Automatic Detection of COVID-19 Infection Using Chest X-Ray Images Through Transfer Learning
2021-04-14EleneFirmezaOhataGabrielMaiaBezerraJoVictorSouzadasChagasAlosioVieiraLiraNetoAdrianoBessaAlbuquerqueVictorHugodeAlbuquerqueSeniorMemberIEEEandPedroPedrosaRebouasFilhoMemberIEEE
Elene Firmeza Ohata, Gabriel Maia Bezerra, João Victor Souza das Chagas, Aloísio Vieira Lira Neto, Adriano Bessa Albuquerque, Victor Hugo C. de Albuquerque, Senior Member, IEEE, and Pedro Pedrosa Rebouças Filho, Member, IEEE
Abstract—The new coronavirus (COVID-19), declared by the World Health Organization as a pandemic, has infected more than 1 million people and killed more than 50 thousand. An infection caused by COVID-19 can develop into pneumonia,which can be detected by a chest X-ray exam and should be treated appropriately. In this work, we propose an automatic detection method for COVID-19 infection based on chest X-ray images. The datasets constructed for this study are composed of 194 X-ray images of patients diagnosed with coronavirus and 194 X-ray images of healthy patients. Since few images of patients with COVID-19 are publicly available, we apply the concept of transfer learning for this task. We use different architectures of convolutional neural networks (CNNs) trained on ImageNet, and adapt them to behave as feature extractors for the X-ray images.Then, the CNNs are combined with consolidated machine learning methods, such as k-Nearest Neighbor, Bayes, Random Forest, multilayer perceptron (MLP), and support vector machine (SVM). The results show that, for one of the datasets, the extractor-classifier pair with the best performance is the MobileNet architecture with the SVM classifier using a linear kernel, which achieves an accuracy and an F1-score of 98.5%. For the other dataset, the best pair is DenseNet201 with MLP,achieving an accuracy and an F1-score of 95.6%. Thus, the proposed approach demonstrates efficiency in detecting COVID-19 in X-ray images.
I. INTRODUCTION
THE COVID-19 pandemic has become a severe health problem being at the center of media cover since December, 2019 [1], [2]. In about 74% of the cases, the COVID-19 causes mild (18%) or moderate (56%) symptoms[3]. However, the remainder of the cases range from critical(20%) to severe (6%) [3]. As of today (2020-04-03), the total number of registered cases is approximately 1 015 667, with 53 200 deaths worldwide and 212 991 cases where a full recovery was achieved. Moreover, the number of active cases is 749 476 [4], [5].
The main symptoms presented by suspected infections include respiratory distress, fever, and cough. The virus may also cause pneumonia in more aggressive infections. Besides pneumonia, the infection can lead to severe acute respiratory syndrome, septic shock, multi-organ failure, and, ultimately,death [6]. Studies showed that men (about 60%) were more affected than women (about 40%), and that there were, up to this point, no significant death rates in children younger than nine years old [2]. Despite being developed, many first world countries have been facing a collapse of the healthcare system due to escalating demand for intensive care units simultaneously [7], [8].
Virus tests take less and less time as new technologies are developed world-wide. The diagnostic of COVID-19 infections involves a chest scan to verify the lung condition, in such a way that, if the patient shows pneumonia in the scans,they are deemed to have a COVID-19 infection. This method allows authorities to isolate and treat affected patients in a timely and affirmative fashion [9].
One of the available methods to detect pneumonia is a computed tomography scan of the chest (CT scan). Automated image analysis based on artificial intelligence is being developed to detect, quantify, and monitor COVID-19 infections, as well as to separate healthy lungs from diseased ones [10]. Ke et al. [11] use the image’s basic characteristics and analyze neural network co-working with heuristic algorithms. The method is divided into the following steps:first, an initial analysis of the possibility of detecting respiratory disease through basic descriptors with a neural network, then, the use of heuristic algorithms for the rapid detection of affected lung tissues, since the possibility of detection is considerable. Poap et al. [12] and Shan et al. [13]developed segmentation studies based on a heuristic and a deep learning method, respectively. These studies seek to segment all-region on a lung that presented infection, isolating the sick region from the rest and thus conduct studies of patterns unique to that region, helping to identify the region in a new sample. Xu et al. [14] aspired to develop an early screening model that was capable of differentiating COVID-19 pneumonia, Influenza-A pneumonia, and healthy lungs using CT scan images and deep learning techniques. A study by Wang et al. [15] developed a deep learning method based on the changes presented in COVID-19 patients’ CT scans that can acquire graphical features and provide clinical diagnosis much faster than waiting for the pathogen test.Following the same basis, Chouhan et al. [16] use different neural network models pre-trained on ImageNet to extract exam characteristics. These characteristics are used to obtain individual classification results for each network. The combination of each network’s results uses the majority vote,so the diagnosis corresponds to the class that achieved the highest number of votes.
X-ray is an imaging technique that is used to investigate fractures, bone displacement, pneumonia, and tumor. X-rays have been used for many decades and provide an astonishingly fast way of seeing the lungs and, therefore, can be a helpful tool in the detection of COVID-19 infections [8],[17]. They are capable of generating images that show lung damage, such as from pneumonia caused by the SARS-CoV-2 virus [18]. Since X-rays are very fast and cheap, they can help to triage patients in places where the healthcare system has collapsed or in places that are far from major centers with access to more complex technologies. Furthermore, there are portable X-ray devices that can be easily transported to where it is needed [18]. CT scans make use of the principles of X-ray in an advanced manner to examine the soft structures of the body. It is also used to obtain clearer images of organs and soft tissues [19]. On the other hand, X-rays use less radiation[20], thus using an X-ray is faster, less harmful, and presents lower cost than a CT scan. Narin et al. [8] proposed an automatic detection of COVID-19 using chest X-rays and CNNs. Apostolopoulos et al. [17] also proposed the automatic detection of the disease but analyzing three classes: COVID-19, common pneumonia, and normal conditions.
In this paper, we propose an automatic system to classify chest X-ray images as from COVID-19 patients or healthy patients using transfer learning with convolution neural networks (CNNs). We performed 144 experiments, which are a combination of 12 CNNs and six classifiers in two datasets.The results show that MobileNet combined with support vector machines (SVM) (Linear) achieved the highest accuracy (98.462%) in one dataset, and the combination DenseNet201 with MLP achieved 95.64% in the second dataset, showing the effectiveness of the proposed approach.
This paper is organized as follows: Section II presents the transfer learning method. Section III describes the proposed methodology of the approach, detailing the dataset, the steps for the feature extraction and classification, and the metrics used to evaluate the approach. Section IV discusses the results. Lastly, Section V presents the conclusion and future works.
II. TRANSFER LEARNING WITH CONVOLUTIONAL NEURAL NETWORKS
Transfer learning is a method that utilizes the knowledge acquired by a CNN from a specific problem to solve a distinct but similar task. This transferred knowledge is used in a new dataset, whose size is usually smaller than the adequate size to train a CNN from scratch [21].
In deep learning, this method requires an initial training of a CNN for a given task, using large datasets. The availability of a sizable dataset is the main factor to ensure the success of the method since the CNN can learn to extract the most significant features of a sample. The CNN is deemed suitable for transfer learning if it is found to be able to extract the most important image features [22].
Then, in the transfer learning, the CNN is used to analyze a new dataset of a different nature and extract its features according to the knowledge acquired in the first training. One common strategy to exploit the capabilities of the pre-trained CNN is called feature extraction via transfer learning [23].This approach means that the CNN will retain its architecture and weights between its layers; therefore, the CNN is used only as a feature extractor. The features are later used in a second network/classifier that will process its classification.
The transfer learning approach is mostly used to work around computational costs of training a network from scratch or to keep the feature extractor trained during the first task. In medical applications, the most accepted practice of transfer learning is to utilize the CNNs that achieved the best results in the ImageNet large scale visual recognition challenge(ILSVRC) [24], which assesses algorithms for object detection and classification in large scales. The use of large datasets for initial training of the network enables high performance in smaller datasets. This performance is linked to various extraction parameters that are typically not allowed as they cause overfitting of the network [25]. That said, feature extraction performed with transfer learning allows a large number of features to be extracted by generalizing the problem and avoiding excessive adjustments [26].
The use of transfer learning also allows the use of the internet of things (IoT) systems to classify medical images.For example, Dourado Jr et al. [27] proposed an IoT system to detect a stroke in CT images. Rodrigues et al. [28] used the system proposed by Dourado Jr et al. [27] to classify EEG signals.
The transfer learning method is used in the feature extraction step for the COVID-19 detection. The process is detailed in Section III-B.
III. METHODOLOGY
In this section, we present the proposed methodology for classifying an X-ray as being of a healthy patient or a patient affected by COVID-19. First, we describe the datasets of images used in this study. Then, we explain the process of feature extraction, which is based on the transfer learning theory. After that, we present the classification techniques applied and the steps of their training process. Lastly, we define the metrics we use to evaluate the results and to compare it to other approaches. Fig. 1 presents the infographics of the proposed approach; each step is explained in the next subsections.
A. Datasets
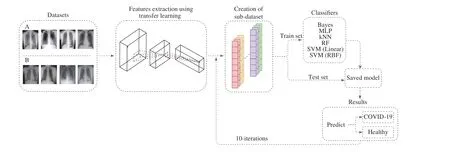
Fig. 1. Infographics of the proposed approach.
In our study, we use frontal-view chest X-ray images. Only posterior-anterior (PA) and anterior-posterior (AP) X-ray views were collected. We divided the samples into two classes: X-ray images of patients diagnosed with COVID-19 and X-ray images of healthy patients. For better evaluation of the proposed method, we built two datasets: Dataset A and Dataset B. Both datasets have the same images for the COVID-19 class, but they have different images for the healthy class. In both datasets, the classes are balanced,consisting of 194 images for each class or 388 images for each dataset.
In Dataset A, the COVID-19 class is composed of 194 chest X-ray images of patients diagnosed with COVID-19, which were collected from different sources [29], [30]. Both these sources were accessed on 2020-03-31. They consist of compilations of X-ray images taken from different papers,databases, and other sources. For this dataset, we collected the set of chest X-ray images of healthy patients from the “Chest X-ray Images (Pneumonia)” challenge available on Kaggle[31]. We randomly selected 194 samples from the X-ray images labeled as “normal” , which correspond to healthy patients. This source was chosen since it has been commonly used in related works that propose methods of detecting COVID-19 in X-rays [17], [8]. However, all the X-ray images from this source are of pediatric patients. Since the X-ray images of the COVID-19 class are mostly of adult patients,we built another dataset with patients of a similar age range.
In Dataset B, as previously mentioned, the COVID-19 class images are the same as in Dataset A. However, due to the age difference between healthy patients and patients with COVID-19 present in Dataset A, we collected chest X-ray images from a different source for Dataset B. For this dataset, we took images from the “NIH Chest X-rays” challenge organized by the National Institutes of Health and available on Kaggle [32].We randomly selected 194 images from the class of “no findings”, which correspond to healthy patients.
All images from the datasets are either in the joint photographic experts group (JPG/JPEG) or in the portable network graphics (PNG) format. The image resolution is varied within the dataset, with resolutions as low as 249 by 255 pixels and as high as 3520 by 4280 pixels. However, all images were pre-processed using the resizing technique. In Table I, we can see the sizes to which the images were resized for each specific CNN. The equipment used to take the X-rays is also diverse and is often not determined; more information about it can be found on the image sources [29]-[32]. We present examples of images from the datasets in Fig. 2. An example of a chest X-ray of a patient with coronavirus disease with PA view is shown in Fig. 2(a) and AP view is shown in Fig. 2(b), an example of a chest X-ray of a healthy patient from Dataset A is shown in Fig. 2(c), and an example of a chest X-ray of a healthy patient from Dataset B is shown in Fig. 2(d).
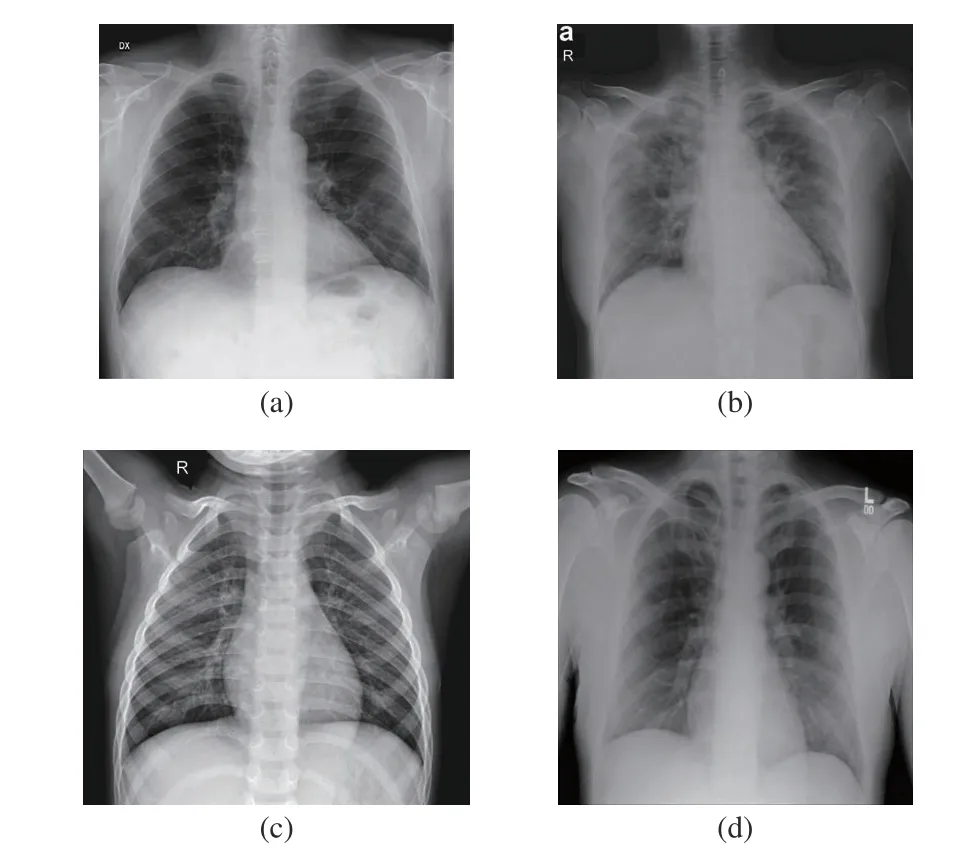
Fig. 2. Samples from the dataset used in this study. (a) X-ray with PA view of a patient with COVID-19; (b) X-ray with AP view of a patient with COVID-19; (c) X-ray of a healthy patient from Dataset A; (d) X-ray of a healthy patient from Dataset B.
In order to have more input data to create a more generalized model, data augmentation was implemented. The augmentation of training set is a widespread technique in the literature [41]. The images were randomly selected to undergo a transformation. The most commonly used transformations for data augmentation are the affine transformations [42]. The affine transformations applied in this study were rotation,change in width, height, and magnification.
B. Feature Extraction Steps
For extracting features from the X-ray images, we use the transfer learning concept discussed in Section II. Firstly, we select different CNN architectures that achieved excellent performance on the ImageNet dataset. Secondly, we choose different configurations, previously trained on ImageNet, from the selected CNN architectures. Thirdly, we remove any fully connected layers from these configurations, leaving only convolutional and pooling layers. These two types of layers are responsible for extracting features from the image, while the fully connected ones are responsible for classifying the features and, consequently, the image. Thus, removing these layers is necessary to turn a CNN into a feature extractor.After this step, the new output of the adapted CNN is a set of features extracted from an input image.
For each CNN configuration, we create a sub-dataset composed of sets of features extracted from each image of the original datasets. In order to build a sub-dataset, we first resize each image according to the input size required by the selected CNN. Then, each resized image is used as input to the CNN,and its set of features is extracted and stored in the corresponding sub-dataset. In Table I, we show all the CNN architectures and their respective configurations used. It is worth noting that InceptionResNetV2 [36] is a hybrid configuration, originated from Inception [34] and ResNet[35]. In Table I, we also present the input image size required by each configuration and the number of features extracted from a single image.
C. Classification Steps
In order to classify the X-ray images, we selected widely used machine learning methods in the literature: Bayes[43]-[45], random forest (RF) [46]-[48], multilayer perceptron (MLP) [49]-[51], k-nearest neighbors (kNN)[52]-[54], and SVM [55]-[57]. In the SVM classifier, we consider the linear and RBF kernels. It is noteworthy that these classifiers are from different types: kNN is instancebased, RF is based on the decision tree method, MLP is based on neural networks, SVM is based on finding an optimal hyperplane, and Bayes is based on probability and statistics.
The classification is performed in three steps: i) model training, ii) model testing, and iii) repetition of processes i)and ii).
Each sub-dataset is composed of features extracted from the extractors presented in Section III-B. These sub-datasets are divided into 80% for training and the remaining for testing.Furthermore, we applied the data augmentation in the training set of Dataset A. The number of images for train and test for each dataset is presented in Table II.
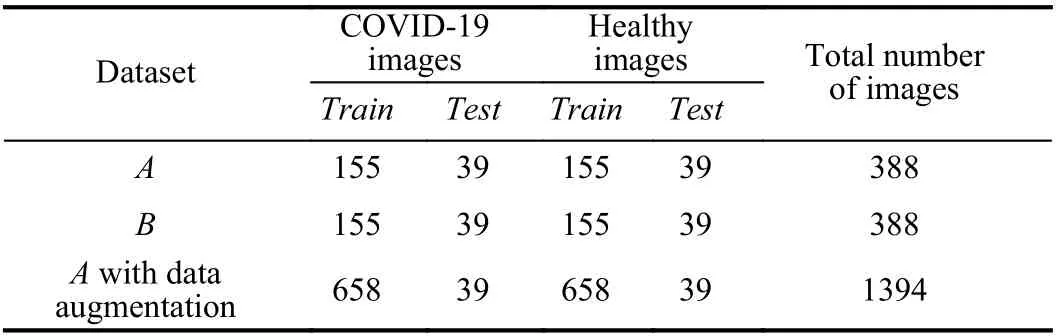
TABLE II DATA SPLIT ACCORDING TO DATASET AND CLASS
1) Model Training: In this step, we use 80% of the subdataset to perform the training of the model. We consider the setup for the hyperparameters presented in Table III to find the configuration of the classifiers on the training set. The classifiers that were configured for a random search perform a 20-iterations search. The hyperparameters for all classifiers,except for the Bayes classifier, are determined after 10-fold cross-validation. Then, each classifier has optimal hyperparameters, which are saved on the computer.
2) Model Testing: In this step, we perform a test in the remaining 20% of the sub-dataset using the saved classifiers.The system determines one class for each sample of the subdataset. In addition, the metrics are calculated in this step.
3) Repetition of Processes 1) and 2): The sub-datasets are randomly divided into other train and test sets. These sets are ensured to be different from the rest by the seed used. Then,we perform ten repetitions of steps 1) and 2).
D. Evaluation Metrics
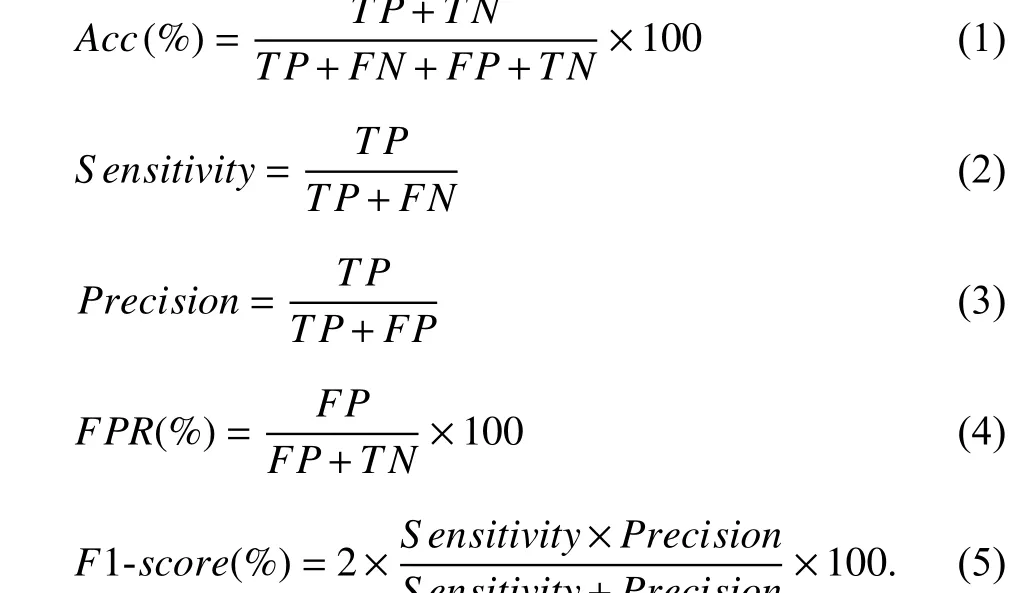
We analyze the results of this paper utilizing the metrics:accuracy (Acc), F1-score , and false positive rate (FPR).Accuracy describes how often the model is classifying correctly. F1-score can be described as the harmonic mean of Sensitivity and Precision; this metric can provide a number that suggests an overall quality of the approach. FPR indicates a rate of healthy patients being wrongly classified. True positives (TP) indicates the number of instances that the model classified the images as COVID-19 correctly. False negative (FN) corresponds to the number of occasions that the COVID-19 images were misclassified as from a healthy patient. False positives (FP) points out the number of times that the model wrongly classified a healthy patient. True negatives (TN) informs the number of healthy patient images that were correctly classified. The equations for Acc,Sensitivity, Precision, FPR, and F1-score are presented on(1)-(5), respectively.

TABLE III SETUP TO SEARCH FOR HYPERPARAMETERS OF THE CLASSIFIERS

TABLE IV METRICS OBTAINED BY CLASSIFYING FEATURES EXTRACTED FROM DATASET A BY DIFFERENT CNN ARCHITECTURES OF THE TOP FIVE RESULTS
In addition to the metrics already mentioned, we also analyze the training, extraction, and test times. The training time represents the length of the period it takes from the beginning of the classifier training to the moment it is ready to perform the classification. Extraction time measures how long the adapted CNN takes to output the attribute vector from the moment it receives the X-ray. Also, the test time is the duration it takes for the classifier to predict the image’s class after receiving its attribute vector. Thus, training time is vital during model building. After this step, the extraction and test times are more relevant. Their sum represents the classification time, which is the period between receiving the X-ray and returning its class.
IV. RESULTS
In this section, we investigate the results achieved by combining the features extracted by CNNs, applying transfer learning, and the classifiers. We executed 72 × 2 = 144 experiments, which are the combination of twelve CNNs and six classifiers in both datasets. The system infrastructure used was an Intel i7, 8 Gb of RAM, with Linux Ubuntu 16.04 system without a graphical processing unit (GPU).
Table IV shows the metrics and their standard deviations of the top 5 results after the 10-iterations of the steps described in Section III-C in Dataset A; the full table can be found in the following link: https://bit.ly/2w9DEpT. We applied Friedman test on the accuracy results of Dataset A to test if there is no statistical difference among the results of classifiers. There is a significant effect at p <0.05 level found for the classifiers(F=30.73, p=0.001). This result is confirmed by the Nemenyi post-hoc test (C D=2.18), which is presented in Fig. 3. We can observe that there is no consistent indication of statistical differences between SVM (RBF), SVM (Linear),MLP, and RF for the 144 experiments evaluated.
Analyzing Table IV, we can observe that all combinations in the top five achieved, reaching a minimum Acc of 98.205%and minimum F1-score of 98.205%. However, the combination that should be highlighted is MobileNet with SVM with the linear kernel, since it reached a maximum Acc of 98.462%, a maximum F1-score of 98.461%, and an FPR of 1.026%.
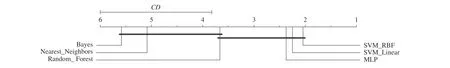
Fig. 3. Result of Nemenyi test for the classifiers on Dataset A.
In the full table of Dataset A provided in the link, we can observe that the CNNs architectures that achieved a minimum of 95% in Acc and F1-score independently of the classifier were: VGG16, DenseNet201, DenseNet169, and MobileNet,showing the effectiveness of the proposed approach. We can also observe that RF achieved the slowest test times independently of the CNN architecture, which is related to its high number of estimators.
In Table IV, we can observe that MobileNet with SVM(Linear) reached a test time of 0.443 ms and an extraction time of 21 ms; then, this time is attractive for real-time implementations, since it would take, approximately 21.443 ms to define if an image is from class COVID-19 or Healthy.Furthermore, although the system infrastructure does not have GPU and its configuration is not high-end, the proposed approach achieved satisfactory extraction and training times,then clinics and hospitals do not need to acquire new equipment for a system to aid in the medical diagnosis.
Table V presents the final confusion matrix of the MobileNet with SVM (Linear). It is a sum of the 10 confusion matrices from the 10-iterations of Section III-C. Therefore, the confusion matrix shows that even after 10-iterations, the combination MobileNet with SVM (Linear) did not have many FPs and FNs. In addition, we can observe on Table IV that this combination reached an FPR of 1.026 %. In a reallife application, this means that not many patients will be misclassified as not infected, then reducing the spread of the disease and allowing them to have the proper treatment. Also,patients without COVID-19 will rarely be submitted to more exams or admissions in dedicated locations to COVID-19,then decreasing the probability of their contamination.
Table VI shows the top five results of Dataset A with data augmentation; the full table can be found in the following link: https://bit.ly/2w9DEpT. We can observe that with data augmentation, the highest Acc only increased 0.5%, with the pair DenseNet121 with MLP, which achieved 98.974%.Nonetheless, the proposed approach with data augmentation reached more combinations with 98% of Acc, showing that the use of the CNN to extract features is effective. Another point that can be observed is that the FPR rates are slightly lower in average than those achieved with the dataset without data augmentation, showing that the model was able to better generalize the problem.

TABLE V FINAL CONFUSION MATRIX OF THE TEST SET FOR THE CLASSIFICATION OF CHEST X-RAY IMAGES AS HEALTHY OR COVID-19 FOR MOBILENET WITH SVM (LINEAR) FOR DATASET A
Since most of the papers that are going to be compared in Section IV-A do not use the same source of healthy X-ray images as Dataset B, Table VII is a summary of the top five results, ordered by Acc; the tiebreaker was the F1-score. The complete results can be found in the following link:https://bit.ly/2w9DEpT.
Analyzing Table VII , we can observe that even though images in Dataset B have similar contrasts and many artifacts in both classes, the transfer learning method combined with consolidated machine learning methods could achieve an Acc of 95.641% and an F1-score of 95.633% through the DenseNet201 architecture with the MLP classifier.
Table VIII presents the confusion matrix for the combination DenseNet201 with MLP for Dataset B; it shows that the errors are balanced. Table IX displays the confusion matrix for the combination DenseNet201 with SVM (Linear)for Dataset B; even though the Acc of this combination is close to that of the combination DenseNet201 with MLP, the combination DenseNet201 with SVM (Linear) is not desirable because more COVID-19 patients are being classified as healthy, which could contribute to the spread of the disease.This difference can also be noted on FPR, where the FPR for DenseNet201 with SVM (Linear) is lower than the FPR forDenseNet201 with MLP. Some distinct characteristics of the exam define the method’s misclassification. For example,exams with high contrast variations and with angulations that do not centralize the critical region, are exams that deviate from the basic standards of Chest X-ray images promoting unsatisfactory results in the classification.
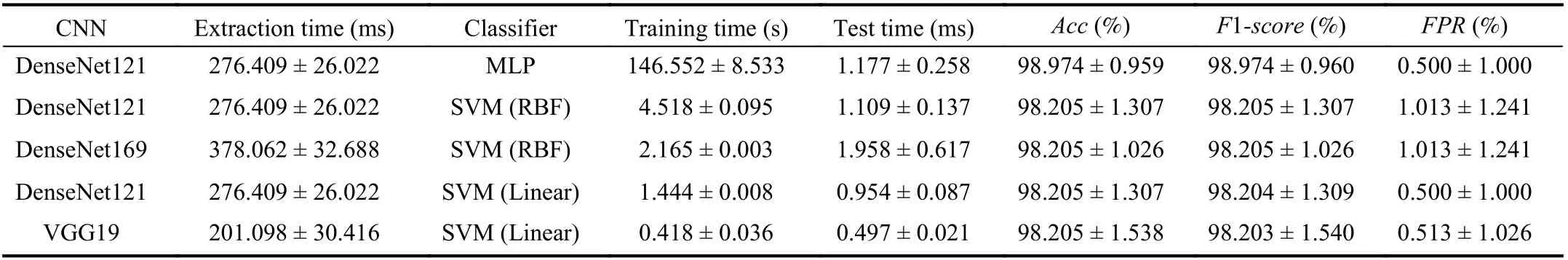
TABLE VI METRICS OBTAINED BY CLASSIFYING FEATURES EXTRACTED FROM DATASET A WITH DATA AUGMENTATION BY DIFFERENT CNN ARCHITECTURES OF THE TOP FIVE RESULTS

TABLE VII METRICS OBTAINED BY CLASSIFYING FEATURES EXTRACTED FROM DATASET B BY DIFFERENT CNN ARCHITECTURES OF THE TOP FIVE RESULTS
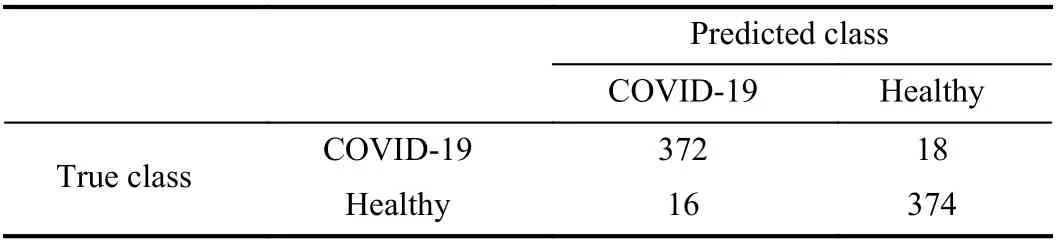
TABLE VIII FINAL CONFUSION MATRIX OF THE TEST SET FOR THE CLASSIFICATION OF CHEST X-RAY IMAGES AS HEALTHY OR COVID-19 POSITIVE FOR DENSENET201 WITH MLP FOR DATASET B

TABLE IX FINAL CONFUSION MATRIX OF THE TEST SET FOR THE CLASSIFICATION OF CHEST X-RAY IMAGES AS HEALTHY OR COVID-19 POSITIVE FOR DENSENET201 WITH SVM (LINEAR)FOR DATASET B
Fig. 4 presents the features extracted by DenseNet201 from both datasets using the t-distributed stochastic neighbor embedding (t-SNE) technique [58]. We can observe that,although DenseNet201 is not in the top five of Dataset A, a hyperplane can separate the classes, which justifies the three combinations with SVM (Linear) in the top five of this dataset. As mentioned in Section III-A, the healthy X-ray images from Dataset A are of pediatric patients, which explains the easily distinguishable cluster formation of the features from this class. In contrast, when analyzing Dataset B, the healthy class features are scattered, which indicates a wider variety of sources in the original dataset. Also, this dataset needs a classifier that can classify non-linear data,which justifies MLP as the best classifier for this dataset.
A. Comparison to Related Works
As shown in Table X, we compare the proposed approach with the studies of Apostolopoulos et al. [17], Narin et al. [8],and Ozturk et al. [59]. Apostolopoulos et al. [17] and Narin et al. [8] used chest X-rays images and the transfer learning method, but they used a second network for the classification process. Furthermore, those papers used the same sources for the creation of the datasets as the proposed approach.
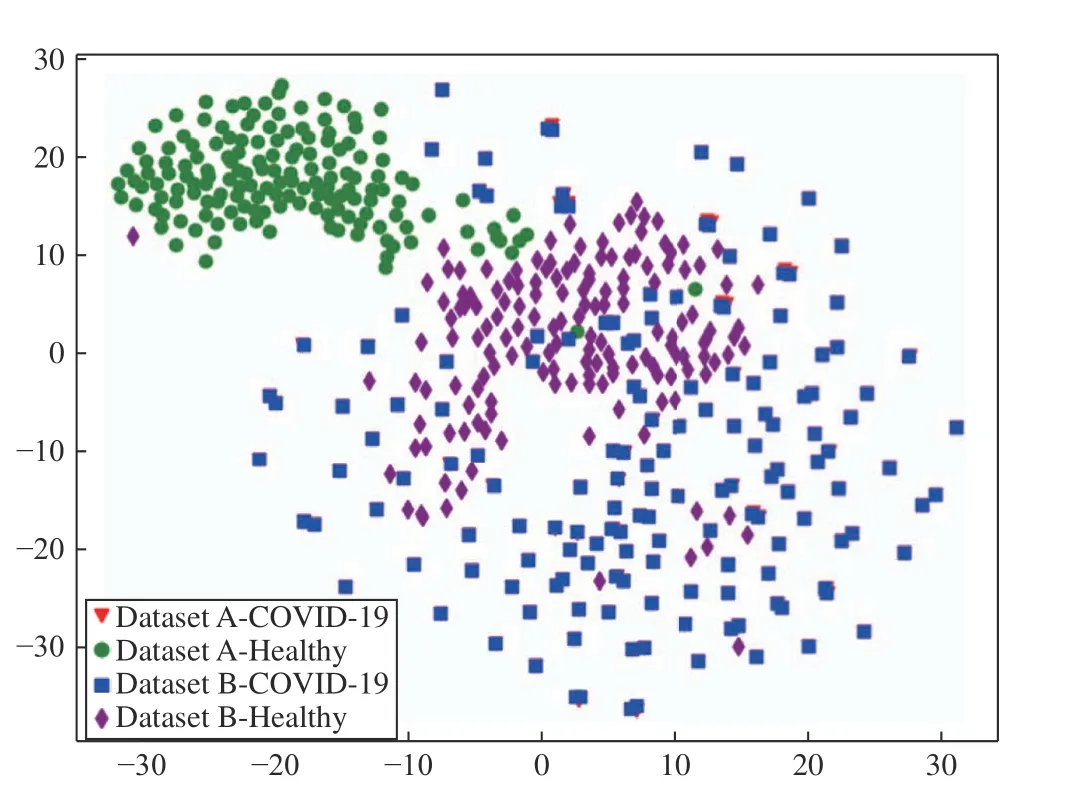
Fig. 4. Visualization of the features extracted by DenseNet201 from both datasets using t-SNE.
Apostolopoulos et al. [17] used images from the datasets“Chest X-ray images (Pneumonia)” [31] and “COVID-19 image data collection” [29]. From these datasets, they selected 50 images of each class, reaching an Acc and F1-score of 98%. However, this dataset is highly selected to be images for educational purposes [60]. In addition, they used a high-end GPU (Tesla K80), which could be impracticable for many clinics, hospitals, and countries. We tested our approach in this dataset, and it achieved 100% in many combinations of CNNs and classifiers.
Narin et al. [8] used a dataset with images from “COVID-19 image data collection” [29], “Chest X-ray images(Pneumonia)” [31], and “COVID-19 X rays” [61], totaling 224 images for the COVID-19 condition, 504 for the normal condition and 700 for common pneumonia. They achieved an Acc of 98.75% when analyzing 2 classes and 93.48% for 3 classes. However, since their dataset is heavily unbalanced,this tends to increase the accuracy.
Ozturk et al. [59] used 127 images from the dataset“COVID-19 image data collection” [29] that were diagnosed with COVID-19. In addition, they used 500 images for normal condition and 500 for pneumonia from dataset “ChestX-ray8”[32]; thus they used the same sources as the Dataset B of the proposed approach. They reached 98.08% in Acc and 95.51%in F1-score, when making a binary classification using a modified CNN based on you only look once (YOLO); thismodified CNN is called DarkCovidNet. Although they achieved an Acc 3% higher, their classes are also unbalanced,which can contribute for better metrics.
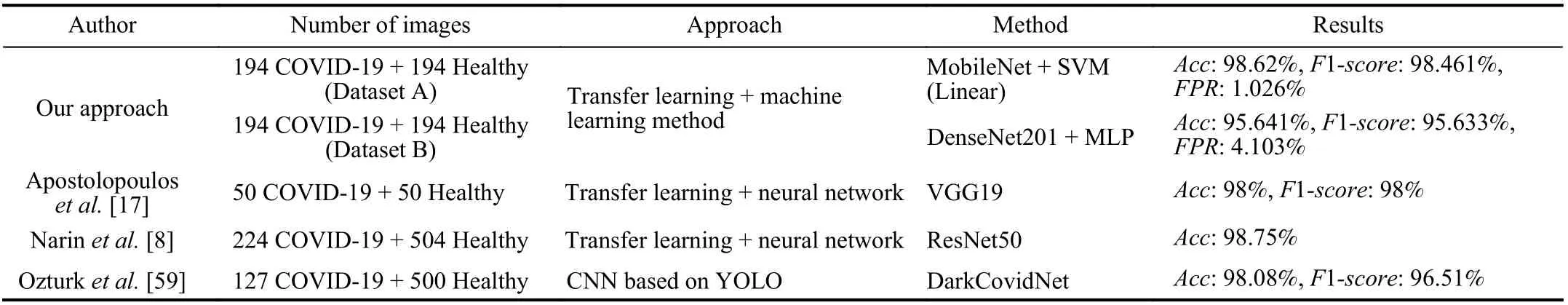
TABLE X COMPARISON TO OTHER METHODS OF CLASSIFICATION OF CHEST X-RAYS IMAGES TO DETECT COVID-19
V. CONCLUSION AND FUTURE WORKS
Early detection of patients with the new coronavirus is crucial for choosing the right treatment and for preventing the quick spread of the disease. Our results show that the use of CNNs to extract features, applying the transfer learning concept, and then classifying these features with consolidated machine learning methods is an effective way to classify Xray images as in normal conditions or positive for COVID-19.For Dataset A, the MobileNet with SVM (Linear) combination had the best performance, achieving a mean Acc of 98.462% and a mean F1-score of 98.461%. In addition, it was able to classify a new image in only 0.443 ± 0.011 ms,proving to not only be accurate but fast as well. For Dataset B,the pair with the best performance was DenseNet201 with MLP, reaching a mean Acc of 95.641% and a mean F1-score of 95.633%. Although it had slightly lower Acc and F1-score,it classified an image in only 0.282 ± 0.154 ms, which is faster than the best combination in Dataset A.
The proposed method has not undergone a clinical study.Thus, it does not replace a medical diagnosis since a more thorough investigation could be done with a larger dataset.Under those circumstances, our work contributes to the possibility of an accurate, automatic, fast, and inexpensive method for assisting in the diagnosis of COVID-19 through chest X-ray images.
For future work, we intend to increase the size of the dataset by adding new X-ray images of patients with COVID-19, as soon as these images are available, and by adding X-ray exams of other lung-related diseases, thus reassuring the efficiency of the proposed approach. Besides, we aim to test the proposed method using an imbalanced dataset. We also intend to integrate our method into a free online platform of image classification, such as LINDA [27]. This way, hospitals and medical clinics around the world would be able to identify diseases in chest X-ray images without the need for building their classification platform. Furthermore, we aim to compare the proposed approach with methods based on fine-tuning,and train a network from scratch.
杂志排行
IEEE/CAA Journal of Automatica Sinica的其它文章
- A Multi-Layered Gravitational Search Algorithm for Function Optimization and Real-World Problems
- Dynamic Hand Gesture Recognition Based on Short-Term Sampling Neural Networks
- Dust Distribution Study at the Blast Furnace Top Based on k-Sε-up Model
- A Sensorless State Estimation for A Safety-Oriented Cyber-Physical System in Urban Driving: Deep Learning Approach
- Theoretical and Experimental Investigation of Driver Noncooperative-Game Steering Control Behavior
- An Overview of Calibration Technology of Industrial Robots
