6BS and 6DS
Thewslgene is on chromosome 6BS.Similarly,I30is on chromosome 6DS (Liet al.2017),the homoeologous chromosome arm of 6BS.The geneI30is between SSR markers6DS-5andXcfd190at genetic distances of 9.1 and 6.4 cM,respectively.In the F2populations ofwsl×CS andwsl×Jimai 22,the SSR markersXwmc494,Xgwm518,Xgpw1079,Xwmc104,Xgwm508,andXgpw7651were closely linked withwsl.Thewsllocus is betweenXgwm518andXgpw7651(Fig.7-C).The physical locations ofXgpw7651andXgwm518on chromosome 6BS,and the markers linked toI30on 6DS were identified by referring to database EnsemblPlants (http://plants.ensembl.org).The collinearity analysis showed thatI30andwslgenes were in the homoeologous region,which indicated that they might be homoeologous genes (Fig.7).

Fig.7 The linkage maps of wsl and the gene collinearity map compared with I30.A,the linkage map of wsl in the segregating population derived from crossof wsl×Jimai 22.B,the linkage map of wsl in the segregating population derived from crossof wsl×Chinese Spring (CS).C,gene wsl is in the region from 88 to 179 Mb on chromosome 6BS.D,the physical map of I30 (Li et al. 2017).In A and B,genetic distances are indicated on the left sides of the linkage groups,and the marker names are shown on the right sides.In C and D,physical locations are indicated on the left sides of the physical maps,and the marker names are shown on the right sides.
4.3.The resistance of wsl to pathogens,and how it impairs leaf development needs further study
The lesion mimic traits are similar to hypersensitive response(HR) of plants in the processes of protection against pathogens.Most of the lesion mimic mutants increase local or systemic resistance to the pathogens before or after the formation of the lesion mimic phenotype (Ryalset al.1996).Therefore,lesion mimic mutants are regarded as ideal materials for studying the disease resistance mechanisms and the pathway of programmed cell death (PCD) of plants(Wuet al.2008).In addition,the trait of white stripe leaf is a typical phenotype of abnormal chloroplast and leaf developments,which severely impacts many agronomic traits such as spike number per plant and thousand-grain weight (Table 1).Currently,it is unclear whetherwslis highly resistant to pathogens and how it impacts leaf and chloroplast development.Therefore,wslneeds to be studied further.
5.Conclusion
We obtained a wheat white stripe leaf mutantwslfrom EMS-mutagenized Guomai 301.The phenotype ofwslis similar to I30,but significantly different from other wheat lesion mimic mutants.Thewslis a novel recessive gene on chromosome 6BS.The geneswslandI30are in the homoeologous chromosome regions of 6BS and 6DS,which suggests that they might be homoeologous genes.The WT geneWSLmust be necessary for wheat leaf and chloroplast development.This discovery and genetic study of the mutantwslprovides a new germplasm for studies of wheat leaf development,especially for studies on the molecular mechanisms of lesion mimic mutants in wheat.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC,31571646) and the Science and Technology Project in Henan Province,China(182102110147).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendixassociated with this paper is available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Receptor-like kinase OsASLRK regulates methylglyoxal response and content in rice
- Construction of a high-density adzuki bean genetic map and evaluation of its utility based on a QTL analysis of seed size
- Effects of temperature and solar radiation on yield of good eatingquality rice in the lower reaches of the Huai River Basin,China
- Difference in corn kernel moisture content between pre-and postharvest
- The effect of elevating temperature on the growth and development of reproductive organs and yield of summer maize
- High plant density increases seed Bt endotoxin content in Bt transgenic cotton
