The effective transfection of a low dose of negatively charged drugloaded DNA-nanocarriers into cancer cells via scavenger receptors
2021-05-12MirzMuhmmFrnAshrfBigChngfiZhngMuhmmFurqnAkhtrAmmrSlmJhnzbMussir
Mirz Muhmm Frn Ashrf Big,Chngfi Zhng,Muhmm Furqn Akhtr,Ammr Slm,Jhnzb Mussir
aLaboratory of Biomedical&Pharmaceutical Engineering of Stem Cells Research,Restorative Dental Sciences,Faculty of Dentistry,The University of Hong Kong,999077,Hong Kong,PR China
bState Key Laboratory of Analytical Chemistry for Life Sciences,School of Chemistry and Chemical Engineering,Nanjing University,Nanjing,210023,PR China
cFaculty of Pharmacy,Bahauddin Zakariya University,Multan,60000,Pakistan
dRiphah Institute of Pharmaceutical Sciences,Riphah International University,Lahore,Pakistan
eDepartment of Pharmacology,Faculty of Pharmaceutical Sciences,Government College University Faisalabad,Faisalabad,Pakistan
Keywords:
Cisplatin(CPT)
DNA-nanowires(DNA-NWs)
HepG2 resistant cancer cells
Scavenger receptors
A B S T R A C T
DNA-nanotechnology-based nano-architecture scaffolds based on circular strands were designed in the form of DNA-nanowires(DNA-NWs)as a polymer of DNA-triangles.Circularizing a scaffold strand(84-NT)was the critical step followed by annealing with various staple strands to make stiff DNA-triangles.Atomic force microcopy(AFM),native polyacrylamide gel electrophoresis(PAGE),UV-analysis,MTT-assay,flow cytometry,and confocal imaging were performed to assess the formulated DNA-NWs and cisplatin(CPT)loading.The AFM and confocal microscopy images revealed a uniform shape and size distribution of the DNA-NWs,with lengths ranging from 2 to 4μm and diameters ranging from 150 to 300 nm.One sharp band at the top of the lane(500 bp level)with the loss of electrophoretic mobility during the PAGE(native)gel analysis revealed the successful fabrication of DNA-NWs.The loading efficiency of CPT ranged from 66.85% to 97.35%.MTT and flow cytometry results showed biocompatibility of the blank DNA-NWs even at 95% concentration compared with the CPT-loaded DNANWs.The CPT-loaded DNA-NWs exhibited enhanced apoptosis(22%)compared to the apoptosis(7%)induced by the blank DNA-NWs.The release of CPT from the DNA-NWs was sustained at<75% for 6 h in the presence of serum,demonstrating suitability for systemic applications.The IC50of CPT@DNA-NWs was reduced to 12.8 nM CPT,as compared with the free CPT solution exhibiting an IC50of 51.2 nM.Confocal imaging revealed the targetability,surface binding,and slow internalization of the DNA-NWs in the scavenger-receptor-rich cancer cell line(HepG2)compared with the control cell line.
1.Introduction
The recently increased cancer prevalence among different age groups from pediatric to geriatric patients is devastating,requiring new tools to be developed for efficient treatment[1-4].Cisplatin(CPT)is a drug of choice for chemotherapy of various types of cancerssuch asprostate,ovarian,cervical,testicular,brain,esophageal,liver,bladder,breast,lung,and head-neck cancers[3].Being a hydrophilic drug,CPT presents a challenge in targeting tumor sites and is potentially cytotoxic to healthy cells.We have designed DNA-based nanocarriers(DNA-nanowires;(DNA-NWs))to effectively load CPT(utilizing its DNA binding capability)and target cancer cells for intracellular CPT delivery[4].We confirmed cancer cell targeting and intracellular delivery of CPT in a scavenger receptor-rich cell line(HepG2)as compared with the cell line that lacks scavenger receptors(CHSE-214 cell line)from the salamander embryo[5].
DNA nanotechnology is an effective approach for achieving nanoscale objects with the desired shape as carriers for drug delivery and bio-imaging[6,7].Watson/Crick base-pairing enables ease of self-assembly of various strands and the attainment of specific shapes.DNA binding drugs,including CPT,can be loaded effectively on compact DNA nano-architecture,owing to their intrinsic DNA-binding ability[8].DNA-based nanomaterials are becoming increasingly crucial as drug delivery carriers due to their nanoscale size precision and control that can be exerted over the shape of the material[9,10].Some drugs have the intrinsic ability to bind with the DNA double helix,such as doxorubicin and cisplatin,which can be effectively loaded onto DNA-based carriers[11,12].
Furthermore,the polyanionic nature of DNA renders internalization in healthy cells difficult[13].As cancer cells have high proportionsofpolyanionic ligand-binding scavengerreceptors,targeting cancer cells is more effective[14].Triangular DNA tiles are synthesized after circularizing the template strand and annealing with staple strands[15,16].Each triangle tile is designed to contain sticky ends at the vertices to combine(with each other),making a compact polymer,resulting in DNA-nanosheets that fold and spin onto itself due to the helical structure and curvature of the DNA to yield compact DNA-NWs[17,18].After tagging CPT with the fluorescein isothiocyanate(FITC)dye,the loading of CPT/FITC onto the DNA-NWs was confirmed by confocal microscopy and UV analysis[19].The mechanism of targeting and internalization of the CPT-loaded DNA-NWs is illustrated in Fig.1.
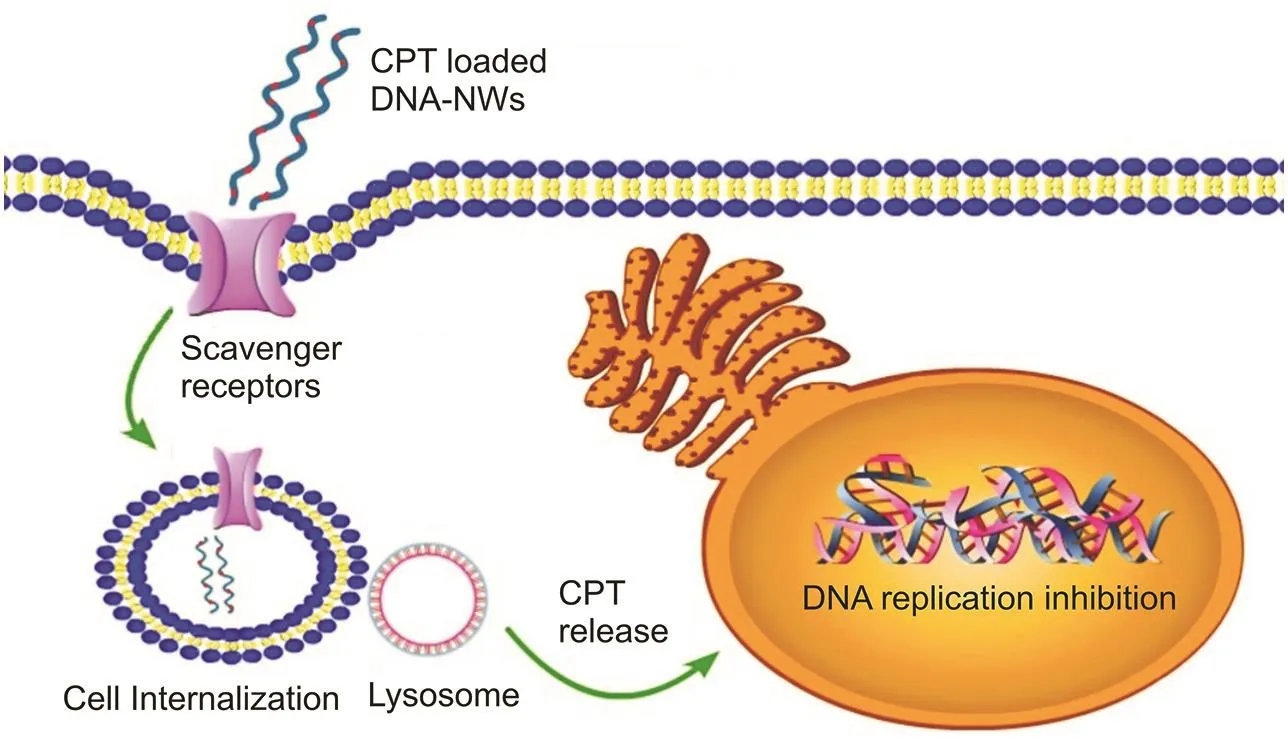
Fig.1.Scavenger receptor-rich cancer cell line(HepG2)shows the rapid internalization of polyanionic DNA-nanocarriers for CPT delivery.
2.Materials and methods
2.1.Chemicals and facilities
DNA-strands with the prescribed parameters and sequences were obtained from Sangon Biotechnology Company,Shanghai,China.Cisplatin(CPT;99% purity)was purchased from Shanghai-ChemicalsCompany,Shanghai,China.MTT-reagent(3-(4,5-dimethylthiazol,2 yl)2,5-diphenyl-tetrazolium-bromide;99% purity)was obtained from Sigma-Aldrich(St.Louis,MO,USA).HepG2 cells were provided by Nanjing University,Nanjing,China.Fast-Scan AFM by Bruker,USA was used for imaging and characterization of nanoparticles.Flowcytometry experiments were performed using a BD/LSR-FortessaX-20 flow cytometer(Becton-Dickinson,San Jose,CA,USA).Ninety-six-well plates were analyzed for UV-Vis measurements using a MultiScan-FC microplate-photometer by Thermo Fisher(Waltham,USA).The microliter volume samples were analyzed for UV-Vis measurements using a Thermo Fisher Nanodrop-2000c spectrometer(USA).Gel analysis was performed using a gel-apparatus by BioRad-Labs(CA,USA).
2.2.Preparation and characterization
The circular scaffold strands(84-NT)were annealed with staple strands to synthesize triangular DNA tiles.Triangular DNA tiles underwent cohesion polymerization via sticky ends at the vertices of the triangles to form DNA-NWs.The following are the detailed steps adopted for the preparation and characterization of DNANWs.
2.3.Triangular DNA-tiles and DNA-NWs
Specific DNA oligo sequences(Sangon-Biotech,Shanghai,China)were purified using the polyacrylamide gel electrophoresis(PAGE)method[20,21].purified oligos were UV-analyzed for concentration measurements using a Nanodrop-2000c.The ordered sequences of different DNA oligos are listed in Table 1.Schematics of the overall synthetic process are shown in Fig.2.
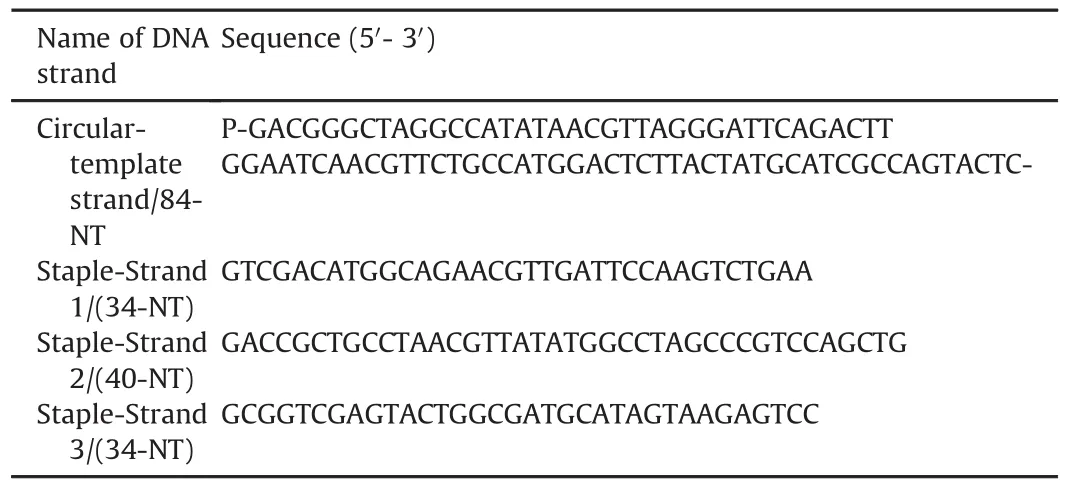
Table 1 Specific sequences of DNA oligos.

Fig.2.A scheme to synthesize DNA-NW and FITC-tagged CPT loading.
2.3.1.Preparing scaffold strands as a circularized template
1.3 检索策略 检索PubMed、Embase、Web of Science,CNKI、VIP、万方和CBM。检索时限均从建库至2017年6月15日,本次检索主要根据以下主题词进行:中文检索词为“针刺”、“电针”、“芒针”、“慢性前列腺炎”、“淋证”、“精浊”“腰痛”“随机对照”等;英文检索词为“acupuncture”“electric acupuncture”“chronic prostatitis”“randomized controlled trail”等。
Scaffold strands were circularized,as previously reported[22].Briefly,a 5′-phosphorylated 50 μM linear scaffold(84-NT)was annealed with a 60μM splint strand(20-NT)such that it was folded to join its two ends together with the help of splint strand[23].The annealing process was performed from boiling to 25°C over 3.5 h[24].After joining the two ends of the scaffold strand with the help of splint strands,the two ends of the scaffold strand were covalently joined together via the ligation process using T4 DNA-ligase(8 μL,350U/μL)at 16°C for 16 h in the presence of freshly prepared TE buffer[25].The ligating enzyme was heat-denatured,followed by exonuclease-1 treatment(8 μL,5U/μL)for 35 min while incubating at 37°C to digest and eliminate linear strands[22].After that,exonuclease-1 was heat-denatured.After rapid re-cooling,50μL deionized form amide was added[26].
2.3.2.Purification of the circularized template
A denaturing-polyacrylamide gel(10%)was used to purify the freshly prepared circularized-template strand.The gel solution was made from a commercially available 3% acrylamide-bisacrylamide mixture in the presence of 4.0 M urea[27].Electrophoresis was performed at 75 V for 4 h in TBE-EDTA buffer at 15°C.The circularized template moved down the gel and gave a sharp band at the designated position under applied voltage and current[28,29].The bands were cut corresponding to the circularized template and macerated in TE buffer at room temperature for 3 days using an orbital shaker to elute the gel’s circularized template,followed by filtration using a syringe filtration assembly[20].
2.3.3.Synthesis of DNA triangles and CPT-loaded DNA-NWs
The circularized template strand was combined stoichiometrically with equimolar staple strands(30μM)in the presence of TE buffer and 10% TAE-Mg buffer with the addition of 3μL of the buffer per 30μL of the final solution.A PCR machine(Thermo Fisher Scientific USA)was used for thermocycling the temperature with a gradual temperature drop from-1°C to-0.1°C every second to attain effective annealing without mismatch.As DNA triangular tiles contain sticky ends,they self-assemble in a robust way to make DNA 2D lattices in the form of sheets that coiled on its(own)axis to form DNA-NWs to act as a drug delivery carrier.FITC-tagged CPT was loaded onto the DNA-NWs using CPT’s binding capability to double-stranded DNA[15].
2.4.Electrophoretic analysis of DNA triangle self-assembly to produce DNA-NWs
Assessing the restriction of the DNA’s electrophoretic movement down the native-PAGE gel(a gel without urea)is a criterion for analyzing successful self-assembly of DNA triangular tiles into a large molecular-weight DNA lattice.Accordingly,the self-assembly of triangular DNA tiles was confirmed by native PAGE analysis using 10μL of the annealed final assembly mixture of the large-sized DNA structure into the 10% PAGE gel to observe the restriction of the electrophoretic movement of the self-assembled DNA-NW samples into gel wells compared with other strands with different lengths and the DNA marker in the other wells of the gel as a control.The process was performed at a controlled temperature(10°C)to avoid heating during the process under the TAE buffer.Different constituting DNA strands corresponding to the final self-assembly of DNA-NWs should not separate or segregate upon applied voltage and current(75 V,20 A).This finding served as an indication of the integrity and strength of the self-assembly of DNA strands and the firm structure of the DNA-NW[17-19].
2.5.Procedure to assess CPT loading onto DNA-NWs
Since the absorption peak of the DNA and the CPT by UV light lies very close to each other(260-280 nm),accurate measurements could be doubtful.An alternative method for UV analysis was used to avoid this potential confound.CPT possesses amino groups in its chemical structure that can be conjugated to the conjugating dyes’carboxylic groups.Hook conjugating dye,manufactured by G-Biosciences(USA)and supplied by Sangon Biotechnology company(Shanghai,China),was successfully conjugated to the DNA-NWs using purification columns.The loading of the FITC-tagged CPT onto the DNA-NWs was determined through UV spectroscopy in relation to the absorption peak of FITC near 500 nm[30].
2.6.AFM to physically characterize DNA-NWs
A Bruker-FastScan AFM(Bruker Scientific,USA)was used for the analysis.A total of 5μL of the final self-assembly(annealed)mixture was placed on the mica surface(freshlycleaved)to let DNA adhere to the mica surface for 5 min.It was followed by washing three times with the ultrapure water.Sample was air blown for drying to obtain a clean sample for AFM analysis.The imaging was performed by selecting three to four different locations,each of 5μm × 5μm areas to estimate the overall size,topology,surface characteristics,and size[31].
2.7.Fluorescence imaging to characterize CPT loaded DNA-NWs
For fluorescence imaging of the DNA-NWs,we loaded FITC-tagged CPT on the DNA-NWs.We analyzed CPT loading under double photon confocal microscopy after making a glass slide[32].
2.8.The quantitative loading efficiency of CPT onto DNA-NWs
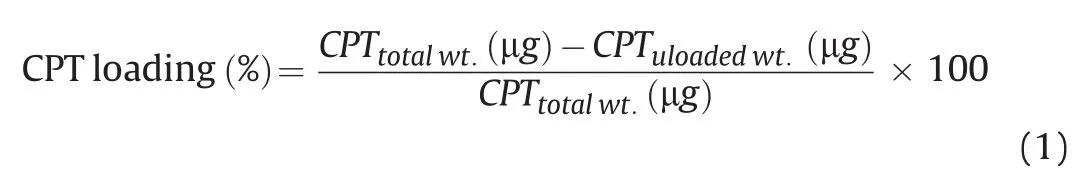
The mixture of CPT-loaded(FITC-tagged)DNA-NWs was incubated for 4 h,and then centrifuged at 14,000 rpm under reduced temperature and pressure to sediment the CPT-loaded DNA-NWs.The unbound CPT-FITC was estimated spectrophotometrically using a Nanodrop-2000c(Thermo Fisher Scientific,USA)from the supernatant mixture using Eq.(1)[22].
2.9.Cell cultures
We used the HepG2 cell line(rich in scavenger receptors)for our targeting experiments and the CHSE-214 cell line(lacking scavenger receptors)from the salamander embryo as a control cell line.Low-glucose Dulbecco’s modified-Eagle’s medium(DMEM;Sangon-Biotechnology Company,Shanghai,China)was used for culturing HepG2 cells.In contrast,high-glucose DMEM(Sangon Biotechnology Company,Shanghai,China)with 10% fetal bovine serum(FBS)(Sangon-Biotechnology Company,Shanghai,China)and 100 U/mL antibiotic mixture(penicillin-streptomycin)was used for CHE-214 cells.Cells were cultured in 5% CO2and 37°C in a humidified incubator(Thermo Fisher Scientific,USA).
2.10.Assessing cell viability,biocompatibility,and cytotoxicity using MTT analysis
2.10.1.Assessing the biocompatibility of the blank DNA-NW(without CPT loading)
To assess the biocompatibility of blank DNA-NW without CPT loading,we performed MTT-assays and flow cytometry experiments.HepG2 cells were cultivated at a cell density of 10,000 cells per well,using low-glucose with 10% FBS and antibiotics supplementation in a 96 well-plate under standard cell-culture conditions(37°C,5% CO2)for a half day.Following the cultivation step,we prepared different concentrations of blank DNA-NWs and placed them into wells,followed by 36-48 h incubation[33].The DNANW-treated adherent cells were washed twice with PBS after the removal of the growth medium.Cells were treated with 5mg/mL MTT reagent,followed by incubation for 3-4 h.The excess unreacted MTT was withdrawn,followed by the addition of 140-150μL DMSO to dissolve the crystalline product while incubating in the dark for 10-15 min.The plates were analyzed for UV absorption at 580-620 nm using a plate reader(Thermo Fisher Scientific,USA).Cell viability was analyzed using Eq.(2)[34].

2.10.2.Anti-cancer activity and cytotoxic analysis for the CPT-loaded DNA-NWs
The MTT assay was repeated to assess the cytotoxic potential of CPT-loaded DNA-NWs,keeping the same DNA-NW concentration as in the blank measurements with different CPT concentrations[34].
2.11.Flow cytometry assay for assessing cell proliferation and apoptosis
For the flow cytometry assay for assessing cell proliferation and apoptosis,HepG2 cells were grown in 6-well plates maintaining a cell density of approximately 1×106cells/well at standard humidified cell-culture conditions(37°C and 5% CO2)for one day.The cultured cells were treated with the CPT loaded DNA-NWs(PI-tagged)for 48 h.The treatment group of cells were compared with the control group of cells receiving blank DNA-NWs(PI-tagged)without prior CPT loading.To make the single cell suspension,the adhered cells were detached by the trypsin treatment for 2 min at 37℃.The pellets of the cells obtained through the centrifugation were washed with PBS twice.The washed cells were then re-suspended in the PBS.The sample was then subjected to a flow cytometry assay(Becton Dickinson,San-Jose CA,USA)for cell counting measurements using parameters set according to the PI dye fluorescence range[4].
2.12.Cell imaging using double-photon confocal laser microscopy
The FITC-tagged CPT was loaded onto the DNA-NWs.Two types of cell lines were selected.The HepG2 cell line was chosen to investigate scavenger receptor-mediated internalization of DNANWs and cancer cell killing after being treated with DNA-NW solution for 12 h.A control cell line(CHSE-214)from the salamander embryo that lacked scavenger receptors was used as a control,which was hypothesized to show no DNA-NW internalization for 12 h[35].
2.13.Statistical interpretations
All of the experiments were reproduced thrice to achieve statistical Significance.
3.Results and discussion
3.1.Confirmation of successful self-assembly of the DNA triangles into DNA-NWs and CPT loading
After the polymerization of DNA-triangular tiles,the resulting giant DNA nanolattices were assessed by gel electrophoresis.Next,we observed the restricted electrophoretic movement of the polymerized DNA nanolattices down the native-PAGE gel(a gel without urea).This movement provides evidence for the successful self-assembly of DNA triangular tiles into a large molecular weight DNA lattice[36].It also provides a clue about the strength and integrity of the resulting DNA lattices,as constituting strands should not segregate under the applied voltage and current down the gel.Therefore,native PAGE analysis gave us an initial clue about the self-assembly oftriangular DNA tiles by observing the restriction of the electrophoretic movement of the self-assembled DNA-NW samples down the gel(Lane 5)compared to other short monomeric strands with different lengths(Lanes 2 to 4),and strands in the DNA marker(Lane 1)moving down the gel to the greater distances[20],as shown in Fig.3A.

Fig.3.(A)Electrophoretic analysis of the self-assembly of DNA triangles through native PAGE.(B)UV peak shift analysis to confirm CPT loading onto DNA-NWs.(C)Percentage of CPT release from the DNA-NWs in the different media.
The shifts confirmed the loading of CPT onto DNA-NWs in the UV peaks of the DNA(260-300 nm),CPT(280 nm),and the FITC dye(500-520 nm)that was tagged to the CPT for loading[22],as shown in Fig.3B.
The CPT release study from the DNA-NWs in the different media was conducted at physiological temperature(37°C)and pH(7.45)to investigate the stability of CPT loading onto DNA-NWs for 5 h,as shown in Fig.3C.The release of CPT was comparable to that of TE buffer(without serum)in the case of FBS as well as cell culture medium(with serum).In all three cases,the release of CPT was less than 75% after 6 h,indicating the stability of CPT loading onto DNANWs[37].
3.2.AFM and confocal characterization of DNA-NWs
DNA is a double-helical form that behaves differently in different designs of DNA nanotechnology structures.As a general rule,our designed DNA triangles were supposed to be arranged into DNA nano-sheets or nanoribbon lattices after self-assembly.However,as shown in the AFM images in Fig.4A,two-dimensional DNA nano-sheets folded onto itself to give three-dimensional DNA-NWs due to the curvature of the DNA[38].
Confocal imaging of the DNA-NWs after loading with the FITC-tagged CPT is illustrated in Fig.4B.We observed the effective CPT loading capability of the DNA-NWs.The AFM and confocal microscopic images revealed a uniform shape and size distribution of the DNA-NWs with lengths ranging from 2 to 4μm and diameters ranging from 150 to 300 nm[24],shown in Figs.4A and B.
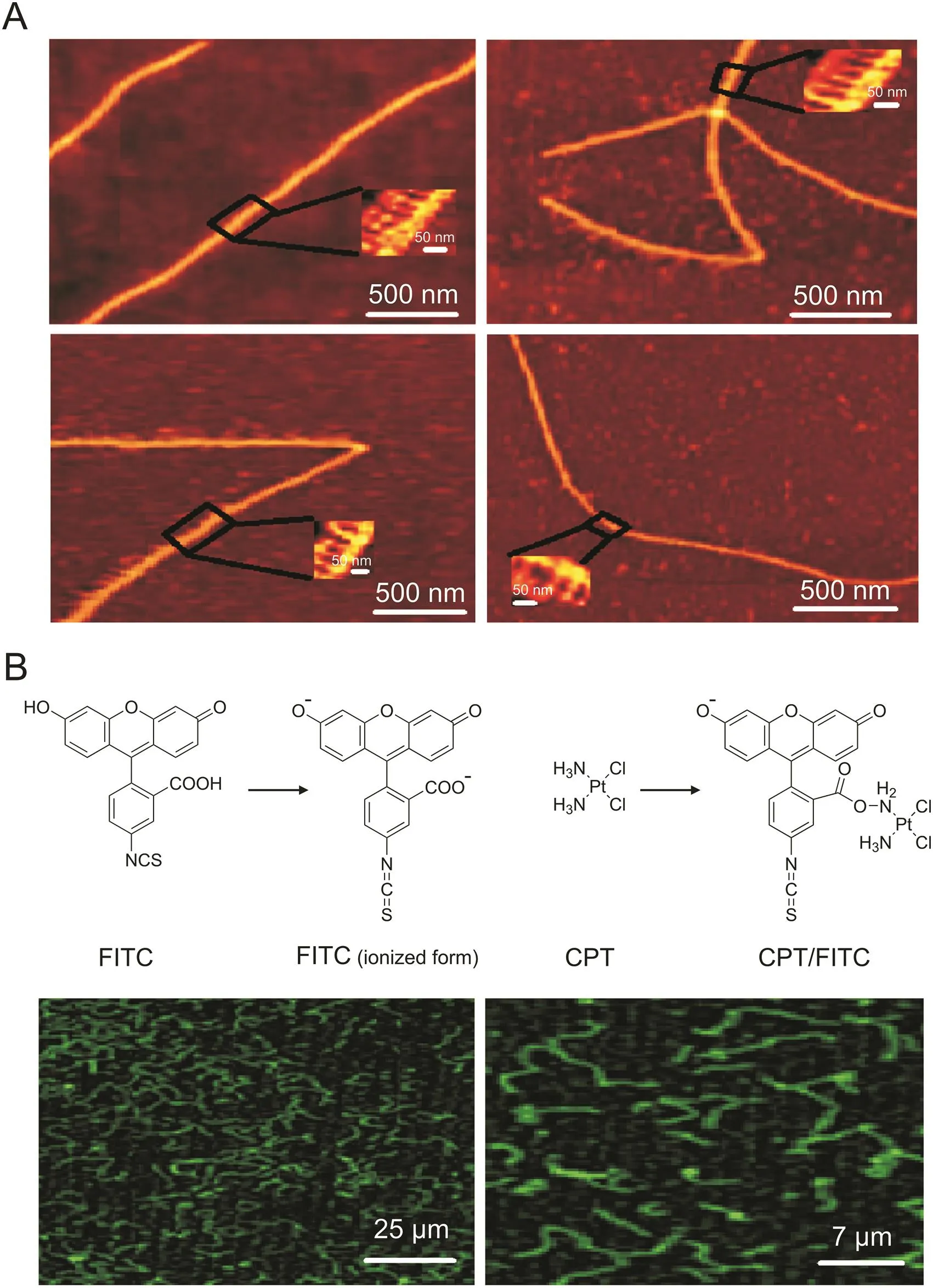
Fig.4.AFM and confocal characterization of DNA-NWs.(A)AFM imaging of DNA-NWs.(B)Confocal imaging of FITC-tagged CPT-loaded DNA-NWs.
3.3.Loading efficiency of CPT
UV absorption of CPT and DNA is close to each other at approximately 260-300 nm,making it challenging to analyze the CPT loading onto DNA-NW through UV spectrophotometry.To confirm CPT loading onto DNA-NWs,CPT was first conjugated with FITC using hook self-conjugating dyes(G-Biosciences,USA)with the purification columns available with the product.CPT has amino groups in its chemical structure,which is tagged with FITC by amide chemistry.Therefore,CPT loading was determined by UV analysis for the absorption of FITC in the range of 500-520 nm.After vacuum centrifugation at 14,000 rpm and 4°C for 10 min of the CPT-loaded DNA-NWs,the unbound CPT in the supernatant was removed.The CPT loading onto DNA-NW settled at the bottom and was measured using UV analysis[39].The efficiency of CPT loading onto the DNA-NWs was calculated as 66.85%-97.35% for different concentrations of the CPT(0.1-51.2 nM)for various formulations(C1 to C10 respectively)(Table 2),keeping the DNA-NW concentration constant(20μM).As a general trend,an increase in CPT concentration from the C1 to C10 formulations resulted in a decrease in the loading efficiency,keeping the DNA-NW concentration constant(30μM).The loading efficiency was observed to be 97.35% in the case of 0.1 nM CPT(C1 formulation)to 66.85% in the case of 51.2 nM(C10 formulation),at room temperature and physiological pH(pH 7.45).This finding might be due to the limited availability of binding sites and exposed pockets for CPT binding provided by consecutive G and C nucleotides,especially near the major and minor grooves of the double helix[19].
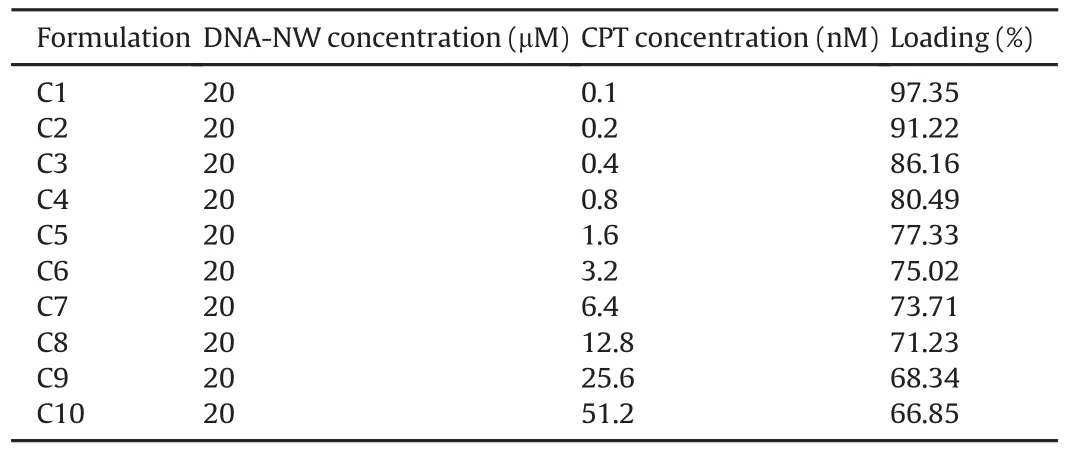
Table 2 CPT loading(%)of various formulations(C1-C10)with different concentrations of CPT.
3.4.CPT docking simulation to bind with the DNA-NW
CPT is an inorganic drug that has excellent DNA-binding capability.We performed molecular docking of CPT binding with the DNA-NWs using autodock-vina/software from the template 3lpv.cif obtained from the protein-databank(https://www.rcsb.org/).Each CPT molecule can form two hydrogen bonds with either A,G,or C.However,the bond of CPT with C is stronger,with a bond length of 1.992 Å,as shown in Fig.5.The bond length of CPT with the G was noted to be from 2.073 to 2.108 Å.CPT can bind to the DNA nanostructure at different positions,including its minor and major grooves.The cavitiesposed bythe G and C nucleotides turned out to be the pockets for CPT deposition,as shown in Fig.5.Hence,CPT incubation with the DNA-NW resulted in effective loading onto DNA-NWs.Thus,CPTexhibited excellent affinity with the DNA-NW,as evidenced by the UV analysis[40,41],as shown in Fig.3B.
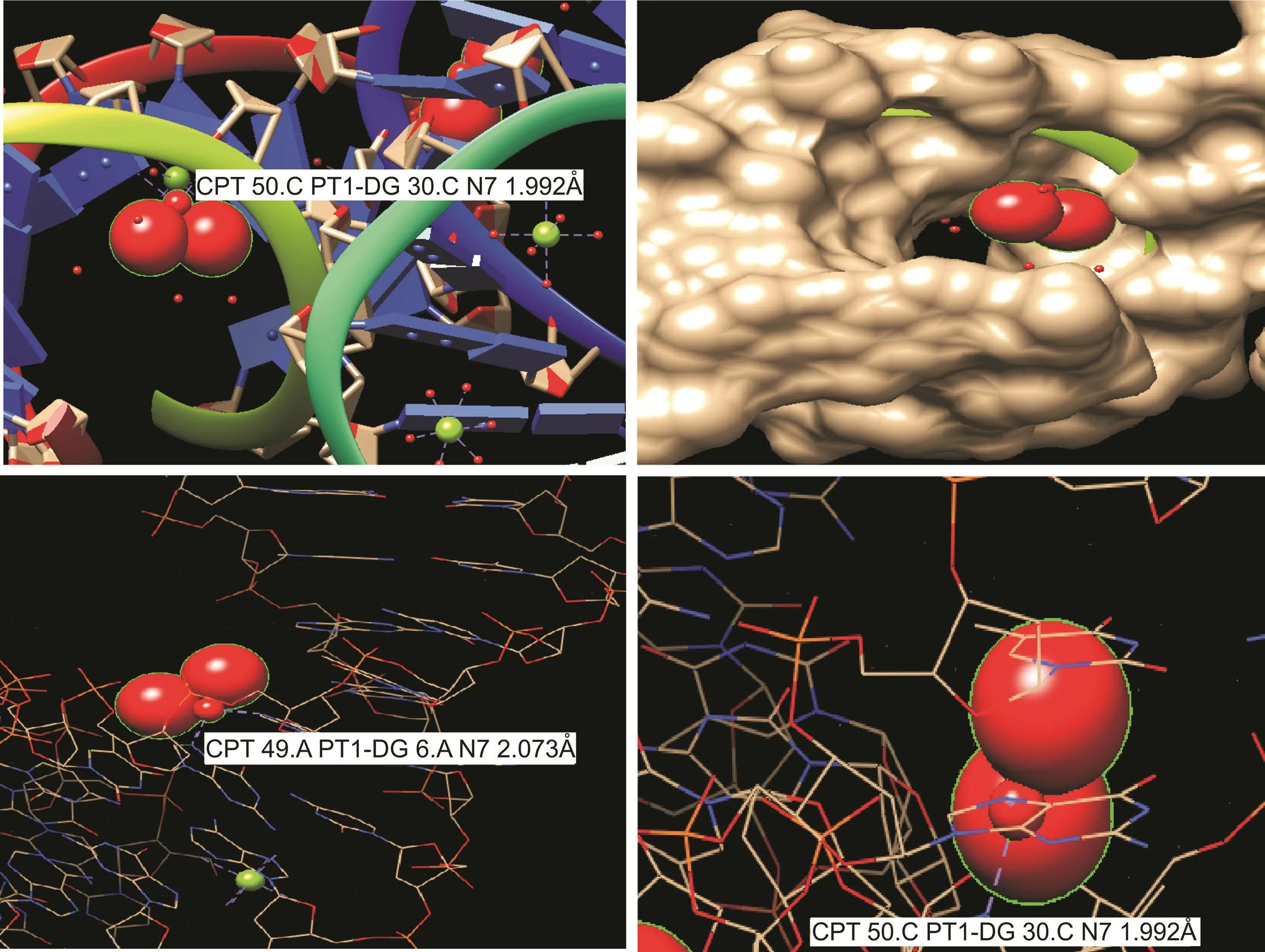
Fig.5.CPT docking simulation to bind with the DNA-NW.Different bond lengths can be seen at different locations between the CPTand the DNA structure.The deposition of CPT in the DNA matrix is visible.
3.5.Biocompatibility and cytotoxicity analysis of blank and CPT-loaded DNA-NW
The MTT test and flow cytometry were used to examine the biocompatibility and cytotoxicity of the blank DNA-NW and CPT-loaded DNA-NW.Various blank DNA-NW concentrations were applied to HepG2 cells for 24-48 h to observe cellular apoptosis.Using the MTT assay illustrated in Fig.6A,the blank DNA-NW was biocompatible from a very low concentration of 10μM to a very high concentration of 640μM.However,the formulations with relatively high CPT(51.2 nM)contents(C10 formulation in Table 2)showed significantly higher cytotoxicity in a targeted manner than the free CPT solution.However,with the lower CPTconcentrations,the cytotoxicity of the CPT-loaded DNA-NWs still significantly surpassed the free CPT solution.This finding might be due to decreased cell internalization of free CPT when used in lower concentrations(0.1-0.4 nM),resulting in slower first-order cell uptake of free CPT.
In the case of DNA-NWs loaded with CPT,polyanionic DNA-NWs showed better cell internalizations,even at extremely low concentrations,due to the binding of DNA-NWs with the scavenger receptors.It can be seen from the MTT results in Fig.6B.The DNANW nanocarrier was efficient in terms of biocompatibility(Fig.6A).It effectively enhanced CPT cytotoxicity at lower concentrations with an IC50of 12.8 nM,compared with free CPT,exhibiting an IC50of 51.2 nM(Fig.6B).Similar findings were observed in the flow cytometry experiments shown in Fig.6C.Empty DNA-NWs were highly biocompatible even at 640μM,with 86% of the cells alive in the Q4 quadrant.These findings were similar to those of the blank solution(PBS),showing 89% of live cells.While CPT-loaded DNA-NW referring to formulation C7 with a CPTconcentration of 6.4 nM caused late apoptosis in 22% of the cells in the Q2 quadrant.This finding suggests enhanced cytotoxicity in the case of CPT-loaded DNA-NWs compared with that of free CPT solution,as evident from the MTT assay results in Fig.6B.However,late apoptotic effects might be due to the time required to internalize CPT-loaded DNA-NWs and its lysosomal digestion and sustained release of CPT in the cells.However,this technique might be useful for in vivo administration to avoid CPT’s cytotoxic effects on healthy cells.Hence,DNA-NW is an efficient nanocarrier in terms of biocompatibility and enhances CPT’s cytotoxicity,even at very low concentrations.DNA-NWswith nanoscale-sized structureslarge enough to load CPTeffectively,and a dense and rigid nanomaterial,were used to load CPT in this study.In addition,dense DNA nanotechnology structures have been reported to be resistant to hydrolysis in body fluids,including blood and serum.Based on our in vitro evaluation,we believe that if applied in vivo,our DNA-NW design might have desired outcomes to target tumor cells without harming healthy cells[34].

Fig.6.Biocompatibility and cytotoxicity assay of blank and CPT-loaded DNA-NW.MTT assay of(A)biocompatibility of blank DNA-NWs and(B)cytotoxicity of CPT-loaded DNA-NWs compared with equivalent concentrations of free CPT solution.(C)Flow cytometry assessment of the biocompatibility of blank DNA-NW and the cytotoxicity of CPT-loaded DNANW.
3.6.Binding and internalization of DNA-NW in cells rich in scavenger receptors
DNA-NWs loaded with CPT(FITC-tagged)were applied to HepG2 cells that are rich in scavenger receptors.As a polyanionic drug carrier,DNA-NWs bind with the scavenger receptors on the cell surface,as illustrated in Fig.7A.DNA-NWs were slowly internalized into cells with the time-dependent sustained or prolonged release of CPT within the cells,as evidenced by the gradual rise of green fluorescence of the FITC-tagged CPTon the cell surface after a 30 min interval as compared with the CHSE-214 cell line that lacks scavenger receptors.Hence,no internalization of DNA-NW was observed,as illustrated in Fig.7B[9].
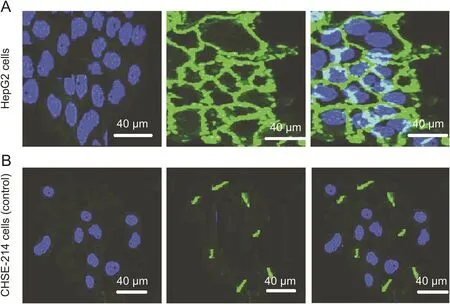
Fig.7.Surface binding of DNA-NW loaded with FITC-tagged CPT to the human liver cancer cell line(HepG2)compared with the control cell line(CHSE-214)derived from the salmon embryo after a 30 min interval,scale bar=40μm.(A)As HepG2 cells are rich in scavenger receptors,DNA-NW binds the cell surfaces followed by gradual internalization into the cells and release of FITC-tagged CPT into the cytoplasm.(B)As CHSE-214 cells lack scavenger receptors,DNA-NW could not bind these cells,resulting in no cell internalizations.
DNA-based nanomaterials are gaining importance as drug delivery carriersbecause oftheirprecise nanoscale size and controlled shape.Some drugs such as dox and CPT have an intrinsic ability to bind with the DNA double helix,which can be effectively loaded onto DNA-based carriers.Furthermore,the polyanionic nature of DNA makes internalization by healthycells difficult[4].As cancer cells have a high proportion of polyanionic ligand-binding scavenger receptors,cancer cells can be effectively targeted[14].DNA-NWs are nanoscale-sized,but big enough to load CPT effectively.DNA nanowires have a dense and rigid topology and are a suitable nanomaterial for CPT loading.Dense DNA nanotechnology structures are reported to be resistant to hydrolysis in body fluids,including blood and serum.Each triangle had 132 base pairs.One structural unit consists of 4 triangles containing 528 base pairs.Each nanowire has 10 to 25 structural units containing 5280 to 13,200 base pairs,with molecular weights of the DNA-NWs(polymer)of 1-4 million[42].
4.Conclusions
Nano-architecture designed using a DNA-nanotechnology approach is receiving the attention of drug delivery scientists.This attention is due to the controllable nanoscale size,geometry,and drugloading and DNA-based vehicles’binding abilities.We designed DNA-NWs ranging in diameter from 150 to 300 nm and a length from 2 to 4μm forloading CPT(loadingefficiency66.85%-97.35%)for anti-cancer resistance therapy.As many cancer cells,such as HepG2 cells,have large proportions of scavenger receptors that are polyanionic,they can be targeted using polyanionic drug delivery vehicles based on DNA nanomaterials.Being polyanionic nanocarriersfor CPT,our DNA-NWs showed better cytotoxic effects and HepG2 cell internalization than the control cells(the CHSE-214 cell line).The apoptosis of the HepG2 cells with the CPT-loaded DNA-NWs was enhanced to 22% compared with the 7% apoptosis in the case of blank DNA-NWs.CPT-loaded DNA-NWs showed sustained release of CPT(75%)for 6 h in the presence or absence of serum(FBS)containing cell culture media is an evidence of its stability in serumcontaining biological media.Hence,DNA-NWs turned out to be very effective nanocarrierin termsofbiocompatibilityand enhancing the cytotoxicity of CPT effectively at very low concentrations(IC50of 12.8 nM)compared to the free CPTexhibiting an IC50of 51.2 nM(see the results in Fig.6).Based on our in vitro evaluation,if applied in vivo,our designed DNA-NWs might produce ideal outcomes in targeting tumor cells without harming healthy cells.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
All the authors acknowledge the State Key Laboratory of Analytical Chemistry for Life Sciences,Nanjing University,China,and the State Key Laboratory of Pharmaceutical Biotechnology,Nanjing University,China,for support.
猜你喜欢
杂志排行
Journal of Pharmaceutical Analysis的其它文章
- Development of a rapid GC-FID method to simultaneously determine triethylamine,diisopropylamine,and 1,1,3,3-tetramethylguanidine residues in an active pharmaceutical ingredient
- Diversity,novelty,antimicrobial activity,and new antibiotics of cultivable endophytic actinobacteria isolated from psammophytes collected from Taklamakan Desert
- Taxifolin stability:In silico prediction and in vitro degradation with HPLC-UV/UPLC-ESI-MS monitoring
- Neuroprotective effects of Ginkgo biloba dropping pills in Parkinson’s disease
- Antimicrobial activity and mode of action of terpene linalyl anthranilate against carbapenemase-producing Klebsiella pneumoniae
- Herb-drug interaction in the protective effect of Alpinia officinarum against gastric injury induced by indomethacin based on pharmacokinetic,tissue distribution and excretion studies in rats
