Biological and molecular characterization of tomato brown rugose fruit virus and development of quadruplex RT-PCR detection
2021-06-02YANZhiyongZHAOMeishengMAHuayuLlULingzhiYANGGuanglingGENGChaoTlANYanpingLlXiangdong
YAN Zhi-yong,ZHAO Mei-sheng,MA Hua-yu,LlU Ling-zhi,YANG Guang-ling,GENG Chao,TlAN Yanping,Ll Xiang-dong
Shandong Provincial Key Laboratory of Agricultural Microbiology,College of Plant Protection,Shandong Agricultural University,Tai’an 271018,P.R.China
Abstract Tomato brown rugose fruit virus (ToBRFV) is a novel tobamovirus firstly reported in 2015 and poses a severe threat to the tomato industry.So far,it has spread to 10 countries in America,Asia,and Europe.In 2019,ToBRFV was identified in Shandong Province (ToBRFV-SD),China.In this study,it was shown that ToBRFV-SD induced mild to severe mosaic and blistering on leaves,necrosis on sepals and pedicles,and deformation,yellow spots,and brown rugose necrotic lesions on fruits.ToBRFV-SD induced distinct symptoms on plants of tomato,Capsicum annumm,and Nicotiana benthamiana,and caused latent infection on plants of Solanum tuberosum, Solanum melongena,and N.tabacum cv.Zhongyan 102.All the 50 tomato cultivars tested were highly sensitive to ToBRFV-SD.The complete genomic sequence of ToBRFV-SD shared the highest nucleotide and amino acid identities with isolate IL from Israel.In the phylogenetic tree constructed with the complete genomic sequence,all the ToBRFV isolates were clustered together and formed a sister branch with tobacco mosaic virus (TMV).Furthermore,a quadruplex RT-PCR system was developed that could differentiate ToBRFV from other economically important viruses affecting tomatoes,such as TMV,tomato mosaic virus,and tomato spotted wilt virus.The findings of this study enhance our understanding of the biological and molecular characteristics of ToBRFV and provide an efficient and effective detection method for multiple infections,which is helpful in the management of ToBRFV.
Keywords:host range,identity,quadruplex RT-PCR detection,phylogenetic tree,symptom,tobamovirus,tomato brown rugose fruit virus
1.lntroduction
Tomato (Solanum lycopersicum) is one of the most popular and economically important vegetable crops worldwide.China is the biggest tomato producer in the world,with an annual production of about 62 million tons (FAO 2020).Viruses are one of the important pathogens severely reducing the yield and quality of tomato (Hanssenet al.2010).Tobamovirusis the largest genus in the familyVirgaviridae(Adamset al.2017).In this genus,four species including tobacco mosaic virus (TMV),tomato mosaic virus(ToMV),tomato mottle mosaic virus (ToMMV),and tomato brown rugose fruit virus (ToBRFV) have been reported to infect tomatoes.Among them,TMV and ToMV are the two most prevalent viruses in tomato production (Liuet al.2019).ToMMV identified in Mexico in 2013 is an emerging tobamovirus infecting tomatoes (Liet al.2013).ToBRFV is a novel tobamovirus first reported in Jordan in 2015 (Salemet al.2016).So far,ToBRFV has been reported in China(Yanet al.2019),Germany (Menzelet al.2019),Israel (Luriaet al.2017),Italy (Pannoet al.2019a),Mexico (Camacho-Beltránet al.2019),Palestine (Alkowniet al.2019),Turkey(Fidanet al.2019),the United Kingdom (Skeltonet al.2019)and the United States (Linget al.2019).
Israeli and Jordanian isolates of ToBRFV can infect plants ofCapsicum annumm,Datura metal,Datura stramonium,Nicotiana benthamiana,Nicotiana clevelandii,Nicotiana glutinosa,Nicotiana megalosiphon,Nicotiana tabacum,Petunia hybrida,Solanum nigrum,Chenopodium murale,Chenopodium amaranticolor,andChenopodium quinoa,but they cannot infect plants ofCucurbita pepo,N.tabacum cvSamsun NN,Solanum tuberosum,andSolanum melongena(Salemet al.2016;Luriaet al.2017).The symptoms of ToRBFV vary by tomato cultivars and environmental conditions,including mosaic,blistering,distortion and narrowing of leaves,and brown rugose and yellow spots on fruits.The fruit symptom is more severe when ToRBFV co-infects with a widespread and seriously harmful tomato virus-tomato spotted wilt virus (TSWV)(Luriaet al.2017;Liuet al.2019).In tomato,three genes,Tm1,Tm2,andTm22,confer resistance to TMV and ToMV(Pelham 1966;Hall 1980),though some isolates of TMV and ToMV can overcomeTm1-,Tm2-and Tm22-mediated resistance (Meshiet al.1989;Weberet al.1993;Strasser and Pfitzner 2007).However,tomato cultivars carryingTm1,Tm2,andTm22genes are susceptible to ToBRFV isolates,with disease incidence close to 100% in some fields (Salemet al.2016;Luriaet al.2017).So far,no tomato cultivar resistant to ToBRFV has been reported.The fruit symptoms,especially brown rugose,make the tomato fruits unmarketable.Therefore,ToBRFV has become one of the most destructive viruses infecting tomato and has aroused vigilance in many countries.Furthermore,like other tobamoviruses,ToBRFV can be transmittedviaseeds and mechanical contacts and by bumblebees,and is stable in soil,clothing,various agricultural tools and glasshouse structures (Broadbent and Fletcher 1963;Gülseret al.2008;Levitzkyet al.2019).Once introduced,ToBRFV is difficult to be eradicated.
Precise detection and early warning for viruses are essential measures to control plant viral diseases.Enzymelinked immunosorbent assay (ELISA) is a common method used to detect viral pathogens.However,with polyclonal antibodies,ELISA cannot distinguish ToBRFV from TMV and ToMV (Luriaet al.2017;Yanet al.2019).Multiplex RT-PCR method has been established and used to differentiate TMV,ToMV,and ToMMV (Kumaret al.2011;Suiet al.2016).However,no multiplex RT-PCR system is available to distinguish ToBRFV from other tomato-infecting tobamoviruses.Mixed infection of ToBRFV and TSWV frequently happens in tomato plants(Luriaet al.2017;unpublished data);therefore,it is necessary to develop a RT-PCR system to differentiate these two viruses.
The current study documented the symptoms and host range of ToBRFV isolate from Shandong Province(ToBRFV-SD),determined the resistance of tomato cultivars commercially available in China,conducted identity and phylogenetic analysis of ToBRFV-SD with other tomatoinfecting tobamoviruses,and finally developed a fast and convenient quadruplex RT-PCR system to distinguish ToBRFV from TMV,ToMV,and TSWV with one RT-PCR test.
2.Materials and methods
2.1.Virus samples
Tomato plants and fruits infected with ToBRFV-SD were collected from Shandong (Yanet al.2019).TMV,ToMV,and TSWV were maintained in tomato plants in our laboratory.
2.2.Host range determination
Seedlings of tested plants were grown at 25°C in a growth chamber.Leaf tissues infected by ToBRFV-SD were homogenized in 0.01 PBS at pH 7.2.At the 3-to 4-leaf stage,five seedlings for each plant species were mechanically inoculated with the crude extracts.At 14 days postinoculation,the presence of ToBRFV in the newly developed leaves was determined by RT-PCR detection.
2.3.RT-PCR and sequencing
Total plant RNA was extracted from the systemic leaves of the tomato plants infected with ToBRFV-SD using TransZol reagent (TransGen Biotech,Beijing,China).Reverse transcription was performed using the HiScript®II Q RT SuperMix Kit (Vazyme,Nanjing,China) as instructed by the manufacturer.The full-length genomic fragment of ToBRFV was amplified by six RT-PCR and cloned into vector pCB301.Three clones for each fragment were sequenced using Sanger sequencing (Sangon Biotech,Shanghai,China).The sequences obtained were assembled using the SeqMan program in DNAStar Lasergene 7.1 (Madison,WI,USA).The names,sequences,and locations of primers used for the amplification of ToBRFV genome were listed in Table 1.
2.4.Sequence identity and phylogenetic tree analyses
Sequences were aligned by using ClustalW multiplex alignment in BioEdit version 7.2.6 (Hall 1999).The identities of the nucleotide sequences and the deduced amino acid sequences were obtained by the sequence identity matrix in BioEdit version 7.2.6 (Hall 1999).Phylogenetic trees were constructed using the Maximum-Likelihood,Neighbor-Joining,and Minimum-Evolution methods that are packaged in MEGA version 10.1.5 (Kumaret al.2018).Nodes with bootstrap values less than 50% were deleted,and those with bootstrap values higher than 70% were kept for 1 000 replicates.
2.5.Quadruplex RT-PCR detection system
Total RNA was extracted and then treated with gDNA wiper enzyme (Vazyme,Nanjing,China).Reverse transcription was conducted by using the HiScript II Q RT SuperMix Kit(Vazyme,Nanjing,China) with random primers.Specific primers were designed according to the conserved sequences of ToBRFV,TMV,ToMV,and TSWV,respectively.The nucleotide sequences and locations of primers in the genome were shown in Table 2.The ubiquitin gene (UBI)was used as a reference.The sizes of expected amplicons for ToMV,ToBRFV,TSWV,TMV,and UBI were 362,604,841,999,and 224 bp,respectively.A 20-μL PCR solution contained 10 μL of 2×TaqMaster Mix (Vazyme,Nanjing,China),0.5 μL of 10 μmol L–1for each forward primer,0.5 μL of 10 μmol L–1for each reverse primer,and 1 μL of cDNA transcribed previously.The mixed system was subjected to amplification in a PCR thermocycler (Thermo Fisher Scientific,Singapore).The PCR condition was 1 cycle of 94°C for 5 min,30 cycles of 94°C for 30 s,53°C for 30 s,and 72°C for 1 min,and 72°C for 7 min.The PCR products were analyzed by electrophoresis on a 1.5% agarose gel stained with ethidium bromide and observed under UV transilluminator (Bio-Rad,Hercules,CA,USA).

Table 1 The names,sequences,and locations of primers used for the amplification of the tomato brown rugose fruit virus (ToBRFV)genome
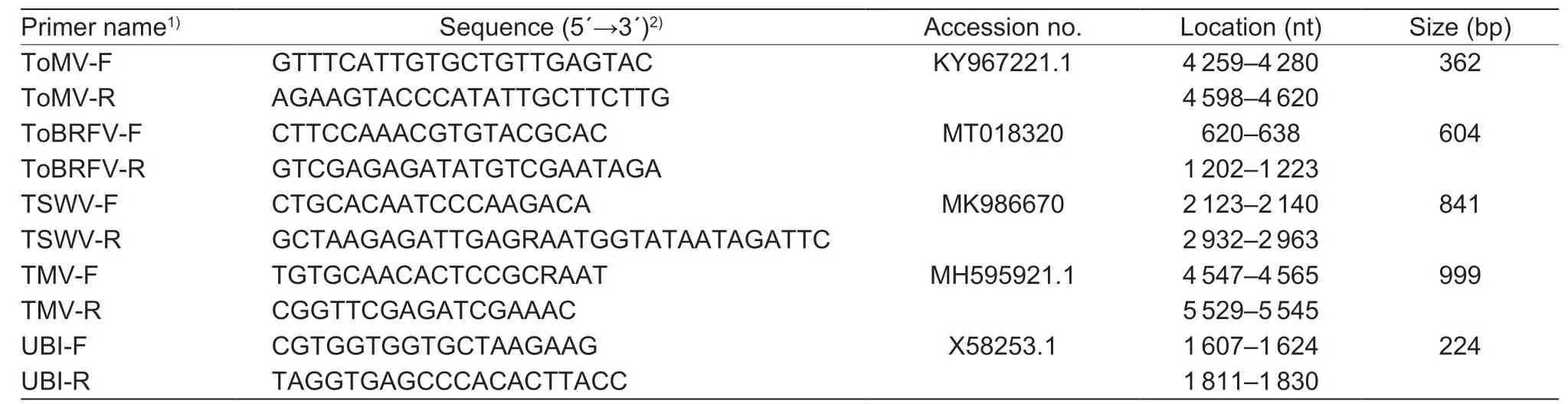
Table 2 The names,sequences,locations of primers used for quadraplex RT-PCR and size of amplified fragments
For sensitivity evaluation,each plasmid containing the amplicon was ligated into vector pCB301.The resultantclones with an initial amount of 0.2 ng were subjected to 10-fold serial dilutions.
3.Results
3.1.Symptoms on ToBRFV-SD infected tomato plants
The tomato plant symptoms induced by ToRBFV were complex in the field.Mild to severe mosaic (Fig.1-A),and sometimes leaf narrowing and rolling were found on tomato leaves.No distinct symptom was observed on the stem,but brown necrosis was observed on the pedicles (Fig.1-B) and the sepals (Fig.1-C).The sepals might become necrotic ultimately.Affected tomato fruits displayed deformities such as sunken ring-shapes in some cultivars (Fig.1-D).A typical symptom was brown rugose necrotic lesions on fruits (Fig.1-E–G).Yellow spots were also found on fruits of some tomato cultivars (Fig.1-H).The fruit symptoms reduced the commercial value of tomatoes.
3.2.Host range of ToBRFV-SD
Extracts of newly developed leaves from tomato plants infected with ToBRFV-SD were inoculated to plants of the familiesBrassicaceae,Cucurbitaceae,andSolanaceae.ToBRFV-SD could infect plants ofC.annumm,N.benthamiana,N.tabacum,S.melongena,andS.tuberosumcv.Kexin 1(Table 3) in the familySolanaceae.At 14 days post-inoculation (dpi),C.annummplantsshowed symptoms of dwarfism and necrosis in inoculated leaves and stems,N.benthamianaplants exhibited leaf yellowing and plant collapse.All the affected plants mentioned above showed positive reactions in RT-PCR detection,whereas plants ofN.tabacumcv.Zhongyan 102,S.melongena,andS.tuberosumcv.Kexin 1 showed no symptom in systemic leaves but were positive in RT-PCR detection.The inoculated leaves ofN.tabacumcv.Samsun NN showed massive hypersensitive lesion (HR) but no symptom on systemic leaves,which were negative in the RT-PCR detection.Plants ofCitrullus lanatus,C.pepo,Cucumis melo,Cucumis sativusin the familyCucurbitaceae,andArabidopsis thalianain the familyBrassicaceaewere asymptomatic and negative in RT-PCR.
The 3-or 4-leaf staged seedlings of 50 tomato cultivars collected from the market were mechanically inoculated with extracts of ToBRFV-SD in a growth chamber.At 14 dpi,systemic leaves of inoculated tomato plants showed symptoms of yellowing,curling,rolling,narrowing,blistering,or mosaic (Fig.2-A).ToBRFV-SD could be detected in all seedlings of the 50 cultivars by RT-PCR(Fig.2-B).
3.3.Genome organization of ToBRFV-SD
The complete genomic sequence of ToBRFV-SD contained 6 392 nt (accession no.MT018320) and had four open reading frames (ORFs) (Fig.3).The 5´-and 3´-untranslated regions (UTRs) consisted of 74 and 201 nt,respectively.ORF1 had 3 351 nt (nt 75–3 425) and encoded a 126-kDa protein.ORF2 had 4 848 nt (nt 75–4 922) and encoded a 183-kDa proteinviareadthrough mechanism.Both the 126 kDa protein and the 183 kDa protein were replicase.ORF3 had 801 nt (nt 4 909–5 709) and encoded the 30 kDa movement protein (MP).ORF4 had 480 nt (nt 5 712–6 191)and encoded the 17.5 kDa coat protein (CP).

Fig.1 Symptoms on tomato brown rugose fruit virus (ToBRFV)-infected tomato plants in the field.A,mosaic and blistering on leaves.B and C,necrosis on pedicles and sepals.D,deformation on fruits.E–G,brown rugose necrotic lesions on fruits.H,yellow spots on fruits.
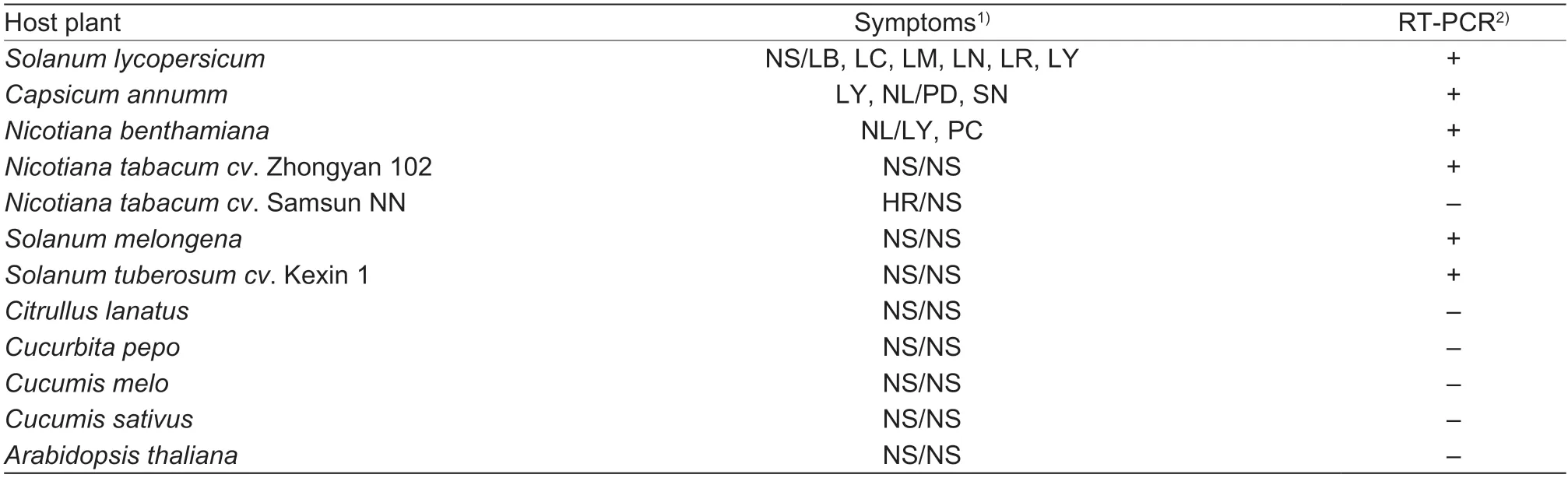
Table 3 Host range and symptoms of tomato brown rugose fruit virus (ToBRFV)-SD
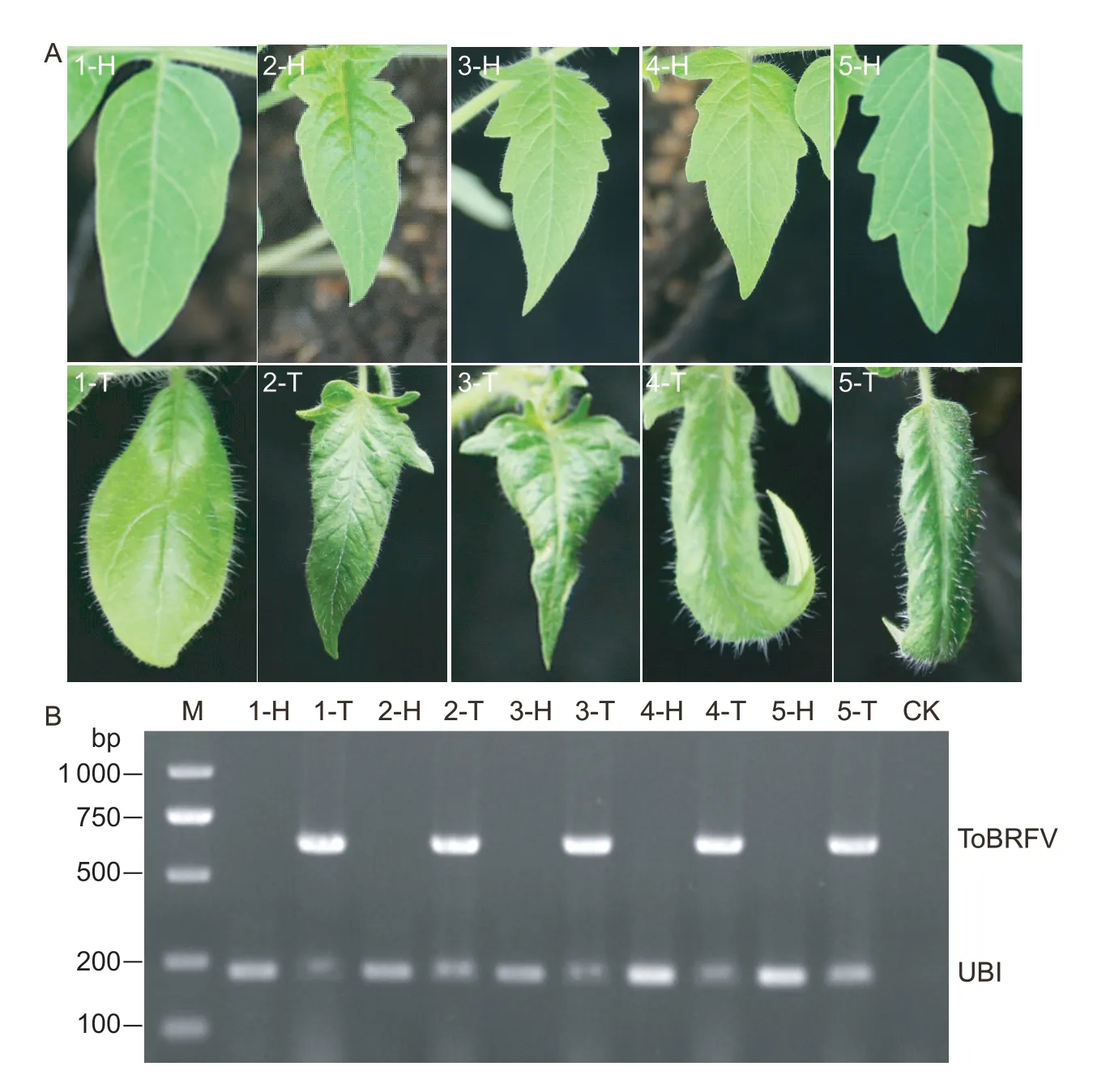
Fig.2 Symptoms on five tomato cultivars infected with tomato brown rugose fruit virus (ToBRFV) in the growth chamber.A,symptoms on five tomato cultivars infected by ToBRFV at 14 days post-inoculation.B,RT-PCR detection of five tomato cultivars infected with ToBRFV.The ubiquitin gene (UBI) was used as an internal reference.M,marker;H,healthy;T,ToBRFV;CK,blank control.
3.4.Sequence and phylogenetic tree analysis
The complete genome of ToBRFV-SD shared nucleotide identities of 80.5–81.9% with that of TMV,ToMV,and ToMMV,but its nucleotide identities with that of other ToBRFV isolates reached 99.6–99.8% (Table 4).For ORFs of ToBRFV isolates,ORF1 and ORF2 were more conserved than other ORFs,with nucleotide identities of 99.6–99.9% and 99.6–99.8%,respectively,and amino acid identities of both 99.6–99.9%;ORF3 and ORF4 were more diversified in that their intra-species nucleotide identities were 99.2–99.7% and 99.3–99.7%,respectively,and amino acid identities (of MP and CP) were 99.2–100% and 99.3–100%,respectively (Table 4).Among the ToBRFV isolates,SD shared the highest nucleotide and amino acid identities with IL from Israel (Table 4).
Phylogenetic trees were constructed with the complete genomic sequences of four tobamoviruses infecting tomato,using Maximum-Likelihood,Neighbor-Joining,and Minimum-Evolution methods.All the methods gave identical results.In the representative tree constructed with Maximum-Likelihood,all the isolates were clustered into four groups corresponding to the four tobamoviruses,in which ToBRFV and TMV formed sister branches,while ToMV and ToMMV formed another common branch (Fig.4).It is necessary to mention that the ToBRFV isolates exhibited a‘star phylogeny’ pattern and shared high identities,indicating that all these isolates had emerged recently (Fig.4).
3.5.Quadruplex RT-PCR for ToBRFV,TMV,ToMV,and TSWV
To establish a fast and convenient method to identify ToBRFV from other tomato-infecting tobamoviruses and TSWV,we firstly screened the optimized primers for each virus (Table 2).In simplex RT-PCR,expected amplicons of 362,604,841,and 999 bp were obtained from tomato plants infected with ToMV,ToBRFV,TSWV,and TMV,respectively(Fig.5-A;lanes 1–4).In duplex RT-PCR,any combination of co-infection with two viruses could be detected successfully from tomato plants (Fig.5-A;lanes 5–10).In triplex and quadruplex RT-PCR,three or four distinctive products were obtained from tomato plants simultaneously infected with three or four viruses,respectively (Fig.5-A;lanes 11–15).
Results of the sensitivity test showed that the lower limits of detection of quadruplex RT-PCR for TMV,ToMV,ToBRFV,and TSWV were all 0.02 pg,corresponding to 1.8×103copies (Fig.5-B).
4.Discussion
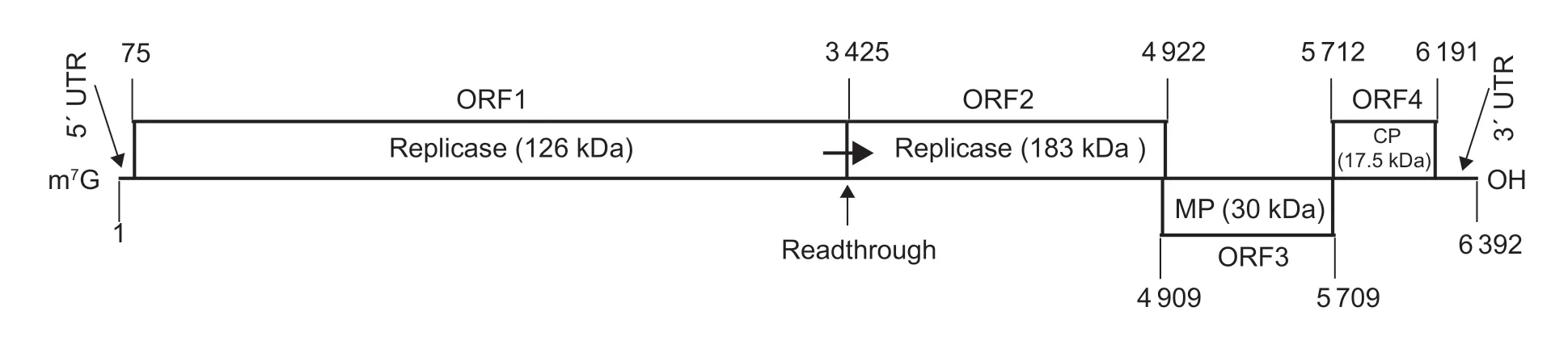
Fig.3 Genomic organization of tomato brown rugose fruit virus (ToBRFV)-SD.The numbers indicated the position of nucleotide.The protein names (size in kDa) encoded by each ORF were listed.The 183-kDa replicase was encoded by ORF2 via a readthrough mechanism.m7G,7-methyl guanine nucleotide;MP,movement protein;CP,coat protein.
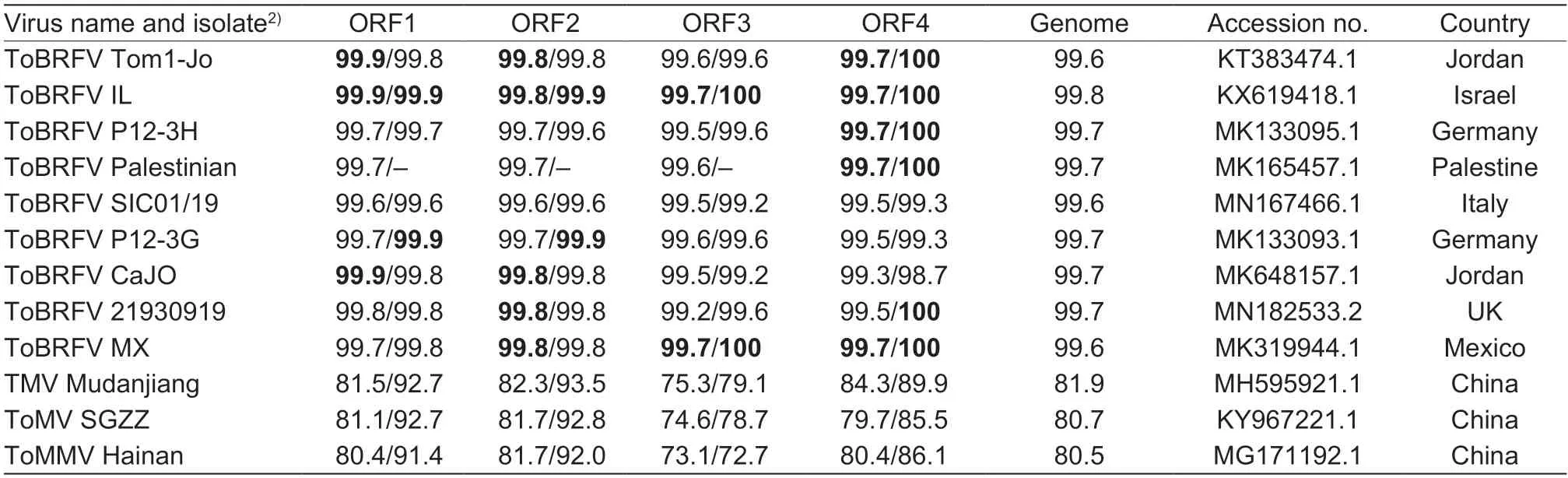
Table 4 Nucleotide and amino acid sequence identities of ToBRFV-SD with isolates of ToBRFV,TMV,ToMV,and ToMMV1)
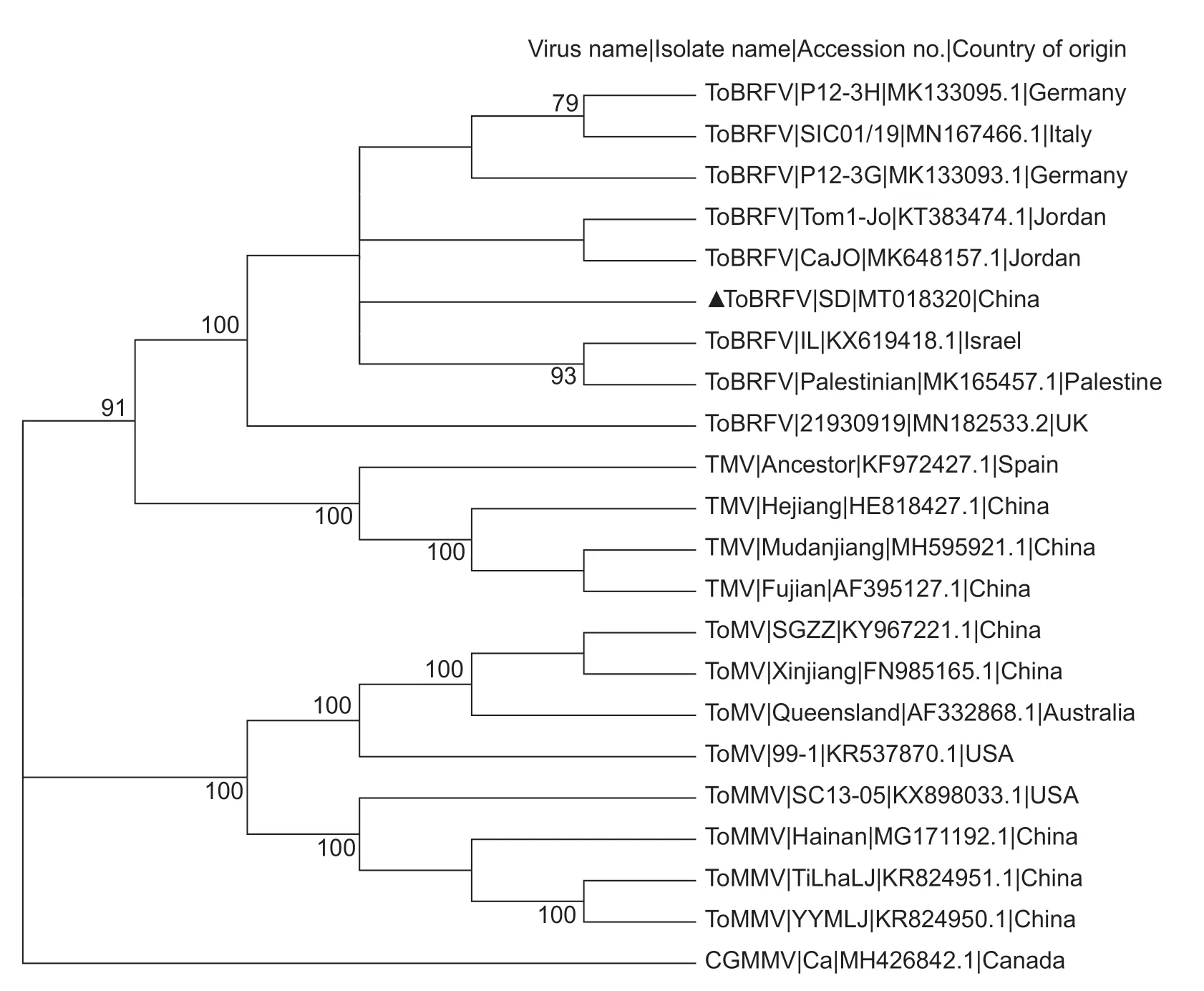
Fig.4 The phylogenetic tree of tobacco mosaic virus (TMV),tomato mosaic virus (ToMV),tomato brown rugose fruit virus (ToBRFV),and tomato mottle mosaic virus (ToMMV) constructed with complete genomic nucleotide sequences using the Maximum-Likelihood method.An isolate of cucumber green mottle mosaic virus (CGMMV) was used as an outgroup.Nodes with bootstrap values less than 50% were cut off.Nodes with bootstrap values higher than 70% were shown.
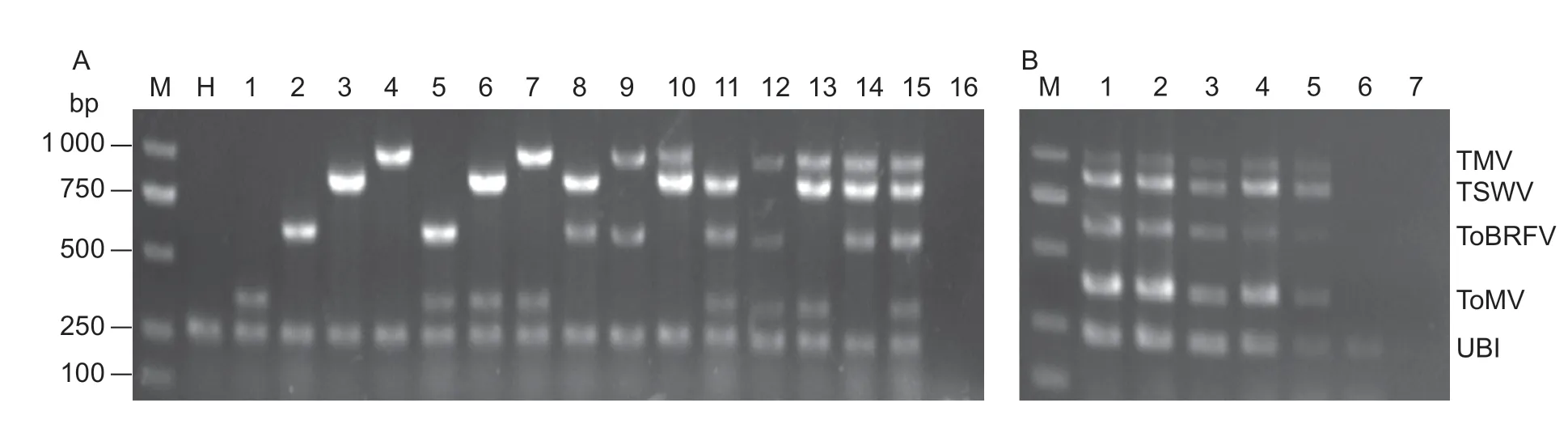
Fig.5 Development of quadruplex RT-PCR detection system for tomato brown rugose fruit virus (ToBRFV),tobacco mosaic virus(TMV),tomato mosaic virus (ToMV) and tomato spotted wilt virus (TSWV).A,specificity of quadruplex RT-PCR.The ubiquitin gene (UBI) was used as an internal reference.H,healthy plants as negative control;1,ToMV;2,ToBRFV;3,TSWV;4,TMV;5,ToMV and ToBRFV;6,ToMV and TSWV;7,ToMV and TMV;8,ToBRFV and TSWV;9,ToBRFV and TMV;10,TSWV and TMV;11,ToMV,ToBRFV,and TSWV;12,ToMV,ToBRFV,and TMV;13,ToMV,TSWV,and TMV;14,ToBRFV,TSWV,and TMV;15,ToMV,ToBRFV,TSWV,and TMV;16,blank control.B,sensitivity of quadruplex RT-PCR.1,200 pg;2,20 pg;3,2 pg;4,0.2 pg;5,0.02 pg;6,0.002 pg;7,0.0002 pg.
We reported here the disease symptoms caused by ToBRFV in Shandong,China.Symptoms are critical to the diagnosis of plant viral diseases.Symptoms of ToBRFV in tomato vary among environmental conditions and tomato cultivars (Luriaet al.2017).In Jordan,ToBRFV causes brown rugose symptom on tomato fruits,and hence the virus was named tomato brown rugose fruit virus (Salemet al.2016).Besides brown rugosity,ToBRFV induced yellow spots on tomato fruits in Israel;interestingly,on cultivar Odelia,ToBRFV produced yellow spots and brown rugose symptoms in two different places (Luriaet al.2017).ToBRFV caused yellow spots on tomato fruits in Germany and Palestine (Alkowniet al.2019;Menzelet al.2019).Similar symptoms,such as deformation,yellow spots,and brown rugose necrotic lesions on fruits,were observed in the current study on tomato plants infected with ToBRFV-SD(Fig.1).To elucidate if the symptoms were caused by a mixed infection of several viruses,we conducted RT-PCR detection and found that the samples we collected from the fields were infected with ToBRFV only.Plants ofN.tabacum cv.Zhongyan102,S.melongena,S.tuberosum cv.Kexin 1 were systemically infected but were asymptomatic (Table 3),indicating that they were asymptomatic hosts for ToBRFVSD.Latent infections were also found in plants ofC.murale,C.amatanticolor,andPentunia hybrida(Luriaet al.2017),all of which are common in the field.Therefore,special attention should be paid to precise diagnosis and detection of ToBRFV to enable early warning.
Planting resistant cultivar is the most effective and efficient method to control viral diseases.ToBRFV infects tomato plants withTm2andTm22resistance genes (Luriaet al.2017).In this study,the 50 tomato cultivars tested were all susceptible to ToBRFV.It was reported that ToBRFV could not infect plants ofS.tuberosum cv.Nicola orS.melongenacultivars Classic and 206 (Luriaet al.2017)However,our results showed that it could infect the tested plants ofS.melongenaandS.tuberosum cv.Kexin 1.Considering the rapid spread of ToBRFV,breeding resistant cultivars is of top priority to the tomato industry worldwide.From another point of view,efficient gene editing using the CRISPR-Cas9 system has been applied successfully(Brookset al.2014).This method may be applied to breed tomato plants that are resistant to ToBRFV when the tomato susceptibility factors are elucidated.
All the ToBRFV isolates formed a star phylogeny (Fig.4),consistent with the fact that ToBRFV was a newly emerging virus.Results of identity analyses showed that ToBRFVSD shared the highest nucleotide and amino acid identities with an isolate from Israel (Table 4).Considering that the importation of Israeli tomato seeds to China is frequent,we suspected that ToBRFV-SD was probably introduced from Israel.
Quick and precise detection is one prerequisite for the early warning and control of plant viral disease.ELISA has a significant advantage in that it can detect a large number of samples at one time.However,all the tobamoviruses have cross-reactions in serological assays (Jacobiet al.1998).Therefore ToBRFV cannot be differentiated from TMV and ToMV with a polyclonal antibody (Luriaet al.2017;Yanet al.2019).A one-step end-point real-time RTPCR assay,using primers targeting the ORF3,has been developed for the detection of ToBRFV (Pannoet al.2019b).That method is rapid and sensitive,but cannot detect other viruses from the samples.Multiplex RT-PCR has the advantage of detecting several viruses in one sample.Kumaret al.(2011) developed a duplex RT-PCR that can detect TMV and ToMV in one reaction.Suiet al.(2016)developed a triplex RT-PCR that can detect TMV,ToMV,and ToMMV.In the current study,a quadruplex RT-PCR method was successfully developed that could distinguish ToBRFV from tobamoviruses,TMV and ToMV,as well as TSWV in a reaction,in which the primers for ToBRFV target nt 620–1 223 in ORF1.It is difficult for non-plant virologists to differentiate the symptoms of ToBRFV and TSWV in tomato fruits.The quadruplex RT-PCR system provides a rapid,sensitive,and cost-effective tool to detect ToBRFV from other tomato-infecting viruses.
5.Conclusion
In summary,ToBRFV-SD induced mild to severe mosaic and blistering on leaves,necrosis on sepals and pedicles,and deformation,yellow spots,and brown rugose necrotic lesions on tomato fruits.It also infected plants ofC.annumm,N.benthamiana,N.tabacumcv.Zhongyan 102,S.tuberosum,andS.melongena.None of the 50 tomato cultivars tested was resistant to ToBRFV-SD.The complete genomic sequence of ToBRFV-SD shared the highest nucleotide and amino acid identities with that of Israeli isolate IL.In the phylogenetic tree constructed with the complete genomic sequences,all the ToBRFV isolates were clustered together and formed a sister branch with that of TMV.The quadruplex RT-PCR system developed here could differentiate ToBRFV from TMV,ToMV,and TSWV in one reaction.Our results will be helpful in the precise detection,early warning,and sustainable control of ToBRFV.
Acknowledgements
This study was supported by the grants from the National Natural Science Foundation of China (31720103912 and 31801704) and the ‘Taishan Scholar’ Construction Project,China (TS201712023).
Declaration of competing interest
The authors declare that they have no conflict of interest.
杂志排行
Journal of Integrative Agriculture的其它文章
- Receptor-like kinase OsASLRK regulates methylglyoxal response and content in rice
- Heredity and gene mapping of a novel white stripe leaf mutant in wheat
- Construction of a high-density adzuki bean genetic map and evaluation of its utility based on a QTL analysis of seed size
- Effects of temperature and solar radiation on yield of good eatingquality rice in the lower reaches of the Huai River Basin,China
- Difference in corn kernel moisture content between pre-and postharvest
- The effect of elevating temperature on the growth and development of reproductive organs and yield of summer maize
