Development and application of microsatellite markers withintranscription factors in flare tree peony (Paeonia rockii) based on next-generation and single-molecule long-read RNA-seq
2021-06-02LlUNaCHENGFangyunGUOXinZHONGYuan
LlU Na,CHENG Fang-yun,GUO Xin,ZHONG Yuan
Beijing Advanced Innovation Center for Tree Breeding by Molecular Design/Beijing Key Laboratory of Ornamental Plants Germplasm Innovation &Molecular Breeding/National Engineering Research Center for Floriculture/Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants of Ministry of Education/Peony International Institute,School of Landscape Architecture,Beijing Forestry University,Beijing 100083,P.R.China
Abstract Tree peonies native to China are a precious crop with ornamental,medicinal and edible oil properties,of which flare tree peony (Paeonia rockii)is one of the most significant germplasms in Paeonia.The development and application of expressed sequence tag-simple sequence repeat (EST-SSR) markers are very valuable for genetic and breeding applications,but EST-SSR resources for the genus Paeonia are still limited.In this study,we first reported the development of SSRs within transcription factors (TFs) in P. rockii based on next-generation sequencing (NGS) and single-molecule long-read sequencing (SMLRS).A total of 166 EST-SSRs containing six nucleotide repeat types were identified from 959 candidate TFs associated with yield,with an average of one SSR per 5.83 unigenes.In total,102 (61.45%) pairs of primers produced amplification products in the two RNA-seq cultivars.Among them,58 (56.86%) pairs of primers from 18 gene families (AP2,bHLH,HSF,etc.) were identified to be polymorphic both in the parents of a linkage mapping population and in eight randomly selected accessions of P.rockii.Further,the 58 EST-SSRs indicated a high level of informativeness with PIC values ranging from 0.32 to 0.91 (mean 0.70) after assessment in 37 tree peony accessions.Transferability studies indicated that the amplification ratio of the 58 pairs of primers ranged from 89.66 to 100% across seven species of Paeonia.In addition,a genetic relationship study was performed in 62 accessions.Cluster analysis using the neighbour-joining (NJ) tree demonstrated that major clusters corresponded to the known pedigree trees.Taken together,these newly developed EST-SSRs have a potential use in the conservation of tree peony germplasm and marker-assisted selection (MAS) breeding.
Keywords:flare tree peony (Paeonia rockii),EST-SSR markers,polymorphism,genetic diversity,transferability,genetic relationships
1.Introduction
Edible oil is an important part of the human diet,which provides abundant amounts of fat and energy for human beings.At present,woody oil crops have become an important source of edible oils for their high nutritional values,stable yields and low cost.Tree peonies,which belong to the genusPaeoniasectionMoutanDC.,Paeoniaceae,are a well-known ornamental and medicinal crop in China and have been widely cultivated all over the world (Cheng 2007).Recently,tree peonies have been considered as a valuable emerging grain and oil tree species,and their seeds have been identified as a novel resource of high-quality edible oil with rich unsaturated fatty acid and α-linolenic acid contents (Liet al.2010,2015).As one of the most representative species of tree peonies,P.rockiihas accumulated a vast amount of genetic variation and shown great potential for mass cultivation as an emerging oil crop in northern China.However,increasing seed yield through efficient breeding has become one of the most important issues.
Some progress has been made in genetic improvement of tree peony through conventional breeding practices,such as hybrid breeding and the phenotypic selection of novel or improved offspring.However,in common with other woody species,the long juvenile phase and complex genetic background make it very difficult to improve breeding efficiency by using traditional breeding processes.In contrast,marker-assisted selection (MAS)is a promising approach in plant breeding,which allows accurate and efficient introgression of the important agronomic trait(s) from a germplasm source to elite lines.It is beneficial for quickly selecting cultivars with desirable traits and shortening the breeding cycle (Lande and Thompson 1990;Burowet al.2019).Hence,the development of suitable molecular markers has become an important foundation for MAS breeding.
Compared with other molecular markers,SSR markers are considered to be the ideal markers for the detection of tree species because of their prominent genetic features such as high polymorphism,co-dominant nature and extensive genome coverage (Varshneyet al.2005;Agarwalet al.2008;Blairet al.2010).SSR markers can be divided into two types according to their sources:EST-SSR and genomic SSR (gSSR).gSSRs are located in both coding and non-coding regions of the genome,and the development process is very complex(Eujaylet al.2004).EST-SSRs are developed based on transcriptional sequences,which is low in cost,highly efficiency and convenient for identifying gene functions(Zenget al.2010).Moreover,as expression TFs,SSR markers derived from genes can provide a much greater degree of resolution,and show both higher versatility within species and the potential for direct gene tagging when compared to those derived from random genomic locations,as they are associated with coding sequences and thus may affect gene expression or function (Varshneyet al.2005;Blairet al.2010;Duet al.2012a;Singhet al.2013;Gonzagaet al.2015;Parthibanet al.2018).At present,SSRs have been widely used in various crops(Boudchichaet al.2018;Boureimaet al.2018;Wang Y Let al.2018).EST-SSRs have also been widely used for genetic diversity analysis,variety identification and map construction of various crops,such as CASeSSR004(Casuarina equisetifolia) and xSEM358b01(Saccharum officinarum) (Xianget al.2018;Xuet al.2018;Torokeldievet al.2019;Ukoskitet al.2019).In tree peonies,the development and application of SSRs are also growing rapidly (Gaiet al.2012;Zhanget al.2012;Wang D Xet al.2013;Wuet al.2014,2017;Caiet al.2015;Penget al.2017).However,only a limited number of SSR loci associated with yield have been developed thus far,which limits their extensive application of functional markers inPaeonia.Therefore,the effective development of ESTSSRs remains an important aspect of MAS breeding,especially for the breeding that can increase the seed yield of tree peonies.
TFs can modulate gene expression,and manipulation of the levels of selected TFs may result in the increased expression of economically useful proteins or the improvement of other agriculturally relevant characteristics such as an increase in yield (Reuberet al.2010).For example,plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres (Nelsonet al.2007).Overexpression of the TFAP37in rice improves grain yield under drought conditions (Ohet al.2009).A wheat CCAAT Box-binding TF increases the grain yield of wheat with less fertilizer input (Quet al.2015).TF molecular markers were originally designed based on WRKY TFs of chili peppers and tomatoes,and subsequently applied to chili peppers.The results showed that this WRKY-based marker system was a fast and simple method which can generate sequence-specific markers for plant gene families (Kimet al.2008).The increasing availability of TF data has also made TFs a valuable resource for genic functional microsatellite marker development.InMedicago truncatula,TF gene-derived microsatellite markers were developed and their cross-species transferability was successfully assessed (Liu W Xet al.2015).One hundred and four SSRs specific to 53MtbZIPgenes were detected and 97 primer pairs were developed.The validation of 30 randomly selected primer pairs in 20 alfalfa cultivars showed that 17 of them were true-to-type SSR loci and highly transferable among alfalfa cultivars(Zhang Z Set al.2015).InPopulus simonii,a total of 31 polymorphic SSR loci were identified in 21 genes of three TF families from 36 unrelated individuals ofP.simoniiby directly sequencing the TF genes expressed under abiotic stresses.The developed gene-based SSR markers of the abiotic stress resistant TF could provide a powerful tool for marker-assisted breeding of new germplasms with desirable abiotic stress resistance inP.simonii(Wanget al.2011).InLiliumspecies,71 SSRs from 31 TF families were idendified,and 14 highly polymorphic SSRs were selected and successfully used to study genetic diversity and population structure in lily accessions(Biswaset al.2018).InP.rockii,this study is the first attempt to identify the SSR markers within the candidate TFs.
Transcriptome sequencing technology is an important way to efficiently obtain functional allelic loci in crops,especially for non-model plants such asP.rockiiwithout reference genomic information.At present,RNA-seq has been widely used to identify EST-SSR markers.InAcrocomia aculeata,a total of 418 EST-SSRs were found to be unique for one transcript and region using NGS(Bazzoet al.2018).Transcriptome sequencing was performed using the Illumina platform and 1 418 ESTSSRs derived from 1 300 unigenes were identified inLarix principis-rupprechtiiMayr (Donget al.2018).InPhyllostachys violascens,18 356 EST-SSR loci were identified in transcriptome data (Caiet al.2019).Illumina sequencing andde novotranscriptome assembly of twoCurcuma alismatifoliacvs.(‘Chiang Mai Pink’ and ‘UB Snow 701’) were used to identify 9 351 EST-SSRs with a distribution frequency of 12.5% of the total unigenes(Taheriet al.2019).In addition,with the development of high-throughput sequencing technology,a growing number of studies have been carried out using SMLRS alone or in combination with NGS technology,which can generate long-read sequences and exponentially vast quantities of sequencing data (Xuet al.2015;Abdel-Ghanyet al.2016;Wanget al.2016).However,only NGS and SLAF-seq technologies have been used most often in tree peonies (Zhouet al.2013;Caiet al.2015;Liet al.2015;Zhang Y Xet al.2015).In this study,we tried to combine the two generations of RNA-seq methods to identify EST-SSRs in tree peonies.
At present,there are no reports on the development and characterization of SSR markers within candidate TFs associated with yield based on NGS and SMLRS inPaeonia.In this study,we characterized the EST-SSRs in terms of frequency and distribution,as well as their informativeness and transferability to related species.Moreover,we assessed the effectiveness of EST-SSRs in revealing the genetic relationships between 62 accessions ofPaeonia.This will lay a foundation for identifying the SSR loci associated with yield in the functional genes ofP.rockiiand further accelerate the process of MAS breeding in tree peonies.It will also be of great significance for the oriented improvement and promotion of the woody oil crop industry.
2.Materials and methods
2.1.Plant materials
The genomic DNA ofP.rockii‘Jing Shun Fen’ andP.rockii‘Fen Mian Tao Sai’ were first used to confirm the amplification specificity of the synthesized EST-SSR primers.The genomic DNA of two other tree peony cultivars (P.ostii‘Feng Dan Bai’ M24 andP.suffruticosa‘Hong Qiao’,which were used as the crossing parents for establishing the mapping population in our laboratory)and eight randomly selected accessions ofP.rockiiwere used to examine the polymorphic microsatellite loci(Appendix A).Then the polymorphism and transferability of EST-SSRs were analyzed in a collection of 62Paeoniaaccessions,followed by an investigation on the genetic relationships of these accessions.The accessions used in this study included 37 tree peonies (sectionMoutan) (five species and 32 cultivars),13 herbaceous peonies (sectionPaeonia) (2 species and 11 cultivars)and 12 intersectional hybrids between tree peonies and herbaceous peonies (Itoh hybrids).
The tree peony species includedP.jishanensis,P.rockii,P.decomposita,P.qiuiandP.delavayivar.lutea(Table 1,1–5) and the herbaceous peony species includedP.sinjiangensisandP.mairei(Table 1,50–51).The remaining 55 cultivars consisted of six distinct cultivar types,including Rockii,Suffruticosa,Lutea hybrid,Itoh hybrid,Lactiflora cultivar,and Herbaceous hybrid(Table 1,6–49,52–62).All of these accessions were at least 15 years old and cultivated with standard agronomic practices in the fields of Jiufeng Forestry Experiment Station of Beijing Forestry University (39°54´N,116°28´E)and Beijing Guose Peony Garden (40°45´N,115°97´E)(Beijing,China).Fresh and young leaves were dried on silica gel and ground into powder with liquid nitrogen.
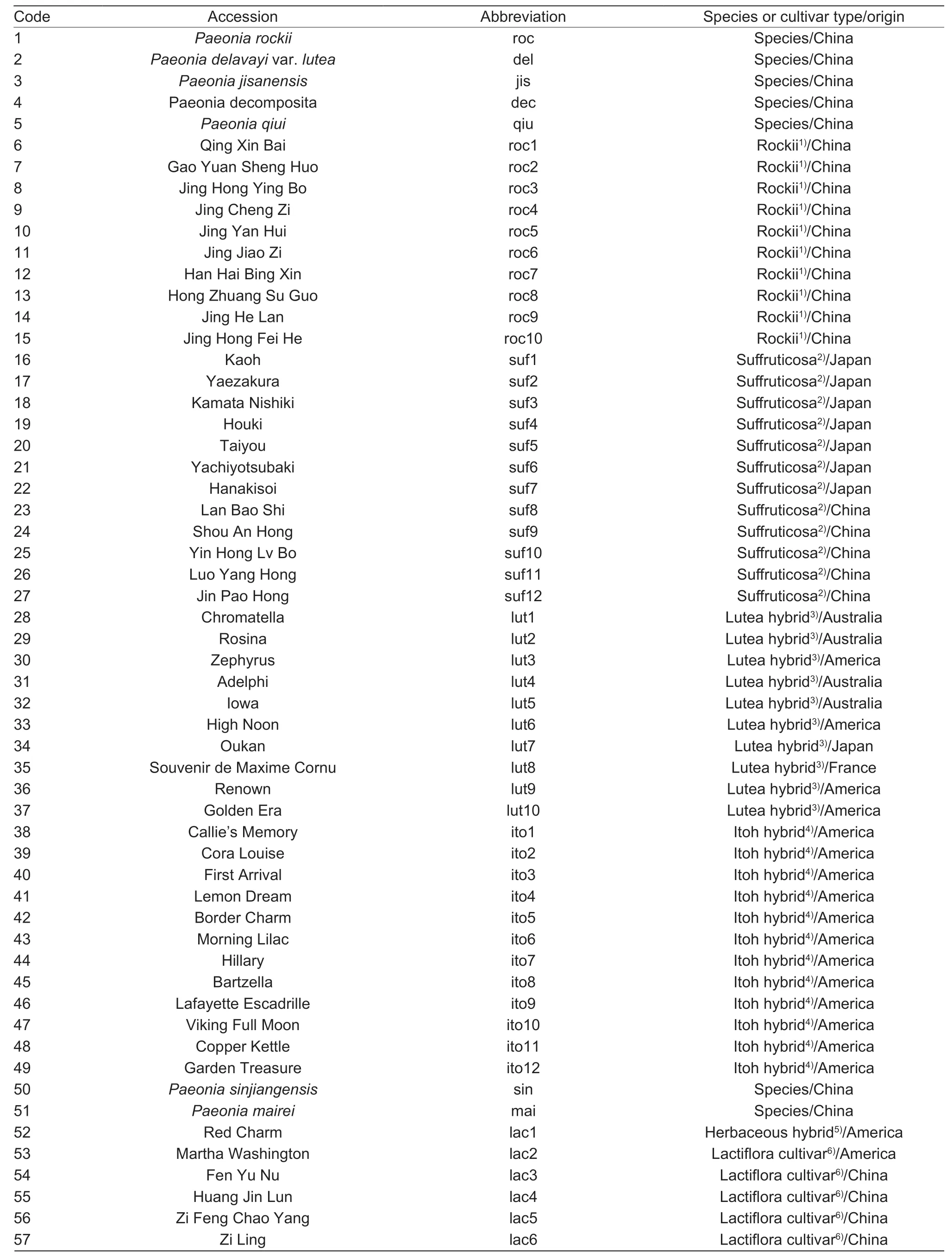
Table 1 The information for the 62 accessions of Paeonia used in this study
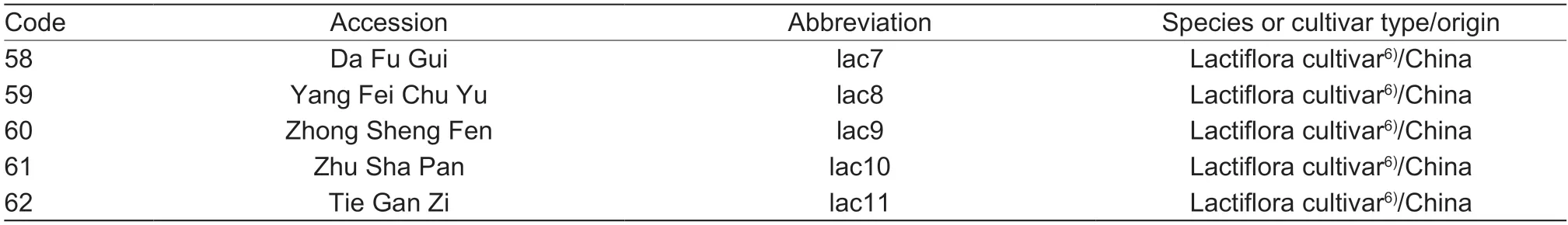
Table 1 (Continued from preceding page)
2.2.DNA extraction,EST-SSR identification and PCR amplification
The genomic DNA of each sample was extracted from 30 mg of leaf powder using the EASYspin Plus Complex Plant DNA Kit (Aidlab Biotechnologies Co.,Ltd.,Beijing,China) according to the manufacturer’s instructions with minor modifications.Then,5 mL of each total DNA sample was assessed by 1% Tris-acetic acid-EDTA(TAE) agarose gel electrophoresis.Simultaneously,1 mL of each total DNA sample was assessed using aNanoDrop ND2000 (A260/A280>1.8,28S/18S>1.0,DNA concentration ≥200 ng mL–1) (Liu Het al.2015).All the DNA samples were subsequently diluted to 50 ng mL–1,which was the working concentration for PCR,and stored at −20°C for further use.
(Continued on next page)
In the previous research,an RNA-seq experiment of flower buds ofP.rockii‘Jing Shun’ Fen andP.rockii‘Fen Mian Tao Sai’ was conducted using NGS and SMLRS methods (Liuet al.2019).The raw data of the sequences have been submitted to the NCBI Sequence Read Archive (SRR9915032,https://www.ncbi.nlm.nih.gov/sra/?term=SRR9915032,and SRR10872586,https://www.ncbi.nlm.nih.gov/sra/?term=SRR10872586).Based on the sequencing data,53 817 annotated unigenes were obtained and the candidate gene families associated with yield were screened.The identification tool program(SSRIT) (http://www.gramene.org/db/markers/ssrtool)(Temnykhet al.2001) was used to search for SSRs within TFs.Primer pairs were designed based on conserved flanking sequences of SSRs using Primer Premier 5.0 Software (Premier Biosoft International,Palo Alto,CA,USA).Primer synthesis was carried out by Shanghai Sangon Biological Engineering Technology (Shanghai,China).
The PCR reactions were carried out in a 10-mL volume containing 5 mL 2×PowerTaqPCR MasterMix (BioTeke,Beijing,China),0.5 mL each primer,3.0 mL ddH2O and 1 mL template DNA.The PCR program was set as follows:5 min at 95°C,35 cycles of 30 s at 95°C,30 s at the appropriate annealing temperature,1 min at 72°C,and 10 min at 72°C,4°C hold.After confirmation of PCR amplification by electrophoresis on a 1% agarose gel,the products were separated by capillary electrophoresis using an ABI3730xl DNA Analyzer (Applied Biosystems,Carlsbad,CA,USA).The reading of polymorphic loci was performed with GeneMarker version 1.80 (Applied Biosystems) using LIZ 600 size standards (Applied Biosystems).Subsequently,Micro-Checker 2.2.3 (http://www.microchecker.hull.ac.uk/) was used to identify and correct genotyping errors (Van Oosterhoutet al.2004).
2.3.Data analysis
The genetic diversity of EST-SSR markers was estimated using GenAlEx version 6.5 (Peakall and Smouse 2006) and POPGENE version 1.32 (Yehet al.1999),including the number of alleles per locus(NA),the effective number of alleles (NE),the observed heterozygosity(Ho),the expected heterozygosity(HE),PIC,the Shannon’s information index (I),and the Wright’s inbreeding coefficient(FIS).The availability of the ESTSSRs was validated using cluster analysis to evaluate the genetic relationships of all accessions.The NJ tree was constructed using the proportion of shared alleles coefficient from the program PowerMarker version 3.25 (Liuet al.2005),in which 13 herbaceous peony accessions were used as an outgroup.The robustness of the dendrogram was subsequently tested using bootstrap analysis with 1 000 replications.
3.Results
3.1.Frequency and distribution of EST-SSR markers
A total of 959 candidate unigenes (TFs) from 21 gene families associated with yield were obtained and used to identify SSR markers based on the RNA-seq library(Appendix B).A set of 166 (17.31%) potential EST-SSR loci were mined from 145 (15.12%) unigenes,of which 124 sequences contained one SSR and 21 sequences contained more than one EST-SSR.On average,one SSR was identified for every 5.78 unigenes.Collectively,the EST-SSRs contained six diverse types of repeats,among which mono-nucleotide repeats were the most abundant type with a frequency of 49.40% (82),followed by tri-(18.67%,31),di-(16.27%,27),complex-(14.46%,24),tetra-(0.60%,1),and hexa-nucleotide repeats(0.60%,1) (Table 2).Among them,mono-and trinucleotide repeats constituted more than 68% of the total EST-SSRs.
Among the 166 potential EST-SSRs,27 major types with mono-,di-and tri-nucleotide repeat motifs were identified.Among the mono-nucleotide motifs,(T/A)nmononucleotides occurred most frequently (accounting for 65.85%),which was also the most dominant motif among all EST-SSRs,followed by (A/T)nmononucleotides(28.05%),(C/G)nmononucleotides (3.66%),and (G/C)nmononucleotides (2.44%).For the di-nucleotide motifs,the most common were (GA/TC) dinucleotides (25.93%)and (CT/AG)ndinucleotides (22.22%),followed by (AT/AT)ndinucleotides (18.52%),(TC/GA)ndinucleotides(18.52%),(AG/CT)ndinucleotides (7.41%),(TA/TA)ndinucleotides (3.70%),and (CA/TG)ndinucleotides(3.70%).Among the tri-nucleotide motifs,the most common were (CAA/TTG)ntrinucleotides (12.90%) and(AGA/TCT)ntrinucleotides (12.90%),followed by (GAA/TTC)ntrinucleotides (9.68%),(GCA/TGC)ntrinucleotides(6.45%),and (TCT/AGA)ntrinucleotides (6.45%).In addition,there were no obvious dominant motifs among the tetra-,hexa-,or complex-nucleotide repeats (Fig.1).
3.2.Development,validity and polymorphic screening,and functional annotation of EST-SSR markers
A total of 166 EST-SSR primers were designed and selected for validation.Among them,102 (61.45%) pairs of primers generated amplification products which were clear and well-sized in the genomic DNA of both RNA-seq cultivars,while the remaining 64 pairs of primers failed to detect PCR amplification products at multiple annealing temperatures (Appendix C).Then all the EST-SSR primers were selected for polymorphic screening in both parents of one mapping population and a random sample of eightP.rockiiaccessions (Appendix A).The results suggested that 58 (56.86%) pairs of primers showed polymorphism (number of alleles≥2),while the remaining 44 (43.14%) pairs of primers were monomorphic.The 58 polymorphic primers included 34 (58.62%) mono-,10 (17.24%) di-,9 (15.52%) tri-,1 (1.72%) tetra-,and 4 (6.90%) complex-motif loci.A combined total of 211 alleles were detected with these markers,with an average of 3.64 alleles per locus.These 58 polymorphic ESTSSRs were distributed in 18 gene families,among which the AP2 (8) gene family contained the largest number of SSRs,followed by the bHLH (6),WD (6),HSF (5),TCP(5),WRKY (4),and GATA (4) gene families,etc.(Table 3,Appendix D).
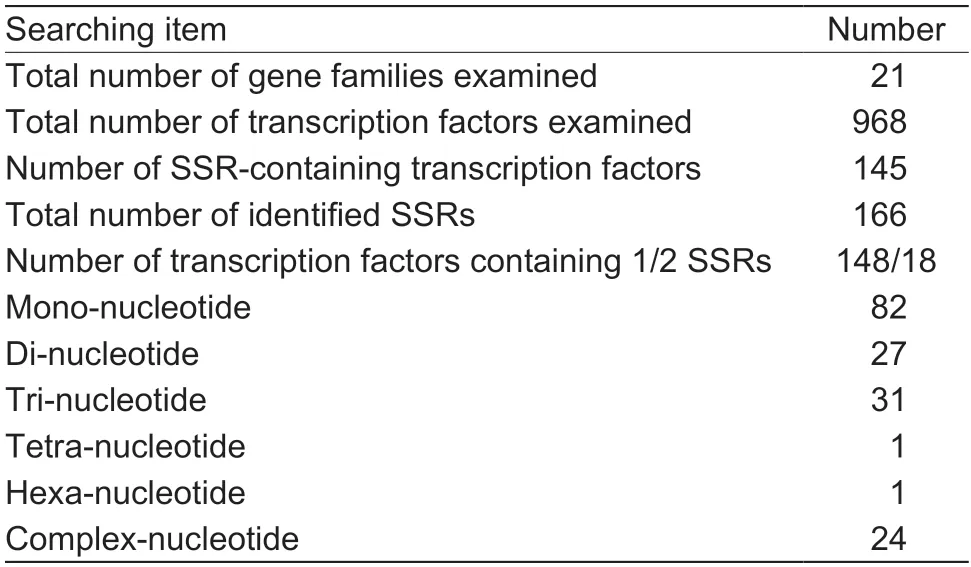
Table 2 Summary of expressed sequence tag-simple sequence repeat (EST-SSR) markers identified within transcription factors in the RNA-seq of Paeonia rockii
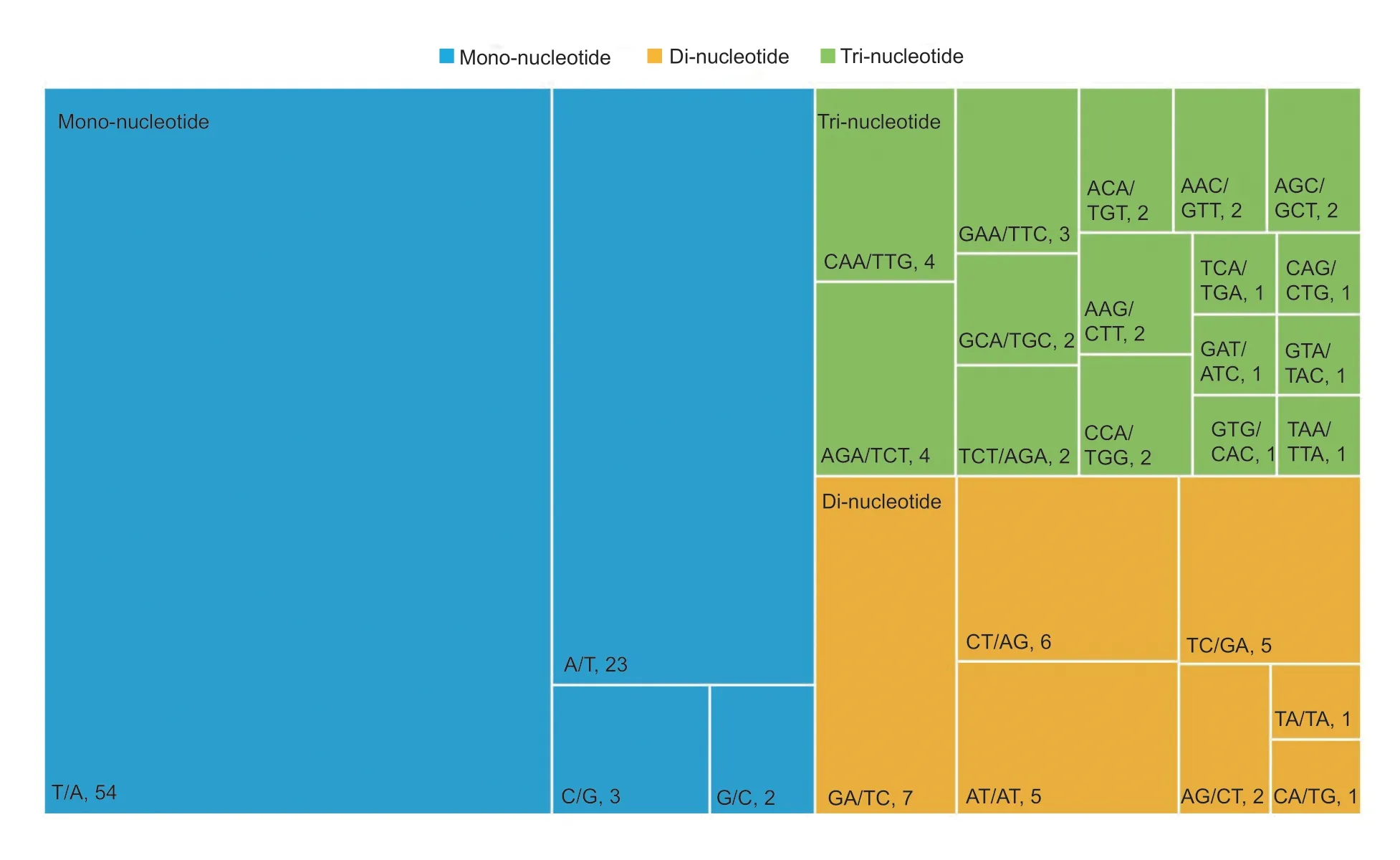
Fig.1 Frequency distribution of mono-,di-and tri-nucleotide SSRs based on motif type and the number of different repeat motifs.
Blastx results indicated that all the 58 polymorphic EST-SSRs obtained functional annotations,among which 24 (41.38%) were aligned to the protein sequences ofVitis vinifera,5 (8.62%) toNelumbo nucifera,4 (6.90%)to each ofTheobroma cacaoandP.suffruticosa,and the remaining EST-SSRs were mainly aligned to the protein sequences of dicotyledons such asRicinus communis,Durio zibethinus,andManihot esculenta(Appendix D).
3.3.Genetic diversity and transferability analysis of EST-SSR markers
When the polymorphism of the 58 EST-SSR markers was further assessed in 37 accessions of tree peonies (Table 1,1–37),we found that 56 (100%) of them showed high polymorphism and 503 alleles were detected.Moreover,NAvaried from 2 (PS123) to 16 (PS73 and PS147),with an average of 9 per locus;NEvaried from 1.63 (PS105)to 11.36 (PS96),with a mean value of 4.51.Ivaried between 0.60 (PS123) and 2.58 (PS96),with an average of 1.63.Horanged from 0.11 (PS105) to 0.97 (PS150 and PS151),with a mean value of 0.57.HEvaried between 0.39 (PS105) and 0.91 (PS96),with an average of 0.73.SinceHowas higher thanHEat seven EST-SSR loci,FISranged from–0.20 (PS150) to 0.80 (PS47),with a mean value of 0.23.The PIC values ranged from 0.32 (PS123)to 0.91 (PS96),with a mean value of 0.70,among which 55 EST-SSRs (all of them except for PS10,PS105,and PS123) showed a high level of polymorphism (PIC>0.5).These results indicated a high level of informativeness within these EST-SSR loci (Table 4).
In addition,when these 58 pairs of primers were used to test the transferability across sevenPaeoniaspecies(Table 1,1–5,50–51),the amplification ratios ranged from 89.66 to 100% (mean 94.09%).Moreover,the 58 pairs of primers revealed a 100% transferability ratio inP.rockii,while the lowest transferability ratio (89.66%) was inP.delavayivar.lutea.Among them,PS25,PS33,PS73,PS159,PS62,PS118,PS50,and PS91 showed varying levels of non-transferability across the tree peony species;PS12,PS50,PS57,PS73,PS91,PS114,PS145,PS151,and PS160 showed various degrees of non-transferability across the herbaceous species;and PS73,PS50,and PS91 showed lower transferability ratios inPaeonia(Appendix E).Overall,the 58 EST-SSRs developed in this study showed a high level of genetic diversity,which can be used to evaluate the diversity and genetic relationship ofPaeonia.
3.4.Evaluation of the genetic relationships among 62 genotypes using EST-SSR profiles
The genetic relationship analysis was performed using 62 accessions ofPaeoniato evaluate the usefulness of 58 newly developed EST-SSR markers.Then an NJ tree was constructed,in which the herbaceous peony accessions were used as an outgroup.As a result,P.lactifloraand the cultivars ofherbaceous peony formed an independent cluster,which was subsequently grouped together with the wild species ofP.delavayivar.luteaandP.jishanensis.Except for ito2 (Cora Louise) and ito5(Border Charm),lut1 (Chromatella) and ito9 (Lafayette Escadrille) respectively formed a separate clade,and all the Lutea hybrids and Itoh hybrids formed independent clusters.Moreover,the cultivars of Suffruticosa and Rockii formed an independent cluster,in which all the floral disks were leathery.Suffruticosa consisted ofP.suffruticosaJapan group (suf1–7) andP.suffruticosaChina group(suf8–12).The wild species ofP.decompositaformed a separate clade and showed close genetic relationships with the cultivars of Rockii.The wild species ofP.rockiiandP.qiuiformed a separate clade (Fig.2).Therefore,the results from the molecular data and dendrogram generated by EST-SSR makers were in good agreement with the traditional classification system inPaeonia.
4.Discussion
4.1.EST-SSR marker development by combining two generations of RNA-seq methods
Transcriptome sequencing technology has proven to be an effective method for obtaining EST sequences,which are essential for mining large numbers of functional SSR markers and novel genes.Currently,large numbers of EST-SSR markers based on NGS have been developed in diverse species (Sainiet al.2019;Sunet al.2019;Tian X Yet al.2019;Torokeldievet al.2019;Ukoskitet al.2019;Zhouet al.2019).This study is the first attempt to combine the NGS and SMLRS methods to develop the EST-SSRs inPaeonia.We identified 959 candidate TFs from 21 gene families which have been reported to have important regulatory effects on yield,including AP2(Liet al.2016),MYB (Zhang Y Xet al.2017),WRKY1)NA,number of alleles per locus;NE,effective number of alleles;I,Shannon’s information index;Ho,observed heterozygosity;HE,expected heterozygosity;FIS,inbreeding coefficients;PIC,polymorphism information content.(Suet al.2015) and other gene families (Hirschet al.2009;Youet al.2011;Heanget al.2012;Kaplan-Levyet al.2012;Rahaieet al.2013;Heet al.2015;Quet al.2015;Chenet al.2016;Manet al.2016;Luet al.2017;Ninget al.2017;Shimrayet al.2017;Guoet al.2018;Srivastavaet al.2018;Renet al.2019;Tian J Get al.2019;Yanget al.2019).The functions of these TFs and the remaining gene families remain to be further verified in future studies.The results showed that the distribution frequency of EST-SSR was 15.12%,higher than that reported in tree peony (Gaiet al.2012;Zhanget al.2012;Wang D Xet al.2013;Wuet al.2017).Compared with other crops,it was also higher thanMedicago sativa(6.26%)andCynodon dactylon(6.4%) (Tanet al.2012;Wang Zet al.2013),but lower thanQuercus palustris(64.69%) (Sunet al.2019).The difference in amplification frequency indicates that the abundance of EST-SSRs varies with the search criteria,the size of the source database and the mining tool used for the SSR search (Liet al.2011).Therefore,we believe that the microsatellite loci developed within genome-wide functional TFs can more comprehensively analyze the contributions of TFs to traits.In conclusion,since the development of TFs-based SSR markers has directly or indirectly accelerated the progress of MAS breeding in plants (Iorizzoet al.2011;Zhaiet al.2014),these developed EST-SSRs will be valuable for the MAS breeding ofPaeonia,especially for high-yield breeding of the oil tree peonies.
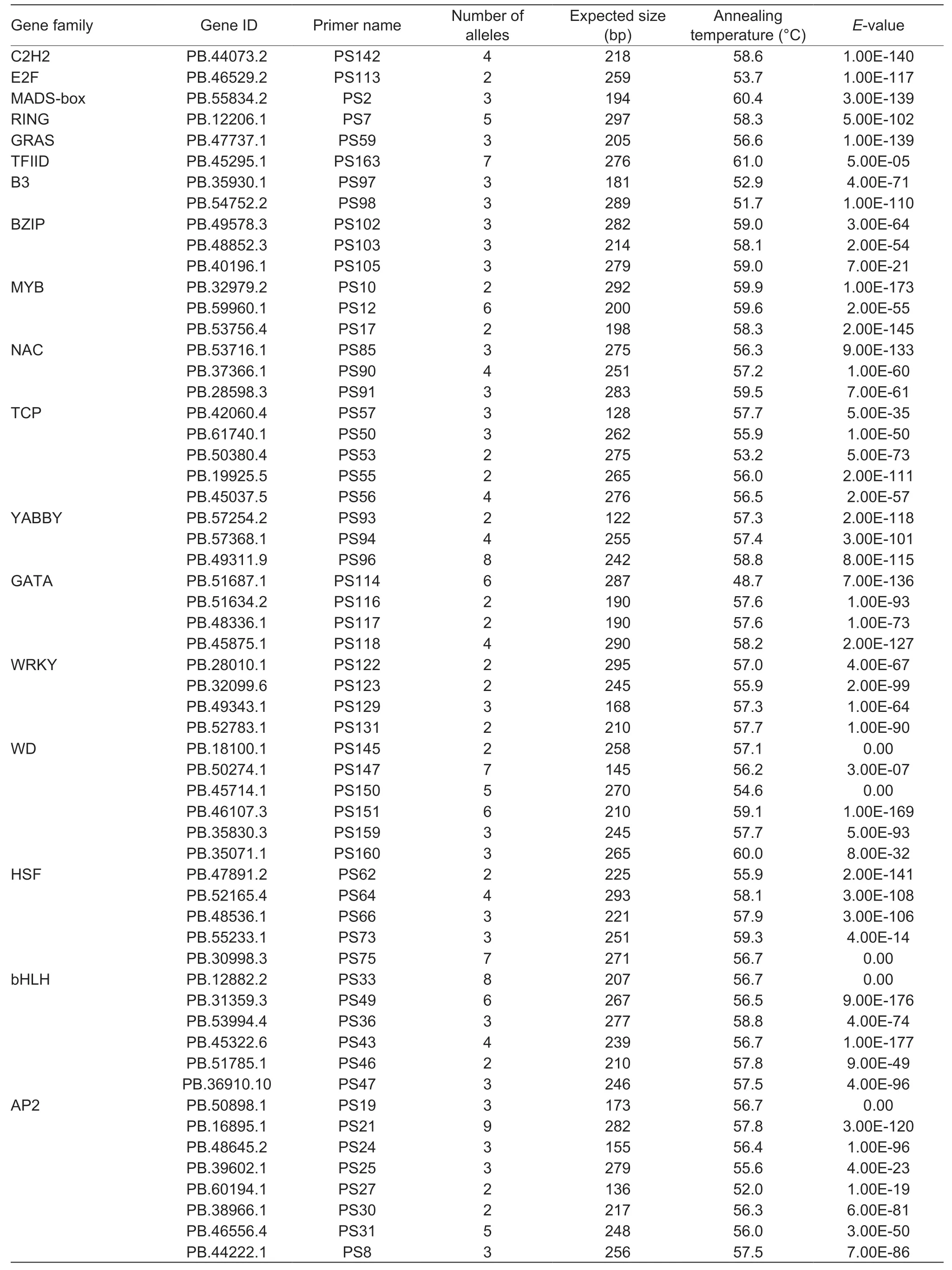
Table 3 Summary information of the 58 simple sequence repeat (SSR) markers within transcription factors obtained by polymorphic screening in the RNA-seq of Paeonia rockii
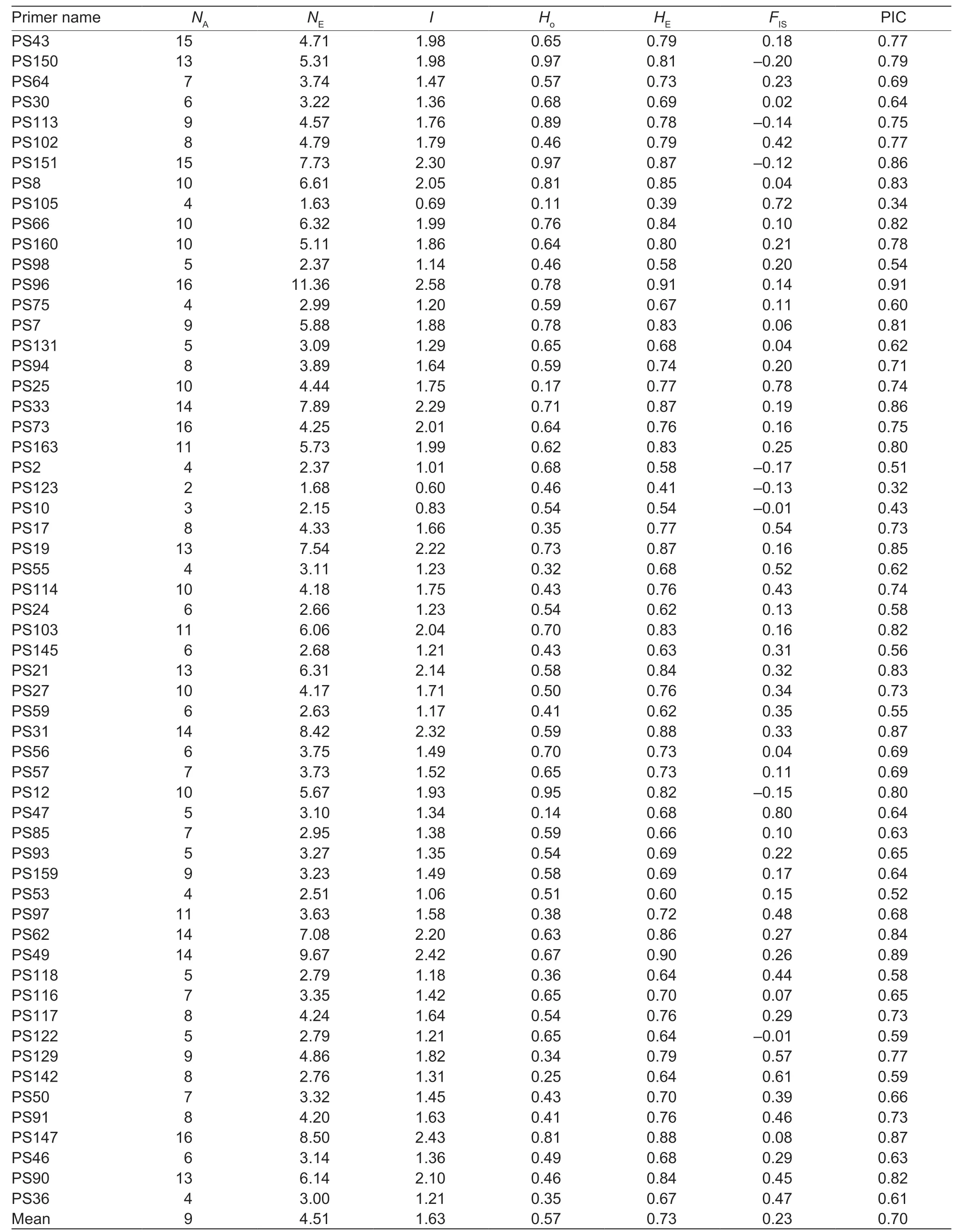
Table 4 Polymorphism parameters of the 58 simple sequence repeat (SSR) markers within transcription factors in the 37 tree peony accessions1)
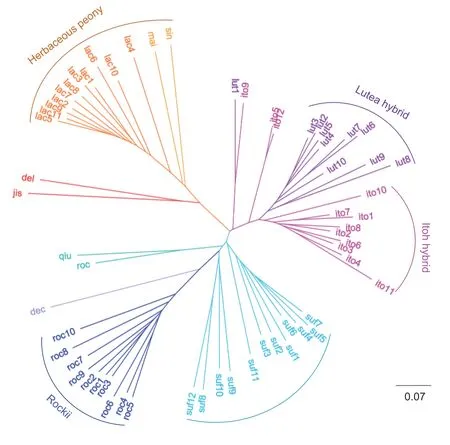
Fig.2 The neighbour-joining tree of the 62 accessions in Paeonia constructed using the 58 expressed sequence tag-simple sequence repeat markers in Paeonia rockii.The information for the abbreviations in the tree is provided in Table 1.
4.2.The characterization of genic SSR markers
Until recently,the types of dominant repeats had usually been different among various plants (Varshneyet al.2005).In this study,mono-nucleotide repeats were the most abundant type and followed by tri-and dinucleotides,which were not consistent with previous reports on tree peony (Gaiet al.2012;Wuet al.2017)and some other plants,such asPopulus tomentosa(Duet al.2012a,b) andPyrus(Zhanget al.2014).We speculate that this difference is due to the exclusion of mono-nucleotide repeat types in screening some other plants,but we have retained them.It may also be due to the differences in the genomic resources (EST,cDNA or gene sequences) from which the markers were derived.For example,studies have shown that di-,tetra-,and penta-nucleotide repeat types are mainly associated with the UTR region,while tri-and hexa-nucleotides are mainly related to the coding sequence (CDS) region(Guptaet al.2014;Wuet al.2014).The EST-SSRs developed in this study are mainly from CDS,so there is an abundance of mono-and tri-nucleotide repeats.In addition,we observed that the most dominant dinucleotide repeat motifs were (GA/TC)ndinucleotides and(CT/GA)ndinucleotides,which was the same as found inRaphanus sativus(Zhaiet al.2014),Zea mays,Oryza sativa,Sorghum bicolor,Triticum aestivum,andHordeum vulgare(Temnykhet al.2000;Kantetyet al.2002).The most prevalent mono-and tri-nucleotide motifs were,respectively,(T/A)nmononucleotides and (CAA/GTT)nand(AGA/TCT)ntrinucleotides,which were not consistent with the results obtained in some other species.We speculate that these differences are due to the fact that the ESTSSRs developed in this study are mainly from CDS.Importantly,we found that mono-nucleotide repeats and(T/A)nmononucleotides were the most abundant types in the association mapping with yield traits ofP.rockii(Liuet al.2020).Therefore,we speculate that mononucleotide repeats based on TFs are likely to play an important role in regulating the yield traits of tree peony.
4.3.The validity and polymorphic screening of genic SSR markers
In this study,61.54% of the EST-SSR primers were amplified in the target genomic DNA,which was higher than the results obtained inP.suffruticosa(47.3%) (Wuet al.2014),M.sativa(30%) (Wang Zet al.2013),andQ.palustris(58.23%) (Sunet al.2019),but lower than inP.tomentosa(70.7%) (Duet al.2012a),R.sativus(78.23%) (Zhaiet al.2014),andGlycine max(68.09%)(Hisanoet al.2007).The results indicate that the amplification efficiency of EST-SSR loci is much higher than that of gSSRs.We speculate that this difference is related to the high conservatism of EST regions in the primer sequences.The lack of successful amplification of certain primers may be caused by fragment insertion or repeated mutation,the presence of introns and RNAseq technology (Sunet al.2019).Moreover,58 (56.86%)polymorphic EST-SSRs were obtained in this study.The polymorphic ratio was higher than those obtained inP.suffruticosa(39.9%) (Wuet al.2014),G.max(12.8%)(Hisanoet al.2007),andIpomoea batatas(41.9%) (Wanget al.2011),but lower than inP.tomentosa(64.4%) (Duet al.2012a).These differences in polymorphism ratios may be related to the number and different geographic origins of samples (Wanget al.2011).In addition,the number of alleles ranged from 2 to 9 (mean 3.64) in this study.We speculate that the relatively low allelic variation may be related to the conservation of genic SSRs in tree peonies.Generally,yield traits are complex quantitative traits regulated by microgenes.In this study,TFs-based microsatellite loci inP.rockiishowed high stability and polymorphism,suggesting that the development of TFsbased microsatellite loci is an effective way to identify regulatory loci for yield traits in woody crops.
4.4.Evaluation of the 58 polymorphic EST-SSR markers
The PIC value can be used to assess the polymorphism level of each EST-SSR marker,and it can be categorised as high (PIC>0.5),moderate (0.25<PIC<0.5) or low(PIC<0.25) (Bosteinet al.1980).In this study,58 primers(except for PS10,PS105,and PS123) exhibited high PIC values,which indicated a high level of polymorphism.This result is different from the report in another tree peony,P.ostii,that EST-SSRs had a lower level of polymorphism compared to the gSSRs (Yuet al.2013).This disparity could be due to the overabundance of heterozygotes in the evolutionary process ofP.rockii,or we may have adopted accessions with a diverse set of genotypes derived from intra-and inter-specific hybridization and wild species.Moreover,the EST-SSR markers identified inP.rockiishowed high transfer rates(94.09%) in the seven species ofPaeonia,which was consistent with the reported results inP.suffruticosa(Wuet al.2014) andEucalyptus(Zhouet al.2014),but higher than the reported range of 10–90% (Ellis and Burke 2007).The results indicate that different species inPaeoniashow a close genetic relationship.Additionally,FISreflects the degree of genetic purification among different species (Wright 1922;Slateet al.2004).In this study,theFISranged from–0.20 to 0.80 (mean 0.23) andFISvalues for eight EST-SSRs were negative,indicating that there was excess heterozygosity in the accessions.We speculate this is related to the fact that the breeding system of tree peonies is predominantly outcrossing and the cultivated cultivars are of hybrid origin (Cheng 2007).
4.5.The application of the developed EST-SSR markers in the genetic analysis of Paeonia
At present,EST-SSR markers have been widely used in studies of genetic diversity,genetic relationships,association mapping and so on (Paritaet al.2018;Xianget al.2018;Agarwalet al.2019).In this study,the 58 EST-SSRs developed were used to evaluate the genetic relationship among 62 accessions ofPaeonia.Cluster analysis in the NJ tree demonstrated that the accessions showed a great consistency with their origins (Yuet al.2013;Wuet al.2014).The genusPaeoniaincludes three sections,namely sect.Moutan,sect.Paeoniaand sect.Onaepia.Tree peonies,which belong to the sect.Moutan,are composed of the subsection Vaginatae and the subsection Delavayanae (Stern 1946).Based on the breeding history almost exclusively through hybridization,the cultivated tree peonies have genetically formed distinct cultivar types like Suffruticosa,Rockii,Lutea hybrid,etc.Among them,intersectional hybridization between the sect.Moutanand sect.Paeoniaforms a new type of group called the Itoh Hybrid.The genetic relationships shown in the NJ tree are basically consistent with the above classification,which verified the validity of the developed EST-SSRs.Moreover,sinceP.rockiiis one of the pedigrees of the Suffruticosa (Yuanet al.2014),the cultivars of Rockii and Suffruticosa form an independent cluster.The wild species ofP.rockiiand the cultivars of Rockii are close relatives to the wild species ofP.decomposita,which is consistent with the reported results (Zhaoet al.2008;Yuet al.2013;Wuet al.2014).It is well known that the ancestors of the Japanese cultivars are mainly from theP.suffruticosaChina group,so they show a very close relationship in the NJ tree.Further,except ito2 (Cora Louise) and ito5 (Border Charm),lut1 (Chromatella) and ito9 (Lafayette Escadrille)respectively form the separate clades,so all Lutea hybrids,and Itoh hybrids form independent clusters in this study.The Lutea hybrids are the crossbred offspring of theP.delavayiiandP.suffruticosa(Cheng 2007;Rogers 1995).Itoh hybrids are the result of the intersectional hybridization betweenP.lactifloraand the Lutea hybrid.In addition,we find some incongruences in a few branches.For example,the wild species ofP.delavayivar.luteaandP.jishanensisform a branch,the Itoh hybrid ito9 (Lafayette Escadrille) and lut1 (Chromatella) form a branch.We speculate that the incongruences observed may be due to the differences of accessions and markers.
4.6.SSR loci within TFs associated with yield
TFs from different gene families play an important role in the regulation of gene expression associated with yield(Sahniet al.2016;Zhang Jet al.2017;Wang Jet al.2018).TFs from the AP2 gene family can control seed mass and seed yield,and enhance tolerances to drought,salt,and freezing,as well as resistances to multiple diseases in the transgenic plants (Jofukuet al.2005;Ohet al.2009;Xuet al.2011).In this study,the AP2 (8)gene family contained the largest number of SSRs,among which PS21 (PB.16895.1) showed the largest number of alleles (9) in the 10 accessions of polymorphic screening and high polymorphism (PIC=0.83) among the 37 accessions of tree peonies.We speculate that ESTSSRs like PS21 from the AP2 gene family are important factors for improving the seed yield or stress response ofP.rockii.bHLH proteins are the second largest class of plant transcription factors and they are involved in grain length and weight (Felleret al.2011).WD proteins are also widespread in plants and involved in regulating seed mass and size (Youet al.2011;Mishraet al.2012).In this study,both WD and bHLH gene families contained six SSR loci,second only to the AP2 gene family.Among them,PS147 (PB.50274.1) and PS33 (PB.12882.2)showed abundant polymorphism alleles (7 and 8) and high polymorphism (PIC=0.87 and 0.86),respectively.We speculate that EST-SSRs identified in the WD and bHLH gene families play an important role in regulating yield traits ofP.rockii.Additionally,the EST-SSR loci identified in all the other 15 gene families (MYB,TCP,WRKY,etc.) showed varying degrees of polymorphisms,but the functions of all the EST-SSR loci still need to be further verified inP.rockii.Taken together,the EST-SSR loci identified from gene families associated with yield provide an important molecular basis for high-yield breeding and association mapping of oil tree peonies.
5.Conclusion
In order to develop the EST-SSR markers ofP.rockii,we first identified 166 SSRs from 959 candidate TFs associated with yield based on the comparative transcriptome data sets.Then the EST-SSRs were characterized in terms of frequency and distribution,validity and polymorphism.Moreover,58 polymorphic EST-SSRs were identified and their transferability was assessed in 62 accessions ofPaeonia.The large set of EST-SSRs identified in this study has remarkably increased the molecular marker repository for tree peonies.It is also of great significance for the application of MAS breeding to increase the yield of oil tree peonies.Perhaps the more interesting questions arising from this research are how the EST-SSRs could represent a powerful tool for genetic diversity analyses,genetic relationship studies and association mapping in tree peonies and related species.In-depth studies are required to answer these questions and to provide insight into the identification of marker-trait pairs in various plants,including tree peonies.
Acknowledgements
We gratefully acknowledge Mr.Cheng Xinyun (Beijing Guose Peony Technologies Co.,Ltd.,China) for his valuable support in maintaining the living plant materials for this study.This work was supported by the National Key Research and Development Project(2019YFD1001500),and the National Natural Science Foundation of China (31471898).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Receptor-like kinase OsASLRK regulates methylglyoxal response and content in rice
- Heredity and gene mapping of a novel white stripe leaf mutant in wheat
- Construction of a high-density adzuki bean genetic map and evaluation of its utility based on a QTL analysis of seed size
- Effects of temperature and solar radiation on yield of good eatingquality rice in the lower reaches of the Huai River Basin,China
- Difference in corn kernel moisture content between pre-and postharvest
- The effect of elevating temperature on the growth and development of reproductive organs and yield of summer maize
