Big Data Analytics in Healthcare — A Systematic Literature Review and Roadmap for Practical Implementation
2021-04-14SohailImranTariqMahmoodAhsanMorshedandTimosSellisFellowIEEE
Sohail Imran, Tariq Mahmood, Ahsan Morshed, and Timos Sellis, Fellow, IEEE
Abstract—The advent of healthcare information management systems (HIMSs) continues to produce large volumes of healthcare data for patient care and compliance and regulatory requirements at a global scale. Analysis of this big data allows for boundless potential outcomes for discovering knowledge. Big data analytics (BDA) in healthcare can, for instance, help determine causes of diseases, generate effective diagnoses, enhance QoS guarantees by increasing efficiency of the healthcare delivery and effectiveness and viability of treatments, generate accurate predictions of readmissions, enhance clinical care, and pinpoint opportunities for cost savings. However, BDA implementations in any domain are generally complicated and resource-intensive with a high failure rate and no roadmap or success strategies to guide the practitioners. In this paper, we present a comprehensive roadmap to derive insights from BDA in the healthcare (patient care) domain, based on the results of a systematic literature review. We initially determine big data characteristics for healthcare and then review BDA applications to healthcare in academic research focusing particularly on NoSQL databases.We also identify the limitations and challenges of these applications and justify the potential of NoSQL databases to address these challenges and further enhance BDA healthcare research. We then propose and describe a state-of-the-art BDA architecture called Med-BDA for healthcare domain which solves all current BDA challenges and is based on the latest zeta big data paradigm. We also present success strategies to ensure the working of Med-BDA along with outlining the major benefits of BDA applications to healthcare. Finally, we compare our work with other related literature reviews across twelve hallmark features to justify the novelty and importance of our work. The aforementioned contributions of our work are collectively unique and clearly present a roadmap for clinical administrators,practitioners and professionals to successfully implement BDA initiatives in their organizations.
I. INTRODUCTION
The advent of healthcare information management systems(HIMSs) is now generating huge volumes of patient-centered,granular-level healthcare data. The high velocity of this data influences the relationship of hospitals and clinics with their patients and necessitates the use of analytics to tap into the needs, attitudes, preferences, and characteristics of clinical entities such as patients and practitioners [1]-[3]. Hence,HIMSs are now required to implement different data deployment, management and analytics strategies with the usage of state-of-the-art big data tools, techniques and technologies in order to utilize and handle the transformation of the heterogeneous healthcare data into valuable and useful insights [4]. In fact, big data is already motivating the use of new architectures to transfer the operational models and data centric architectures of HIMSs [5], [6]. Also, big data in healthcare is rapidly changing with the advent of system development approaches that are highly compatible with widely distributed systems, particularly non-relational NoSQL technology for big data ingestion, storage, management,querying and analysis, e.g., through the use of MongoDB’s and Apache Hadoop’s ecosystems [7], [8].
The process of analyzing big data, or big data analytics(BDA) can tackle large volume, high velocity data streams enabling personalized medicine, which provides physicians with a more comprehensive (in-depth) understanding of an individual’s health. For instance, BDA can be applied to improve diagnostic treatment decisions amidst unaided human inference [9], [10]. The focus on the potential benefits of BDA has never subsided in research papers, technical blogs, and videos, motivating researchers to design solutions to address the aforementioned issues [11]. However, BDA has presented challenges in multiple business domains in the last decade.There is considerable hesitation to invest in big data technologies due to lack of standardization, a rapidly-evolving technology stack, complicated architecture design, a skill set which is difficult to learn, high resource and cost requirements, and data management, storage, access and analysis challenges. Another issue is the lack of a standard protocol of communication between the BDA team and the business side; the BDA team typically does not have enough background knowledge of business domain to model the analytics as per business requirements and the business side does not have the appropriate analytics knowledge(algorithms, technology stack, etc.) to tune and guide the BDA results according to personal needs. In fact, Gartner estimated that 85% of big data and BDA projects were failing in 2019 due to aforementioned issues [12]. BDA applications in healthcare are also (currently) plagued by these issues.
In this paper, we thoroughly investigate the domain of BDA applications in the healthcare sector, particularly with respect to patient care because a majority of healthcare big data sources are related to patient care, as are the majority of research works related to BDA for healthcare. Our intention is to provide a roadmap to clinical practitioners for BDA applications in healthcare. Previously, researchers have applied data science, business intelligence and data warehousing techniques to enhance patient care [13]-[19].These applications, although useful and numerous, are created with considerably limited and small datasets and their usability in the presence of big data cannot be guaranteed.They are also not sufficient to justify clinical use [20]-[22].Big data is far more complex, varied, and voluminous and requires different data management tools and technologies to obtain better insights as compared to traditional data miningbased analytics. Considering the rapidly expanding big data space and the importance of patient care, it becomes important to clearly investigate and determine the exact BDA applications in this domain, their achieved benefits and the difficult challenges which need to be addressed for further research in this area.
Our vision of a roadmap in this paper is comprehensive and unique and based on the following requirements. We initially need to define the characteristics of big data as applicable to healthcare; it is generally known that HIMSs integrate,manage and synchronize big data which is characterized by 4 V’s(volume, velocity, variety, value) at a general level [23]. We need to understand the meaning of these 4 V’s in the context of healthcare, and also check their compliance with the target dataset. Rapidly-expanding and powerful NoSQL technology has alone solved many of the big data management problems since 2007, particularly through the use of Apache Hadoop and its ecosystem [7], [8]. Hence, we need to investigate and describe the current NoSQL applications in healthcare with academic research or other types of online content, and also highlight the benefits which have been achieved with these applications. We then need to determine the exact challenges being faced by the healthcare big data community, both with or without the application of these NoSQL data stores. In fact,a roadmap needs to be presented which solves these challenges in a concrete way by highlighting the untapped potential of NoSQL databases for the healthcare sector. For this, guidance needs to be provided particularly with respect to the implementation architecture for healthcare BDA.Designing a software architecture for BDA is complicated due to numerous analytical tasks which need to interact with each other over a complicated and large technology stack. Some guidance is provided by the lambda and kappa architectures but these have serious limitations [24]. The newly introduced zeta architecture [25] solves these issues and in our opinion, is an ideal solution for healthcare big data companies if it can be properly formalized. An architecture proposal also needs to be coupled up with a success strategy, because many BDA projects have failed in recent years due to lack of strategic direction in leading BDA projects [3].
We address the aforementioned requirements for our roadmap specification through two main research questions(MRQ1 and MRQ2). We define MRQ1 as follows:
1) MRQ1: What is healthcare big data, and how has it been analyzed in research using BDA applications, and what challenges and benefits do these applications have in assisting patients, doctors, physicians and other medical practitioners?
To answer MRQ1, we divide it into the following four subresearch questions (SRQs):
a) SRQ1: Do healthcare datasets exhibit the characteristics and properties of big data? (answered in Section IV-B)
b) SRQ2: What are the challenges identified in research literature in applying BDA to healthcare? (answered in Section V)
c) SRQ3: What are the applications of BDA in healthcare in research literature specifically in regards to NoSQL technologies? (answered in Section VI)
d) SRQ4: What are the benefits of BDA applications in healthcare? (answered in Section VII)
MRQ2 builds upon the results of MRQ1 and we define it as follows:
2) MRQ2: Can the evolving NoSQL technology solve the current BDA challenges, what is the most relevant BDA architecture for such a solution, and what are the strategies by which it can be ensured that this solution will be successful in clinical and medical industries?
To answer MRQ2, we divide it into the following three SRQs:
a) SRQ5: What is the potential of the state-of-the-art and rapidly evolving NoSQL technology stack in addressing the challenges in BDA applications to healthcare? (answered in Section VIII)
b) SRQ6: How can BDA architecture incorporating NoSQL and other big data technologies be used as a guidance for future BDA implementations in the healthcare sector?(answered in Section IX)
c) SRQ7: What are the practical strategies which can be employed by healthcare professionals to ensure successful execution of this BDA architecture? (answered in Section X)
The remainder of the paper is organized as follows. In Section II, we describe the methodology for our systematic literature review and describe the relevant background on big data in Section III. In Section IV, we describe the important dimensions of healthcare big data along with big data characteristics extracted from the relevant literature (SRQ1).In Section V, we identify and classify the challenges in the relevant literature (SRQ2), and in Section VI, we describe all relevant NoSQL applications for a BDA healthcare setting(SRQ3) followed by the identified benefits in Section VII(SRQ4). In Section VIII, we identify the potential benefits of NoSQL databases to improve healthcare BDA applications(SRQ5), followed by our proposal of the Med-BDA architecture for BDA healthcare in Section IX (SRQ6) and success strategies in Section X (SRQ7) to allow practitioners to implement these improvements in their organizations. In Section XI, we compare the contributions of our work across twelve hallmark features with other related literature reviews pertaining to BDA healthcare and finally conclude our paper with future research directions in Section XII.
II. RESEARCH METHODOLOGY
To answer SRQ1-SRQ7, we conducted a systematic literature review focusing on the following research domains:healthcare analytics, big data applications in healthcare, BDA applications in healthcare, NoSQL healthcare applications,and NewSQL healthcare applications. NewSQL is the preferred type of NoSQL databases in industry because they provide ACID guarantees like with relational databases [7],[8]. Our search queries (described later on) are based on more popular terms related to these domains. We have selected these domains to include the complete set of big data technologies in the market. Of particular interest to us are the more popular and successful solutions like Apache Hadoop and MongoDB, along with the cloud solutions of Amazon(AWS) and Microsoft (Azure) [26]. We targeted all types of academic research content as well as non-research content(e.g., technical blogs and company websites). For the research content, we selected Google Scholar which is the most comprehensive search for computer science content along with four other well-known sources, i.e., IEEE, Springer,Elsevier, and ACM. Content from remaining sources (Wiley,Taylor & Francis, etc.) was retrieved by Google Scholar,which indexes content from all other computer science-related sources through mutual contracts [27]. Healthcare research content is also indexed by Google Scholar, e.g., the US National Library of Medicine (www.ncbi.nlm.nih.gov) [28].We focused on research from 2005 onwards, but did not ignore the more historical content if we deemed it essential.We selected Mendeley due to its increased usage and better features to manage our citations after a survey of other tools[29]-[33]. To retrieve the non-research content, we used the Google search engine.
We adopted the following three-step methodology to filter out the relevant subset of research articles from our Mendeley database. In the first step, we filtered articles based on their titles, i.e., the extent to which these titles matched our selected research domains. In the second step, we filtered the first-step articles based on their abstracts, and in the third step, we filtered the second-step articles based on their research content(after reading the first 2 pages). Following are the six basic search queries: “big data”, “NoSQL”, “NewSQL”, “big data tools”, “big data techniques”, and “big data analytics”. We combined each of these queries with “healthcare” and then with “healthcare analytics”, giving us a total of 18 queries. We considered these queries generic enough to extract content related to our sub-research questions, i.e., challenges,applications, architecture, benefits, potential, and success stories of healthcare big data.
The results of our article filtration methodology are shown in Table I. Title filtration gave us 260 articles, out of which we filtered 150 after abstract filtration, and finally, 99 articles after text filtration which we use to answer our seven subresearch questions. Also, Table II shows the distribution of our 260 title-filtered articles with respect to digital sources;the majority of articles were retrieved by Google Scholar (70)while IEEE provided the minimum number of relevant papers(33), with both ACM and Springer providing 55 odd articles.Finally, Google Search Engine retrieved 4 relevant technical blogs with our 18 search queries which were all retrieved in title-filtration stage. In Table III, we show the distribution of content type for our 99 selected articles; majority of these are published in journals (74) while conference and other publishing methods have a reduced frequency comparatively.In Table IV, we show the distribution of these 99 articles with respect to SRQ1-SRQ7; here, parentheses represent repetition as a given article could be answering multiple sub-research questions. Articles discussing BDA healthcare challenges are the most frequent, followed by applications, big data characteristics, benefits and potential of BDA for healthcare.Articles focusing on the use of BDA architectures or presenting success strategies are least frequent, and none of them propose any architecture or present a roadmap. Also, the year-wise distribution of the 99 articles is shown in Fig. 1,which shows a well-defined peak in publications from 2011 to 2014 corresponding to a spark of interest in BDA applications brought about by the increasing popularity of several NoSQL databases, particularly MongoDB (introduced in 2010), Redis(2009), Apache Hadoop (2007 onwards), Apache Spark(2014) for speeding-up Hadoop along with AWS cloud services (2009 onwards). This is proved at least by the use Hadoop and MongoDB in our extracted papers. However,since 2017 onwards, academic research has apparently dwindled due to the complicated nature of healthcare data and the BDA process. Such a trend has also been seen in the telecommunications sector [24]. The academic and corporate healthcare companies then apparently need the comprehensive roadmap presented in this paper to solve their BDA implementation issues and extract value from datasets. To drill-down further, we present the break-down of 260 papers(filtered through title) with respect to distribution of search queries over digital sources in Fig. 2 (with six basic queries),Fig. 3 (with six queries combined with healthcare (HC)), and Fig. 4 (with six queries combined with healthcare analytics(HA)). All four technical blogs were retrieved with “Big Data HA” search query in title-filtration stage. Some of the important insights we can derive from these figures are given below:
1) The hyped terms “big data” and “big data analytics” have been used most frequently by authors and were retrieved in the majority of relevant content, while “NoSQL”, “NewSQL”,“techniques”, and “tools” retrieved relatively less relevant articles.
2) The distribution of content seems uniform across all digital sources for the terms “ big data” and “ big data analytics”.
3) The term “healthcare” is more commonly-used by authors (and retrieved more relevant content) as compared to“healthcare analytics”.
4) The large body of papers retrieved with “big data” and“big data analytics” discuss more generic topics like big data characteristics, challenges, benefits, etc., but do not present any roadmap or concrete NoSQL-based application to enhance and motivate research in this domain; this has been done to a limited extent in papers retrieved with other keywords.
5) Overall, it is apparent that research on NoSQL applications of BDA to healthcare, and on solutions to their implementation problems through big data tools and techniques are limited.

TABLE I ARTICLE FILTRATION RESULTS

TABLE II DISTRIBUTION OF TITLE-FILTERED ARTICLES WRT DIGITAL SOURCES
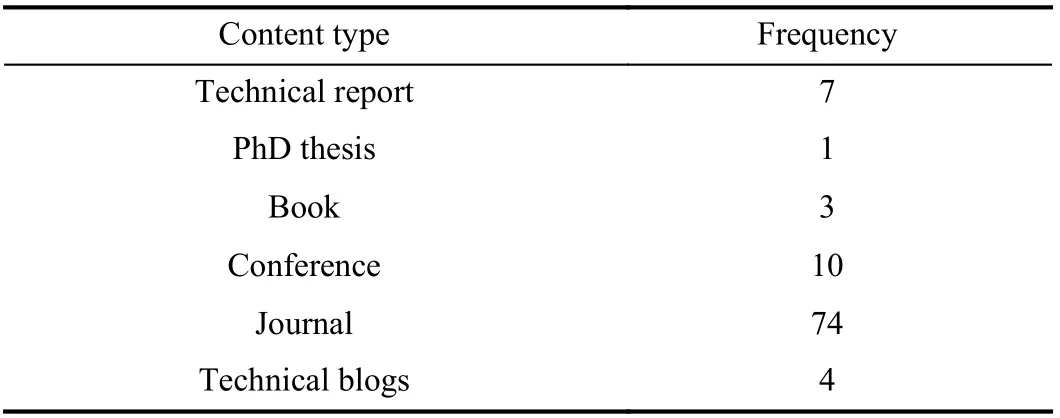
TABLE III DISTRIBUTION OF 99 SELECTED ARTICLES WRT CONTENT TYPE

TABLE IV DISTRIBUTION OF 99 SELECTED ARTICLES WRT SRQ1-SRQ7(NUMBERS IN PARENTHESES REPRESENT REPETITIONS)
III. BACKGROUND ON BIG DATA
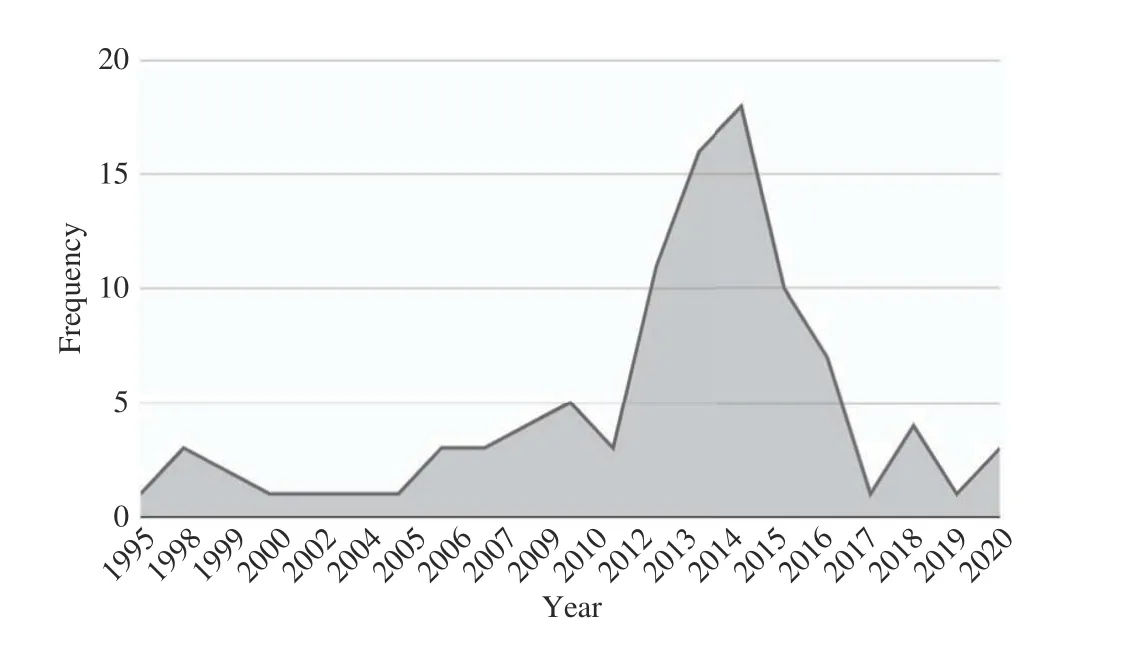
Fig. 1. Year-wise distribution of selected 99 articles.

Fig. 2. Digital source distribution for six basic search queries.
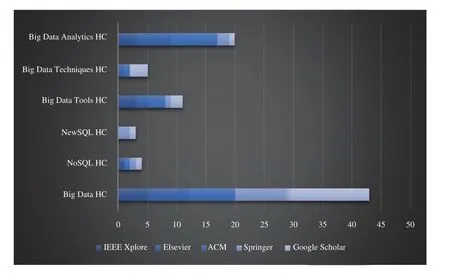
Fig. 3. Digital source distribution for six basic search queries + healthcare(HC).
Ever since the emergence of the Internet, the volume of corporate data has been increasing; nowadays, processing terabytes of data on a daily basis is common practice in retail,financial, healthcare and other representative sectors. The rise of social networking platforms has increased the size of big data further into petabytes and exabytes particularly in the case of E-Commerce companies like Amazon, Google, and Yahoo [34]. Although solutions for big data are evolving rapidly, a large amount of effort is still needed to standardize them in a global scale [35]-[38]. For instance, one recommendation for healthcare is to view data-driven approaches as tools to facilitate understanding of these clinical entities not as a disruptive process [39].
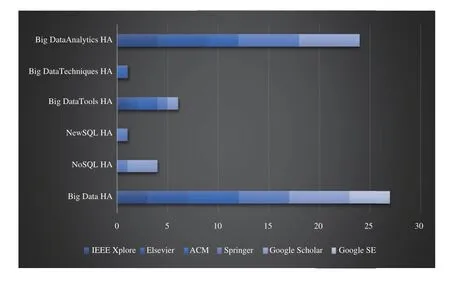
Fig. 4. Digital source distribution for six basic search queries + healthcare analytics (HA).
A. Big Data: Definition and Characteristics
Francis Diebold first formally introduced big data as “the explosion in the quantity (and sometimes, quality) of available and potentially relevant data, largely the result of recent and unprecedented advancements in data recording and storage technology. In this new and exciting world, sample sizes are no longer fruitfully measured in “number of observations”,but rather in, say, megabytes. Even data accruing at the rate of several gigabytes per day was not uncommon” [40]. In this paper, we consider four standard characteristics of big data,i.e., the well-known “3 V’s” (volume, variety, and velocity)presented by [41] and value. Volume refers to size of data(e.g., in terabytes), velocity refers to the incoming data speed(streaming or batch), variety refers to different data types(relational, images, text, videos, etc.) and value refers to insights derived through BDA. These 4 V’s are the essence to validate the measurement of data characteristics of any big data use case [42]-[44]. An excellent classification of big data is given in [5] as follows:
1) Big Data Properties: Specifying the challenges that can occur due to the presence of 4 V’s.
2) New Data Models: Specifying the need for novel types of models for big data which can deal with common issues such as referral integrity, provenance, data linking, and data lifecycle implementation.
3) New Analytics: Specifying the need for BDA based on data science and real-time analysis mechanisms.
4) Infrastructure and Tools: Specifying the need for novel infrastructure mechanisms related to network, data storage,high performance computing, along with tools for heterogeneous multi-provider services integration, data processing, data-centric security models (for trusted infrastructure), and data-centric service models.
5) Source and Target: Specifying the novel breed of input and output tools for big data that deal with data sources capturing high speed/velocity data generated from a variety of smart sensors, delivery of data to consumers, actionable systems, and implementation of visualization techniques.
B. Big Data Storage
Relational databases (Oracle, MySQL, SQL Server, etc.) are not capable of handling data-intensive and large-scale applications, that involve the storage and management of huge volumes, and high frequency reads and writes efficiently, and at the same time flexibly cope with different types of data models and support heterogeneous data [45]-[47]. Even though relational databases like PostgreSQL and others have tried to address these challenges, they are not efficient and effective compared to standard big data storage, which typically called NoSQL data stores [48], [7], [8]. NoSQL technologies have been able to solve a majority of data management, data storage, data processing, and data querying problems in BDA initiatives. They hence comprise a crucial component of BDA architectures. NoSQL stores having a global impact are MongoDB, CouchBase, Cassandra, Neo4J,Redis, Amazon’s NoSQL DynamoDB and Microsoft Azure’s NoSQL CosmosDB along with Apache Hadoop and its ecosystem [8]. Stores like MongoDB, Redis, and CouchDB can store and query huge volumes of streaming big data along with optimizing query latencies. Almost all standard NoSQL stores now satisfy the ACID requirements of relational databases allowing users to always view and query complete,updated data. Such NoSQL stores are also known as NewSQL stores and also work in a distributed scenario, e.g., MongoDB and Redis [48], [49], [7].
Hadoop is a state-of-the-art solution to address the complex issues of BDA initiatives, primarily because it brings forth a whole ecosystem of tools and technologies to solve these issues. It brings forth a shared-nothing and distributed architecture, which spawns new cluster nodes to address increased storage and computational power requirements. The addition of a node is a seamless process with all processing and data distribution among multiple nodes occurring transparently. It performs ingestion, wrangling (cleaning),storage, querying and analysis on big data (peta-byte scale)both effectively and efficiently, through fault-tolerance on low-cost, commodity storage and servers. A key feature of Hadoop is that it separates data storage from data processing[50]. Hadoop has 4 components: 1) Hadoop Common comprising common libraries, 2) HDFS which is a default storage and file system as well as the communication backbone between nodes, 3) Hadoop YARN (Yet Another Resource Negotiator) which manages resources for running Hadoop’s MapReduce tasks, and 4) Hadoop MapReduce which is the programming paradigm for processing data stored in HDFS.
Hadoop’s ecosystem is shown in Fig. 5 and comprises opensource projects maintained by Apache Software Foundation.HBase is Hadoop’s persistent database which runs atop HDFS while Hive is Hadoop’s warehouse which can be used to execute SQL queries over HDFS data and store this data in the form of tables (much like relational stores) [51]. Through Pig,one can easily write programming code related to Hadoop MapReduce for data processing and Mahout is used to perform Machine Learning on HDFS data. Sqoop is used to fetch data from relational and other sources and insert it into HDFS while Zookeeper is used to maintain coordination between the nodes (its required for any Hadoop cluster setup).Chukwa and Flume allow ingestion of big log data files and their monitoring, while Ambari and Hue (not shown) are used to monitor the health of Hadoop’s cluster. Avro is a data serialization format for storing HDFS data on a hard disk and Oozie can create work flows for Hadoop jobs. In fact, Hadoop is not suitable for interactive and iterative jobs due to the overhead of iterative reads and writes to and from the disk[52], [53]. Spark [52] is another Apache project, developed to overcome the shortcomings of Hadoop. It avoids most of the disk inputs/outputs operations by executing processes in the memory when possible. Spark processing has been shown to be at least 100× faster than MapReduce.
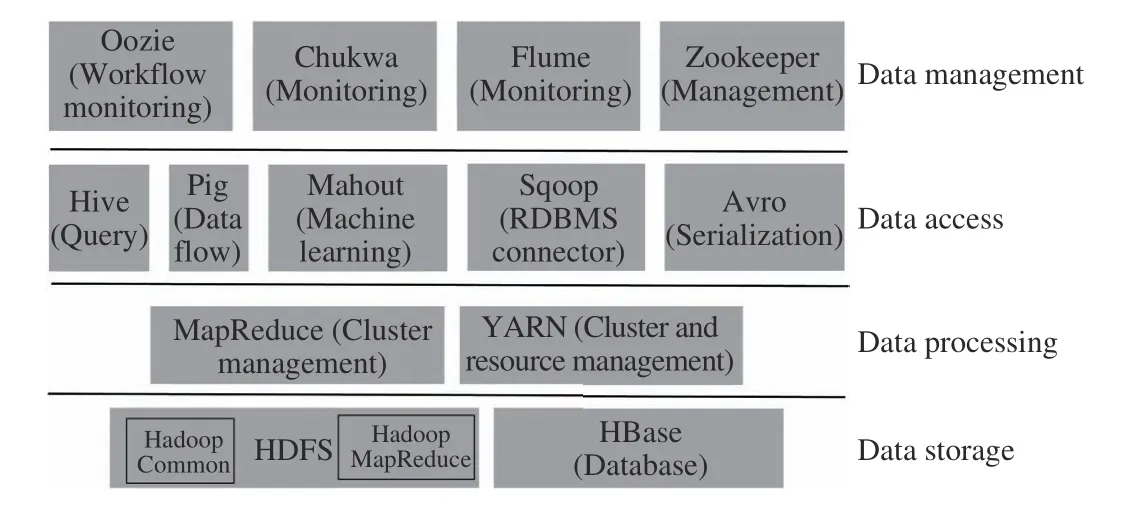
Fig. 5. Hadoop components and ecosystem.
Here is how Hadoop handles the 4V’s requirement:
1) Volume: Hadoop splits large-volume data and dataprocessing between multiple data nodes (slaves). With the increase in the processing workload or data volume on each individual data node, the data can be split by adding more data nodes.
2) Velocity: Hadoop can store streaming data in HDFS and avoids, or at least postpones, the latency from storing it directly into a relational database.
3) Variety: Hadoop has no specific variety requirement for storing data on HDFS; it is able to store data as operating system files without prior processing or checks. Any variety of data can hence be stored and there is no need to understand and define the data structure beforehand.
4) Value: The Hadoop ecosystem allows big data organizations to use these multiple technologies in a standard architecture to derive value or insights from big data. There have been many such experiments across different corporate domains in the last decade and in this paper, we mention those related to healthcare.
C. Big Data Analytics (BDA)
Data analytics can extract unknown patterns and trends,hidden correlations, and other useful information from data(value). BDA does the same with big data. BDA results enable analytics professionals, predictive modellers and data scientists to analyse and examine huge volumes of data sets containing variety of data forms that may be undiscovered by traditional analytics processes [54]. BDA can lead to competitive advantages, improved operational efficiency,better service, more effective new opportunities and other benefits.
The BDA process is far more comprehensive and complicated due to the presence of 4Vs. Traditional analytical methods for RDBMSs cannot always be successfully applied over big data. Following are three differences identified from our research literature [4], [55]:
1) Data Structure: Structured data in an RDBMS is processed after collecting and organizing it with sophistication and ease. Big data, on the other hand, cannot be integrated and organized easily due to its heterogeneity and its streaming,real-time nature.
2) Sample of Analysis: Conventional data analysis uses a pre-designed question approach, in which a sample is selected from the existing known population and the process of analysis begins after posting research question(s). This process is based on collection of data to seek appropriate answers to those questions. In BDA, data sizes are large if velocity and variety are both high, so acquiring the right sample cannot be always guaranteed. Thus, it operates on both known and unknown populations.
3) Nature of Data: Traditional analytics performs analysis on data at rest, i.e., data residing statically in a database. BDA also handles streaming and high velocity data.
Within the last decade or so, these differences have given rise to novel research directions focused on creating new analytics algorithms, techniques, frameworks, models, tools,architectures and designs particularly targeted only for BDA.The most notable have been the design and use of a large variety of NoSQL databases [4], [56], which can be categorized into three categories: a) computational and processing, e.g., MapReduce and Apache Spark; b) storage,e.g., HDFS; and c) Analytics, e.g., Apache Mahout and Apache Hive [56], [57].
IV. BIG DATA IN HEALTHCARE
Over the last decade, the use of electronic medical records(EMRs) by the medical staff, physicians and doctors has grown substantially. Most EMR usage is focused on patient care [10], [58]. Medical science research is bringing smart wearable devices and new technologies for patients, which generates big data from the continuous monitoring of vitals by the doctors in real-time, in an attempt to reduce the frequency of physical patient visits to the hospital [59].
A. Data in Healthcare
HIMSs are rapidly becoming a necessity at a global scale.Physicians, doctors, patient care staff, and management staff have started to use HIMSs regularly. An HIMS provides an interface between patient care and medical researchers. It becomes inevitable especially in a medical emergency. The mainstay of HIMSs are databases to store patients’ related data including treatment plans, prescriptions, and diagnoses[60]. HIMS is a patient care data intensive system. There are many processes in an HIMS generating new data every minute at the same time (see Fig. 6). These processes are producing multiple records for each patient of same data. Along with patient care referenced data, new derived data is also created[61].

Fig. 6. Data generators for an HIMS.
An HIMS can have many data sources, on the basis of which we can classify the different healthcare data types as follows [62]-[65]:
1) Clinical Data, e.g., measurements of clinical judgements,fluid intake-output, vital signs and clinical examination(including Boolean questions such as “Does the patient use any drugs? Did the patient previously undergo any surgical operation? Did any family member of the patient suffer a specific disease?”).
2) Administrative Data, e.g., patient admissions, number of beds available, and rate of usage of a medical equipment.
3) Finance Data, e.g., data related to medical insurance,patient fees, adjustments, and diagnoses-related group costing.
4) Medical Imaging Data, e.g., test results of Ultrasound-Mammography, magnetic resonance imaging (MRI),computer tomography (CT), positron emission tomography(PET), and Radiography.
5) Laboratory Test Data, e.g., Protein Blood test results,Urine test results, Enzyme, and Blood Sugar test results.
From the above, we can deduce the following modes of clinical data collection.
1) Oral Collection, e.g., when patient provides responses to oral questions regarding patient history. Oral data can be registered on paper, or fed into HIMS or fed directly into a handheld device [66].
2) Manual Collection, e.g., check up of blood sugar using stick, Blood pressure, Respirations per minute, Fluid outtake(with a catheter), or physical examination by the medical doctor [67].
3) Autonomous Collection, e.g., laboratory and medical imaging results, along with smart patient monitoring data which are also stored autonomously. Images are usually compressed with simple lossless and near-lossless methods and usually require large storage space. Standards used for storage and transmission include picture archiving and communication system (PACS), digital imaging and communications in medicine (DICOM).
B. Big Data Characteristics in Healthcare
BDA is all about the integration, valuation, management,synchronization and analysis of high volume, variety and velocity of data [23]. Big data is particularly defined by its heterogeneous nature [21] and has changed the culture of doing research, management and business, from a data perspective. Existing traditional data analysis approaches cannot cope with the frequency of change, variety and increase in size of data. Therefore, the architecture of the traditional RDBMS requires essential evolutions [68].Considering the healthcare domain, the implementation of quality patient care services requires a better strategic relationship with patients. Strategic patient relationships management uses technology, processes, techniques,information and medical staff, as components of the BDA process. However, this process is highly affected by the heterogeneous, high growing volume of in-motion patient care data. Research work has primarily identified our 4 Vs(volume, variety, velocity, and value) regarding healthcare big data, as a result of initial processing and big data classification, [69]-[73] (see Fig. 7). We now discuss these V’s in the context of healthcare as follows:
1) Volume: The volume of healthcare data is growing rapidly; currently it is more than 500 petabytes, which is expected to spiral up 50-times to 25 000 petabytes in 2020.The major source of this data are HIMSs, which are generating new data every minute at the same time [65], [70].Particularly, medical imagery has much to contribute to volume. The enhancement in the quality of medical images has resulted in increase of image resolution. Hence, the size of medical images (previously not more than several Kilobytes/Megabytes) is now ranging from Megabytes to Gigabytes. Another major contributor to data volume is the need to store patient history in EMRs, due to which the size of an EMR can easily reach up to Gigabyte scale. For research purposes, a number of providers’ organizations are retaining patient masked data for an indefinite period [70], [74].
2) Variety: The variety of patient care data is directly linked to the data types mentioned above, i.e., clinical, administrative,finance, medical imaging, and laboratory testing. There is also unstructured data as text notes from nursing and clinical staff,along with videos, images and information from monitoring equipment and smart wearable sensors, all creating a wider variety of data types and formats [75]. As popularity of healthcare gadgets grows, data from these streams are expected to integrate patient care data in the near future. It is a complex challenge to combine these diverse types of data to diagnose accurately and prescribe the best treatment and cure for a specific patient. To resolve this, healthcare industry is already moving towards big data and analytics [74].
3) Velocity: Healthcare data can be either recorded manually by medical staff or autonomously through smart sensors. The former doesn't have much velocity and is typically used by data warehouse and analytics solutions in “batch” mode. This cannot compete with the real-time high-frequency and highvelocity sensor data, which is driven by the growing use of smart sensors, high-resolution medical images and video.Real-time data applications, such as early detection of infections and drug discovery could be helpful in the reduction of mortality and morbidity of patients and it could also be helpful in the prevention of hospital outbreaks [75]. In fact, high velocity can easily overwhelm HIMS’s ability to store and analyse streaming data [76]. BDA has revolutionized healthcare analytics with its capability to perform real-time analytics on high velocity data. Velocity is also proposing different prospects to enhance outputs by integrating the several activities of the healthcare value chain like integration between wards and laboratories in an operationally viable bench-to-bed paradigm [77].
4) Value: The real driver for using big data in healthcare is ultimately the identification of valuable information which can potentially improve patient care [78]. For this, healthcare industry is focusing on operational efficiencies and business process enhancements. The latter aims to reduce fraud, waste,and costs by applying more efficient approaches for service delivery, data analysis, management, and integration. The former aims to discover new techniques of providing patient care while efficiently allocating healthcare services [79], [80].Data-driven healthcare organizations are shifting from conventional monitoring reports to discovery of insights to overcome traditional ineffectiveness and develop smoother workflows for better coordination among healthcare staff and patients and improved patient care [20], [22], [81].

Fig. 7. The 4 V’s big data identified in healthcare research literature.
V. CHALLENGES IN HEALTHCARE BDA
We have identified five challenges being faced by healthcare industry in application of BDA. These are shown in Fig. 8. We describe them as follows.
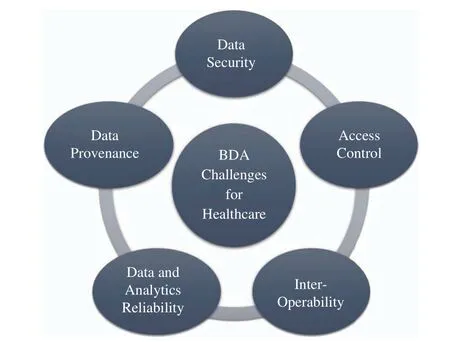
Fig. 8. The Challenges in Application of Big Data Analytics to Healthcare.
A. Confidentiality and Data Security
The misappropriation of patient healthcare information may lead to issues in the patient’s employment and/or insurance coverage [39]. This adverse effect is directly correlated to confidentiality risks and data access [82]. Ignorance of privacy on medical and scientific data may result in public data which can be accessed openly. This confidentiality challenge would require changes in legislation involving healthcare delivery [83]. Legislative changes with regards to data security and confidentiality could provide a more flexible framework that could be helpful in the adaption to BDA technologies [84]. However, analytics on sensitive patient care data is still a challenge with the adaption of BDA in the datadriven health sector [85].
B. Granular Access Control
Granular access control in healthcare enables patients and hospital medical users’ responsibilities, privileges, rights and roles to be set such that users related to the hospital are given privileges only to their relevant data or functional area of the system [86]-[90]. Ensuring high level of usability and security to access relevant piece of data is an often-cited challenge in BDA application to healthcare [91]-[93]. The specific problems with granular access control are as follows:
1) Successfully tracking the privacy policy integrity,
2) Successfully tracking user access,
3) Difficulty of keeping track of secrecy/security policies and requirements in a cluster-based big data environment,
4) Keeping track of multiple users in a cluster-based big data ecosystem,
5) The risk of privacy invasion when different user types(patients and healthcare professionals) access different components of the big data ecosystem simultaneously [94],and
6) The successful implementation of mandatory access control with proper application of secrecy/security requirements [95].
C. Interoperability
Interoperability between the different healthcare data types in order to achieve some healthcare strategic vision is a major challenge [43], [96], [97]. This challenge demands an agreement on common data sets, developing common interfaces, recording health information, and defining quality healthcare standards policies, languages and clinical standards[98], [99]. In the presence of multiple components and their different users, it remains unclear as to how one can enhance big healthcare data interoperability across the different data sources and types [100].
D. Data and Analytics Reliability
Maintaining the reliability of data and BDA results is another core problem in application of BDA to healthcare[98], [101], [102]. We have seen the different data types which can be generated in the healthcare domain, the different modes through which the data can be collected, and the different methods of storing this data. Along with this is the problem of high data velocity and integrating data variety[61], [63], [66], [67]. These complex dynamics can potentially decrease reliability of data and analytics results due to the following situations [96], [98], [101]-[104]:
a) There is an increased chance of an erroneous data entry in the manual mode (through humans).
b) The data integration process can remain unoptimized due to high data diversity occurring at high velocity.
c) Different components of HIMSs may be managing data at different volumes and velocities, making the BDA process heterogeneous.
d) The pre-BDA extract transform load (ETL) process, i.e.,cleaning of dirty data and developing an understanding of healthcare data lake, can turn out to be very complicated and inefficient,
e) Due to the difficulty of data integration, it could be required to learn different BDA models for different HIMSs components/data sources, hence increasing the complexity of the overall process (lesser efficiency and more BDA models to maintain)
f) If data is to be sampled for BDA, it is complicated to acquire representative samples from high velocity data streams.
g) The BDA models operating on streaming data lakes are potentially inaccurate due to inappropriate sampling or frequent change in patterns; these models need to be then learned at a lesser velocity which can itself compromise the final BDA outputs.
h) In a data pipeline based on 3 V’s, incorporating a permanent BDA infrastructure with a traditional analytics pipeline is a time-consuming activity potentially requiring technical trade-offs/compromises.
i) Considering the large number of BDA techniques, tools,and algorithms available, it could be time consuming to select the right personalized BDA solution, particularly in the case of non-availability of BDA experts. If this search is not guided by extensive experimentation, BDA results will be incorrect and/or unreliable.
j) Inadequate training of healthcare staff in the use of BDA can lead to sub-optimal performance, hence minimizing BDA benefits.
E. Data Provenance
Data management and provenance is another challenge for BDA applications in healthcare. Effective coordination of multiple departments in the health sector to use big data is a complex task [105]. The segregation of duties is not similar to operational systems in big data. It is unclear how responsibilities in healthcare big data systems are divided across other relevant bodies of healthcare. Improved healthcare data management is necessary for effective data usage to facilitate access [20], [22]. Some vulnerabilities related to big data storage are consistency, data provenance,confidentiality, and integrity. Malfunctioning infrastructure of big data applications is a major threat to data integrity. In the applications of big data, the provenance meta-data is similar to meta-data. It contains the provenance for the infrastructure of big data itself. The complexity of the provenance information contained in the metadata of the big data system increases with the growth of volume of data. There is a wide variety of sources to collect big data. The paramount importance in this environment is that how much trustworthy the data is.Protection of the provenance meta-data can be effective in the verification of multiple data sources [43].
VI. BIG DATA APPLICATIONS TO HEALTHCARE
In this section, we will describe the applications of BDA to healthcare extracted from the results of our systematic literature review. These applications are centered around four NoSQL types, i.e., key-value stores, columnar stores,document stores, graph stores, and hybrid stores. We will describe each type with an healthcare example and then describe the research works using that NoSQL type. Based on an analysis of NoSQL applications to healthcare [106], we extract the following important NoSQL properties of interest to healthcare:
1) Scaling Out: Scaling horizontally from tens to thousands of nodes for storing and processing ever increasing volumes of EMRs.
2) Automated Scaling: Autonomous scaling out of EMR data in case a node capacity or user query hit ratio crosses some threshold.
3) Reliability: Reliability and fault-tolerance of BDA process is achieved through replication of EMR data in distributed data execution mode.
4) Data Model Options: Flexibility in choosing the data model to cater for structured, semi-structured and unstructured EMR data streams.
5) CAP Theorem Compliance: Ensuring either availability of EMR data to the queries, or the consistency of this data, in the face of data distribution (partitioning).
6) Compliance with Eventual Consistency: In case EMR data consistency is compromised at run-time, there is a standard guarantee that it will eventually become consistent at some later point of time.
7) NewSQL Compliance: If healthcare administrators are strict on both consistency and availability, then NewSQL solutions can offer both, along with complete compliance with ACID properties; in essence, this is RDBMS-based EMR mapped onto big data.
8) Optimized Query Execution: Most of the NoSQL/NewSQL solutions have personalized query execution engines, which would remain optimized for EMR data with 3 V’s.
9) Cost-Effective: The standard big data solutions (e.g.,Hadoop ecosystem and MongoDB) are open-source and hence, would incur zero purchase cost for a BDA healthcare infrastructure.
A. Key-Value Stores
Key-value stores are databases which are based on the keyvalue model, in which values are mapped corresponding to keys, i.e., a given value is given identity through its key. A snapshot of a key-value store from the healthcare domain is shown in Fig. 9. This store consists of three databases, i.e.,patient, practitioner, and diagnosis. The unique key of a Patient DB is the patient’s medical record number (MRN),and the values contain the first and last names, age, and the list of symptoms. Two values for patients John Buck and Jack Owen are shown. The key for the Practitioner DB comprises the doctor’s employee number (EMN) and values contain the doctor’s department and the respective list of consultation clinics. The key for a diagnosis DB comprises a diagnosis number (local to the hospital), and values comprise the list of laboratory tests executed along with the list of symptoms.“Breaking up” table-based data in this way into a multitude of flexible, lightweight key-value pairs leads to remarkably better query response times as compared to traditional RDBMSs [55].

Fig. 9. A snapshot of key-value store from healthcare domain.
Authors of [107] addressed the issue of inadequate collaborative patient care by applying and developing ontology and its corresponding rules by designing a crossdomain, reusable, evidence-based knowledge base. They design a clinical context model for the u-healthcare domain.This model stores data in the form of a key-value store,integrates data from diverse mobile platforms, and is formalized as a set of ontologies. On a similar note, contextual information (CI) is applied by [108] to develop a healthcare model based on ontology. The basic data structure in CI are key-value pairs. Value in CI is used as an environment variable. The proposed ontology for healthcare includes service systems in several spaces, e.g., office, home, etc., and several devices, e.g., computer, mobile devices, etc. This ontology has been implemented in ubiquitous environments for personalized healthcare services. Finally, in [109], the author presents a framework for integrating key-value stores within a typical HIMS architecture. The core benefits of the efficient key-value approach is patient monitoring, clinical predictions, and corresponding simulations, all done in realtime. Another benefit is the scalability of the framework to include more hospitals, and the offering of the framework on the cloud.
B. Columnar Stores
The idea of columnar stores was initially conceived by Google and implemented in their BigTable columnar store[110]. In a columnar store, a single table is dynamically distributed over a cluster. There is no stringent requirement of avoiding null values (as in RDBMS). Columnar stores can be easily quite sparse, with each new row having a different schema. So, a single column can remain empty across thousands of rows and there is no storage cost for these null values. Columns can also be combined together to form column families. The model is scalable in that columns can be removed or added at run-time, with millions of columns also possible in a single store. For performance reasons, these columns are sorted on the disk, minimizing random access.Overall, the storage efficiency is enhanced in columnar stores[111]. The disadvantage is the update operation. While in RDBMS an update of tuples with a foreign key can be enough, a column-oriented big data database may require an update of all values in a column for all records.
A snapshot of a healthcare-based columnar store is shown in Fig. 10. Here, we form two column families, one for the patient’s residence location (City, Province) and the other for the patient’s vitals (blood pressure (BP) and body temperature(Temp)). We notice the sparsity of data, and also that its not necessary for a family data to be fulfilled completely. “PJB”and “Punjab” represent the same province (Prov.) but this flexibility of using different strings is allowed. The surgery column (Surg.) lists the surgery type given to a patient and it is obviously sparse as a sample patient sample gets surgeries generally. The Satis column, represents the patient satisfaction as acquired by a survey, is also sparse as not all patients provide responses. It is possible to add hundreds to thousands more columns related to a patient’s EMR.

Fig. 10. A snapshot of columnar store from healthcare domain.
To predict and efficiently manage the patient’s disease, the authors in [112] propose a patient-customized healthcare system based on Hadoop with text mining (PHSHT). PHSHT consists of a text mining-based Hadoop module (TMHM), a medical data collection module (MDCM), a disease management and prediction module (DMPM), and a disease rules creation module (DRCM). These modules operate as follows:
1) MDCM: It stores healthcare big data in HBase, which is divided into both structured and unstructured entities.
2) TMHM: It converts unstructured data to structured form through text mining, and distributes it collectively with other structured data in HBase.
3) DRCM: It uses conditional probability set theory (CPST)to generate rules associating the relevant set of patient’s EMR attributes with the diagnosis.
4) DMPM: It provides customized patient medical services by comparing a patient’s family history, patient’s current status and patient’s information with the disease rules stored in the DRCM. The most important service of DMPM is its capability of efficient prediction of a patient’s disease based on patient’s current and historical status [112].
A more concrete work on the use of HBase in healthcare is presented in [113]. According to the authors, EMR data is typically spread out in different Excel files or related applications. These files can grow in volume and it is difficult to query them collectively. They also contain diverse data.Authors then motivate HBase to integrate these files, and show a simple experimental run of their idea on a 2-node cluster. These runs demonstrate the efficiency of HBase with respect to Excel file queries. Moreover, in [114], the authors implement a complicated POC for a Canadian healthcare project to load 30 TBs of healthcare data to an HBase cluster with all base processing done by MapReduce. The authors show that data ingestion and loading with HBase takes a long time (one month), the MapReduce process has high limitations, and it is difficult to establish a schema for complicated healthcare data. However, the authors highlight key-value databases as the best solution for healthcare data.
C. Document Stores
In document stores, the smallest atomic data storage unit is a document, which are semi-structured and comprise a collection of key-value pair data [65], [115]. Every document has its unique identifier and serializes tabular or object-based data by encoding it in semi-structured formats such as java script object notation (JSON), binary SON (BSON), and XML. In this regard, document stores are better than RDBMSs or object databases on many fronts. They consume lesser storage, offer more efficient query access, and have a highly flexible schema.
A snapshot of a document store for the healthcare domain is shown in Fig. 11. It comprises three different databases, i.e.,Patient, Practitioner, and Diagnosis. We show several documents for each database. A document is equivalent to one row of an RDBMS table. Notice the following:
1) It is not necessary that every attribute (key) is recorded in the same way in a given document, e.g., names and symptoms are recorded differently in the patient DB as are clinics in practitioner DB.
2) It is not necessary for documents in a DB to follow the same schema. Specifically: a) attribute relationship status(RelSt) is included in second document in patient DB but not in the first, b) attribute Designation is recorded in second document in practitioner DB but not in first or third, c)attribute Clinic is not recorded in third document and d)attribute prescription criteria (PresCrit) is included in second document in diagnosis DB but not in first one.
In [65], the authors create and test two databases using open source Apache CouchDB which is a well-known document store. First, they loaded 1 949 753 images in a larger database through Ruby scripting, while complying with the digital imaging and communications in medicine (DICOM) standard.Then, they used CouchDB for database replication and attachment management in a distributed fashion. Authors applied the MapReduce paradigm to execute queries over CouchDB, which demonstrated efficiency in information retrieval as compared to RDBMS scenario.
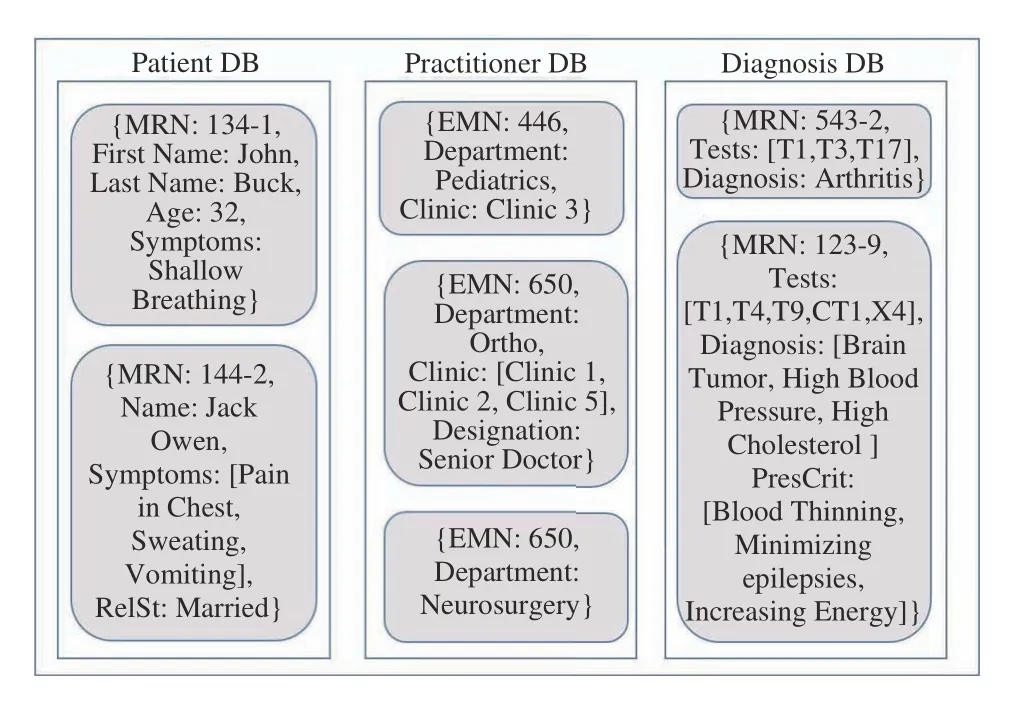
Fig. 11. A snapshot of a document store from healthcare domain.
D. Graph Stores
In a graph database, data is represented in the form of a graph (either directed or undirected). Data is associated with each node and each edge. A snapshot of such a scenario for the healthcare domain is shown in Fig. 12. Here, the nodes represent two patients named Dave and Sharon, and a practitioner named Kate. Practitioners are identified by EMN and patients by MRN. An undirected edge between Dave and Sharon shows similarity: both live in USA and were diagnosed with cancer in 2016. The two directed edges show that Kate treated Dave and Sharon: Dave passed away(Mortality) after one year of treatment (Time), while Sharon recovered after three years.
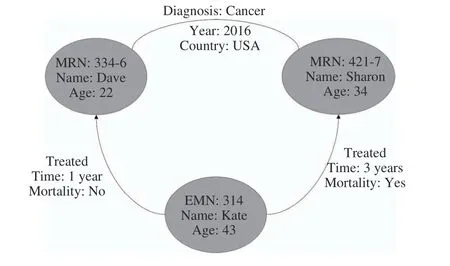
Fig. 12. A snapshot of a graph store from healthcare domain.
Physicians, patients, hospitals, insurance companies, and other healthcare agencies have investigated the usefulness of a graph database in the healthcare domain [116]. In [117], the authors propose and implement a social network data structure to integrate the variety of healthcare data and facilitate the practitioners in their treatment processes. They show average performance results over basic graph operations, i.e.,betweenness, centrality, closeness, and eccentricity. Also, in[118], a web-based graph database is implemented to illustrate the flowchart of a complicated cancer treatment, along with some basic statistical graphs, e.g., cancer incidence per age group and per gender.
Moreover, authors of [119] have compared an application of Neo4J graph database1https://neo4j.comand mySQL RDBMS. They propose transformation rules for mapping a normalized RDBMS schema to Neo4J. Results over two queries demonstrate the efficiency of Neo4J over mySQL. Authors also present an efficient implementation decision support to the medical experts designed rules by analyzing the whole big data system. For analysis and the integration of medical reports,[120] presented a graph database framework called Gpf4Med to introduce an effective and efficient healthcare research tool.The framework was based on the architectural design and implemented BDA by taking exponential growth of data into consideration.
E. Hybrid Stores
The term “hybrid” in the NoSQL domain implied the use of more than one NoSQL store in combination. However, the method of this combination is not clearly defined and was left to the user. In academic research, there are four articles which employ this definition of hybrid. Specifically, in [121], the authors implement a proof-of-concept (POC) for a Czech healthcare center to manage healthcare big data through the Vertica NoSQL hybrid. The four step BDA process followed by authors include data management, data storage, data analytics, and data visualization. Primarily, execution time for querying TBs of data is reduced while increasing the number of Vertica nodes to 5. In [122], the authors implement a POC to benchmark a hybrid architecture of MonogoDB, HBase and Cassandra on e-health clouds for an industrial project based in India. The primary components are a query interface, query administrator (which converts queries to MapReduce code),and translators for the hybrid NoSQL arrangement. Authors execute some basic queries on the cloud to validate the query efficiency of this hybrid. In [123], the authors implement a cloud-based POC comprising a hybrid of MongoDB,PostgreSQL, and Neo4j for specific healthcare data types,within the context of an Indian project for data portability between clouds. The FHIR standard2http://www.hl7.org/implement/standards/fhir/)is used for prototyping the selected data and the authors present some basic execution results to validate the approach. In [124], the authors implement a POC to compare the performance of three NoSQL databases, i.e., BaseX, eXistdb, and Berkeley DB with CouchBase. They validate the superior performance of CouchBase for high-end big data workloads.
F. Gaps in BDA Applications to Healthcare
In summary, we derive the following gaps and limitations regarding applications of big data solutions to healthcare domain:
1) The frequency of practical BDA implementations using NoSQL data stores in published research is limited; there are only 13 such articles.
2) The standard NoSQL technology stack currently comprises several tens of successful solutions which have not been employed in published research, e.g., Redis, Riak,Aerospike, BerkeleyDB, Apache Cassandra, and many tools of Hadoop’s ecosystem [8], [125]-[128].
3) Highly successful in-memory NoSQL technologies like Apache Spark and Redis have not been used and have nor had their potential fully realized in BDA healthcare applications.
4) None of the research works propose a formalized healthcare architecture for BDA, e.g., using a lambda, kappa or zeta architecture.
VII. BENEFITS OF BDA APPLICATIONS TO HEALTHCARE
In this section, we mention the specific benefits of BDA applications to healthcare identified from our research papers.These benefits can be particularly realized if heterogeneous healthcare data can be successfully converted to knowledge[4]. BDA can enhance patient care, decision-making and healthcare planning [67], and identify best practices and effective treatments [129]. Nurses also benefit from big data,since nursing care is related not only to the assessment of the patients’ clinical needs, but also to understand and focus on the psychological and social problems of the patient [130],[131]. The BDA benefits are illustrated in Fig. 13 and summarized below:
1) Better Healthcare: BDA empowers medical professionals to improve quality of life, cure diseases, avoid preventable deaths and predict epidemics. It can reduce medical errors and improve healthcare outcomes [9].
2) Better Patient Care: BDA revolutionizes patient care by identifying infections swiftly and suggesting the right treatments to patients. It also promotes personalized care to specific patients. This can be helpful to the patients to effectively manage their health such as medication adherence,diet, exercise, etc [132].
3) Better Medical Care: BDA can help hospitals and clinics to store, digitally collate and analyze its patients’ conditions related data to receive the best medical care. Through smart devices, patients can be monitored and treated irrespective of locations. This provides better 24/7 medical care and is similar to having medical staff in every patients’ room [132].
4) Better Healthcare Value: BDA can effectively reduce the costs of processing and storing of healthcare data and then apply sophisticated big data techniques to transform that patient centered data into valuable outcomes [78], [133].
5) Better Care Delivery: BDA can be helpful in preventing duplication of treatment and unnecessary laboratory tests by instantly accessing and tracking the patient’s medical history to determine the patient’s condition progress. This on-time coordination of the patients’ records can be used to increase effectiveness and efficiency of care delivery. In emergencies by delivering patients’ related information at the right time BDA provides better healthcare delivery [79], [134].
VIII. POTENTIAL OF NOSQL APPLICATIONS TO HEALTHCARE
NoSQL technologies have been able to solve a majority of data management problems and have had a global impact. It is imperative to enhance NoSQL applications to healthcare big data. We conducted several Google searches to verify that the same idea is being recommended in various industrial blogs and commentaries, described as follows:
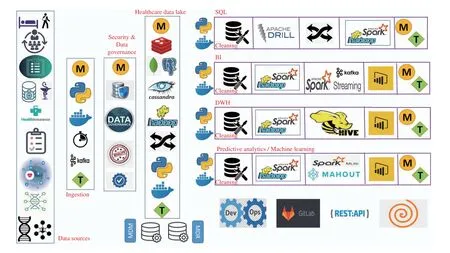
Fig. 13. Med-BDA: A state-of-the-art BDA architecture for healthcare.
1) In [135], technical analyst Martin stresses the importance of using MongoDB, Hadoop and Neo4J graph store to manage and store healthcare big data. He particularly mentions unstructured, geo-spatial and sensor healthcare data, and stresses the need to ensure success in BDA initiatives through careful selection of the NoSQL data stores.
2) The company MarkLogic has successfully implemented its proprietary NoSQL database to solve healthcare big data problems of the American Psychological Association [136].MarkLogic stresses that NoSQL is necessary as RDBMSs are now incapable of handling healthcare big data.
3) The MongoDB company lists its successful applications to store and query healthcare big data on its website [137].According to MongoDB, “Healthcare companies rely on MongoDB to address a broad variety of use cases while at the same time meeting compliance standards and improving healthcare outcomes”. Some use cases are 360-degree patient view, population management for at-risk demographics, and lab data management and analytics.
4) The CouchBase company has implemented an architecture which uses its NoSQL database to solve healthcare data management problems [138]. According to the company, this database is suitable for healthcare due to high data availability, robust connections between health mobile devices, best-in-class performance, flexibility, security,regulatory compliance and scalability.
5) In [139], the authors conduct a POC to store and query representative electronic health records (EHRs) in MongoDB,in the context of a healthcare project in Botswana. They propose a MongoDB schema for EHR and mention the benefits of this implementation for healthcare sector (e.g.,efficiency of query execution, scalability, reduced costs, load balancing, etc.).
6) In a technical report for a healthcare project in Romania[140], the authors concretely define the limitations of RDBMSs and motivate the use of NoSQL databases to solve the data management problems for patient monitoring data.They propose the use of SimpleDB, CouchDB, and MongoDB as document databases, Voldemort, Riak, Scalaris,Memcached as key-value stores, and HBase and Cassandra as wide columnar stores. They also propose MySQL Cluster,VoltDB, Clustrix, ScaleBase, NimbusDB as scalable relational systems which can, to some extent, solve healthcare BDA problems.
IX. MED-BDA: A STATE-OF-THE-ART BDA ARCHITECTURE FOR HEALTHCARE
In our opinion, the core reason for limited SQL applications for healthcare BDA is the lack of a standardized architecture primarily because: 1) the more well-known lambda and kappa BDA architectures are both complicated and expensive to implement, 2) the state-of-the-art Zeta architecture solves the issues of lambda and kappa but there is no guidance on how to implement it for the healthcare sector, 3) a rapidly expanding NoSQL technology stack makes it difficult to decide on a particular store, 4) there is a lack of available expertise to handle the complicated configuration of NoSQL stores and their programming within a BDA architecture, 5) there is a need for extensive trial-and-error to tune the usage of NoSQL data stores. To solve these issues, we propose a layered BDA architecture for healthcare big data which we label as Med-BDA (shown in Fig. 13) and in the next section, we define success strategies on how to ensure a successful BDA initiative with Med-BDA. It is important to stress that the application of Med-BDA requires a BDA requirement from the clinical managers. This requirement could be related to one or more BDA specifications, namely: data assessment and data quality management (under the umbrella of data governance), SQL-based querying, business intelligence, data warehousing or predictive analytics (machine learning).Following are the hallmark characteristics of Med-BDA:
1) State-of-the-Art Technology Stack: We have designed Med-BDA based on a thorough research of NoSQL and other big data tools and technologies with respect to their performance (read, write, query), scalability, ease-of-use,successful applications, user acceptability, limitations, and community support3A group of graduate students participated in this activity over a period of 3 months. For the sake of brevity, the details are outside the scope of this paper.We also used our previous knowledge of designing BDA architectures, e.g., our work done for the telecom sector [24].
2) Comprehensive: Med-BDA is designed for all types of healthcare analytics and BDA applications.
3) Zeta Architecture: Med-BDA follows the state-of-the-art Zeta BDA architecture proposed by MapR technologies,which solves the limitations of the historical lambda and kappa BDA architectures and enhances efficiency, resource utilization and NoSQL tool management [25].
4) Python: Med-BDA architecture development is completely based on Python language, which is the top-most big data programming language currently, according to the popularity of programming language index [141].
5) Hybrid Database: Med-BDA employs a hybrid database which combines several NoSQL stores and a relational store under one access mechanism (detailed below). To the best of our knowledge, this is the first proposal of a hybrid for healthcare and is necessary to cater for the complicated and diverse nature of healthcare data.
6) Data Governance: Med-BDA is the first BDA architecture to incorporate the requirement of data governance, a rapidly-expanding technology which ensures data quality, security and management throughout the organization and is a top-most analytical trend in 2020 [142], [143].
7) Meta-Data: In Med-BDA, we record the relevant metadata at each layer, depicted by the icon labeled “M” (with yellow background), as per requirement of data governance practices.
8) Master Data: Med-BDA also implements master data management, which is a critical activity to maintain a clean,updated and ubiquitous version of the most important data in an organization.
9) Micro-Services and Containerization: In the context of Zeta, Med-BDA uses micro-services implemented with containerization. Containers are small, light-weight software components with pre-installed functionalities. A containerized BDA architecture has numerous containers interacting with each other (called orchestration) in a plug-and-play fashion,which greatly enhances resource optimization and reduces time and cost of running the architecture. In Med-BDA, we have proposed the use of well-known Docker tool for containerization (whale icon) [144]. Each unique software component and process in Med-BDA, specifically data store,data ingestion, data governance and security process,analytical engine, data query and processing technologies and visualization tools, will be running in its own Docker container. All Docker containers within Med-BDA will coordinate with each other using either Docker Swarm,Docker Compose or Kubernetes (see [144] for more details).
10) DevOps: Development in Med-BDA follows the DevOps technology, allowing continuous development,testing, integration and live testing, all coordinated through the well-known GitLab software (a project of GitHub).DevOps is the de-facto standard in an architecture development process and is globally applied [145].
In terms of the above features, we now describe Med-BDA’s layers as follows (in order of analytical data flow):
1) Data Sources: This layer comprises all potential healthcare data sources, namely (from the top), in-patient, outpatient, human resource, EHRs, all types of medical databases, pharmaceutical, health insurance, patient surveys,IoT-related (smart devices), bioinformatics, genomics and social networking data.
2) Ingestion: This layer ingests data from Data Sources,based on the BDA requirement. Ingestion APIs should be developed in Python. The meta-data to be recorded includes name of connected sources, the time and schedule of ingestion, amount of data ingested, etc. Apache Kafka is the de-facto standard for ingesting data streams, and we recommend the same. The ingestion activity will initially pose configuration issues (as is standard for any open-source tool usage) but with tuning (represented by the icon “T” on green background), the issues will be solved. Tuning represents change of parameters and ingestion methodology.
3) Security & Data Governance: This layer implements the required data security practices, e.g., to anonymize the data, as required by clinical regulatory authorities. This is part of a data governance initiative which initially assesses the quality of the data and then implements standard rules throughout the clinical organization to improve the current quality and ensure that errors in data and analytical processes do not occur in the future [143].
4) Healthcare Data Lake: This layer inserts the audited and secure healthcare data into a hybrid database, which forms our data lake. The implementation of a lake is now standard practice in BDA [146]. For our hybrid, we propose the use of MongoDB, Redis and Apache Cassandra (running on Hadoop)as NoSQL stores and PostegreSQL as the relational store, the latter being the best relational store for BDA and used extensively by Amazon Web Services cloud. Redis can also be used as a caching service. Due to the use of Hadoop, it is also possible to execute data warehousing through Apache Hive and faster processing through Apache Spark during analytics later on. We are confident that all types of healthcare data (to be used for BDA) can be accommodated in our hybrid database. For instance:
a) Complicated and high volume EHR data can be stored in Cassandra to cater for the scalability requirement and provides excellent query performance.
b) The IoT healthcare data can be stored in MongoDB which has many successful real-time analytics use cases.
c) Bioinformatics and genomics data can be stored in Redis as key-value pairs as there are long and complicated value patterns.
d) Pharmaceutical data is more regular in nature (drug research, supply chain, sales, etc.) and can be stored in PostegreSQL.
In this layer, we recommend implementation of master data management (MDM) practices which are associated with data governance [143] along with the meta data repository (MDR).Note that meta-data will be stored in this layer extensively along with fine-tuning of the hybrid database to all the incoming data ingestion streams. Although the tuning process can get lengthy (e.g., see [114]), our choice of the data stores is expected to solve these issues more efficiently.
5) SQL: This layer allows execution of SQL queries on clean and processed data (ETL) from the lake. For this, we propose Apache Drill which is the best choice to execute SQL queries on NoSQL and relational data stores. SQL layer could also require the use of Spark (over Hadoop) for processing before SQL is applied. Meta-data is recorded and tuning for Drill and Spark usage will be required.
6) BI: This layer allows execution of Business Intelligence queries on clean and processed data (ETL) from the lake. For BI tool, we recommend using PowerBI, the leader in Gartner’s magic quadrant for BI in 2020 [147]. PowerBI can be applied to both batch data (using Spark over Hadoop) or streaming data (using Spark Streaming over Hadoop). Meta-data is recorded and tuning for Spark usage will be required.
7) DWH: This layer allows execution of data warehousing queries on clean and processed data (ETL) from the lake. For this, both Hive and Spark can be used. Meta-data is recorded and tuning for Spark and Hive usage will be required. DWH outputs (multi-dimensional aggregate values) can be viewed in PowerBI.
8) Predictive Analytics: This layer allows execution of machine learning on clean and processed data (ETL) from the lake. For this, both Apache Mahout (using MapReduce over Hadoop) and MLLib (using Spark over Hadoop) can be used.Meta-data is recorded and tuning for Mahout or MLlib usage will be required. Machine Learning outputs (prediction accuracies, etc.) can be viewed and analyzed in PowerBI.
We note that all client interaction through PowerBI dashboards will occur with REST APIs which are standard methods of communicating between GUIs and back-end architectures. The orange spiral (bottom-right) implies that the above data flows are repeatable for different types of analytics. So, given an analytical requirement, a subset of the pool of Docker containers can be used to set up an analytical pipeline on-the-fly and this pipeline can be terminated once the requirement is complete. It can be easily verified that Med-BDA is state-of-the-art and potentially more effective,efficient, cost-effective, and comprehensive as compared to other published BDA architectures for healthcare [114],[121]-[123].
X. SUCCESS STRATEGIES FOR BDA IN HEALTHCARE
In this section, we will lay down a road map to achieve a successful BDA implementation for healthcare based on our systematic review and the Med-BDA architecture. The roadmap consists of the following steps:
1) Understanding the Healthcare Domain: The initial task of the BDA team is to understand the particular healthcare process, i.e., acquire the domain knowledge. For instance,EHR analysis requires BDA team to understand the in-patient,out-patient and other treatment-related processes. To gather this knowledge, the team can use YouTube, online blogs and also domain experts (healthcare managers). This activity should last from 2 weeks to 1 month.
2) Specifying the Problem Statement: The next task of the BDA team is to interact with the clinical domain experts to extract the exact problem(s) which needs to be solved and document it. This will serve as the crux for the whole project.This step is necessary because sometimes, even the experts are not sure about the requirements as the BDA team. Multiple discussion sessions on this topic will evolve the specific problem(s). This activity should be completed within 2 weeks.
3) Identify and Understand Data: The next task is to identify the relevant data sources for solving the above problems and understand the current schema of this data,particularly the names of the attributes. For this, the BDA team will need active communication with the IT department which typically manages the database. The output is a data glossary document which will assist the team in later stages of data usage. This activity should last from 1 week to a maximum 3 weeks, and is followed by data ingestion.
4) Data Governance Application: The healthcare organizations need to implement a data governance initiative which is managed through the creation of a council comprised of all the business managers, IT managers and BDA managers in the organization. The council ensures enterprise-wide data security and regulatory requirements, data assessment for data entry errors (missing values, incorrect values, duplicated values, outlier values), and implementation of corrective rules at all data entry points to ensure autonomous removal of these errors and other related data issues in future data. Data governance is particularly important in the context of the general data protection regulation (GDPR), an EU regulation on data privacy which is now being adopted globally at a rapid pace since its introduction in 2019 [143].
5) Maintaining the Technology Stack: The big data technology stack has been evolving rapidly in the last 15 years. Our proposal of Med-BDA is based on the current standard stack. To achieve success, this needs to evolve based on the latest, well-known and most usable tools (always determined over several comparison parameters).
6) Automatic Update of BDA Results: The BDA results from Med-BDA are not a one-time execution output. Data sources will keep on pumping big healthcare data into Med-BDA. So, for each analytics process in Med-BDA, a pipeline in Python should be developed which autonomously takes the new batch of data and executes the whole BDA process until visualization in PowerBI or Apache Drill. One application is autonomous machine learning where a predictive model autonomously updates itself to cater for new training data,while maintaining the predictive accuracy.
7) Investment in Hardware vs Cloud: If allowed by regulatory bodies, medical institutions should implement Med-BDA on the cloud (AWS or Azure). This will save them the hassle of buying hardware (server machines) and maintaining a complicated network consisting of tens of Docker containers interacting with each other in different ways. Otherwise, Med-BDA will be implemented in-house over dedicated server machines (at least 3-5) along with NAS(Networked Attached Storage) as backup. This in-house setup is definitely more expensive than a cloud-based installation.
8) Investment in BDA Skillset: One of the major reasons for BDA project failures has been the lack of relevant skill sets.The medical institution and/or the BDA vendor should invest in developing a team with core BDA skill set, specifically,expertise in Python, Linux platform development, Hadoop Ecosystem and NoSQL store installation and usage, and architecture development skills.
It is important to mention that all major BDA healthcare challenges presented in Section V can be successfully addressed by Med-BDA and our success strategies, specified below:
9) Confidentiality and Data Security: Med-BDA’s security and data governance layer allows implementation of data anonymization, security requirements and regulatory compliances to protect the patients’ treatment, insurance and other clinical data. The activities in this layer will be automated and the governance team will monitor these activities on a regular basis.
10) Granular Access Control: Providing data and information access control to clinical employees, at any level of data security, is another feature of the security and data governance layer. Software programming is used to automatically execute the access rules whenever an employee logs on to the system. In fact, all data security practices can be managed extremely effectively with the right data governance tools and team (for details, refer to [143]).
11) Interoperability: Med-BDA’s hybrid database allows interoperability between different healthcare data types, by storing this data variety in different NoSQL databases and controlling them through a unified interface. We have already mentioned that this is fast becoming a practice in BDA applications [146].
12) Data and Analytics Reliability: All issues in maintaining reliability of data and analytics in healthcare BDA (listed in Section V-D) can be now solved through Med-BDA’s technology stack. For this, we have divided them in the following headings:
a) Data entry errors: Manual data entry errors are completely eliminated through application of automated, hardcoded data rules determined by the governance team after an initial data assessment activity of healthcare database.
b) Data integration: The data integration process can be managed effectively at the ingestion and data lake layer. Basic data cleaning before data integration occurs at ingestion layer,and the integrated schema (for hybrid database) is programmed in advance so ingested data is automatically integrated within the hybrid at high data speeds. The use of NoSQL allows more flexibility in data integration than relational databases, allowing a single BDA pipeline to cater for healthcare data integrated in different ways within the same database [8].
c) Data comprehension: Our hybrid database allows different NoSQL query engines to run concurrently over different healthcare data types, hence providing enhanced data comprehension more efficiently as compared to traditional technologies. Python’s programming modules is then used to further explore and understand the data, e.g., through the standard Pandas and Numpy libraries [141].
d) Data sampling: The tuning process at the ingestion layer can determine the right sample of data from real-time or near real-time data streams, due to the use of Apache Kafka, the capability of NoSQL databases to store streaming data, and containerization. In fact, testing of these samples also occurs at high speed (not in the traditional batch-based fashion).
e) Infrastructure and technology stack: Our proposal of Med-BDA solves this problem through the use of a wellresearched, effective, efficient and previously successful technology stack and architecture.
f) Inadequate training: We mention developing the BDA skillset within the clinical organization as a success strategy which could involve several employee trainings, e.g., on Docker, NoSQL databases, and containerization.
13) Data Provenance: To ensure data provenance, we record meta-data at all data activity points in Med-BDA and the choice Med-BDA’s technology stack solves all data and analytics reliability requirements.
We note that, currently, there are many different types of BDA problems in healthcare, e.g., related to patient care,pharmacy, health insurance, IOT-related (e.g., body area networks), bioinformatics and genomics. The Med-BDA architecture is generic enough to be applicable to each of these problems, particularly due to the plug-and-play nature of its zeta architecture. For each application, the roadmap defines the application process and Med-BDA provides the implementation details. In other words, the technology stack for any type of healthcare BDA application will remain exactly the same as we have proposed in Med-BDA.However, we cannot generalize this to other domains (finance,telecommunications, retail, agriculture, etc.) because the data management dynamics of each domain is unique and requires a unique, tailored architecture (e.g., see [24] for a proposed BDA architecture for telecom industry).
XI. COMPARISON TO RELATED LITERATURE REVIEW AND COMMENTARY PAPERS
In this section, we compare the hallmark features of our work to the following nine (9) selected literature review and commentary papers of BDA applications to healthcare:[148]-[151], [103], [152]-[155]4To the best of our knowledge, this list is complete as of June 2020.. The hallmark features of our work are as follows:
1) Systematic Literature Review (SLR): This feature recordswhether the compared paper is an SLR (or not). In our papers,a non-SLR paper is a commentary paper. Since our work is an SLR, we need to compare it against both SLRs and commentary papers. Note that an SLR applies a more effective and robust method of paper extraction than a commentary paper, which does not execute any research methodology.

TABLE V COMPARISON OF RELATED REVIEW AND COMMENTARY PAPERS TO OUR WORK; SLR = SYSTEMATIC LITERATURE REVIEW; CHRT =CHARACTERISTICS OF BIG DATA; BNFT = BENEFITS OF BDA; APP = BDA APPLICATIONS; CHLN = CHALLENGES OF BDA APPLICATIONS; PTNL = POTENTIAL OF BDA APPLICATIONS; LIM = LIMITATIONS AND GAPS OF BDA; SS = SUCCESS STRATEGIES OF BDA INITIATIVES; DT = BIG DATA TYPES; PROCESS = BDA PROCESS; ARCHITECTURE = BDA ARCHITECTURE; NOSQL = NOSQL DATABASES
2) Big Data Characteristics (CHRT): This feature records whether the compared paper determines big data characteristics in healthcare data or not. We have answered this through a formal research question (SRQ1).
3) Benefits of BDA Applications (BNFT): This feature records whether the compared paper investigates the benefits of BDA applications to healthcare (or not). We have answered this through a formal research question (SRQ4).
4) BDA Applications (APP): This feature records whether the compared paper extracts papers related to BDA applications to healthcare (or not). We extracted applications through a formal research questions (SRQ3) and categorized our applications in following dimensions of NoSQL (Section VI): scaling out, automated scaling, reliability, data model options, CAP theorem compliance, eventual consistency,NewSQL compliance, optimized query execution, and costeffectiveness.
5) Challenges of BDA Applications (CHLN): This feature records whether the compared paper investigates the challenges of BDA applications to healthcare (or not). We have answered this through a formal research question(SRQ2).
6) Potential of BDA Applications (PTNL): This feature records whether the compared paper investigates the potential of BDA applications to healthcare (or not).
7) Limitations of BDA Applications (LIM): This feature records whether the compared paper investigates the limitations and gaps of BDA applications to healthcare (or not). We specifically mention them in Section VI-E.
8) Success Strategies of BDA Applications (SS): This feature records whether the compared paper investigates the strategies of ensuring a successful BDA initiative in healthcare domain (or not).
9) Data Sources of BDA Applications (DT): This feature records whether the compared paper deals with BDA applications across all possible healthcare data sources (or not). In our work, we are dealing with all the datasets.
10) Process of BDA Applications (Process): This feature records whether the compared paper proposes and discusses a BDA process for healthcare (or not).
11) Architecture of BDA Applications (Architecture): This feature records whether the compared paper proposes and discusses a BDA architecture for healthcare (or not).
12) NoSQL Applications (NoSQL): This feature records whether the compared paper discusses BDA applications with respect to NoSQL databases (or not). This feature is important, considering that big data has to be stored in NoSQL databases, which itself has a strong impact on the ensuing analytics (Section III-D).
In Table V, we compare our work with the nine selected papers across the hallmark features. Our proposal of Med-BDA is unique in that none of these papers has proposed any standard BDA architecture, although some of them list steps for a BDA process. Note that in Med-BDA, we also define the process to be followed along with the architecture. Also, only[155] proposes the use of NoSQL stores for healthcare BDA besides our work, showing that the other works are not perfectly aligned with the latest trends (as shown in Section VII).The word “L” in Table V means “limited” ; for columns BNFT, CHLN, and Process, this means that the benefits and challenges are defined superficially and for APP, it means that no formal attempt was made to extract all application-related papers (true for 75% of the papers). Also, only two works are SLRs (like our work) with the rest being simple commentary papers with no formal review methodology. The first [153]reviews only 22 papers as compared to our 80, with applications limited to predictive analytics for healthcare operations and supply chain management. The second [154]reviews 65 papers as compared to our 80 with applications limited to machine learning, cloud-based, heuristic-based,agent-based, and hybrid mechanisms. This paper contains long tables difficult to interpret in one go. Big data characteristics, its potential for healthcare and big data types are all mentioned most frequently across all papers. The limitations of BDA research are mentioned in only three works while success strategies are mentioned in only two works. Overall, we have proved that this paper combines a set of hallmark features which have not been collectively addressed in any previous paper5A detailed discussion of the nine compared papers is outside the scope of this work; we invite the reader to go through these papers for more required information..
XII. CONCLUSION AND FUTURE WORK
The healthcare sector is now facing a deluge of big data from multiple heterogeneous sources. BDA is hence a core requirement to treat patients effectively and run all datarelated clinical operations smoothly. However, the BDA process is complicated to a large degree and has seen much failure in different industrial sectors due to increased costs, a skill set which is difficult to acquire, rapidly expanding technology stack, and increased management overhead. We have previously tried to present a roadmap which solves these issues to some extent for the telecommunications sector [24].In this paper, we have adopted a more aggressive approach to solving these issues for the healthcare sector through a systematic literature review. We verify big data characteristics for healthcare, identify the current BDA challenges, extract and review the BDA applications based on NoSQL databases,identify their limitations, motivate the further use of NoSQL databases to solve all BDA challenges, propose a state-of-theart BDA architecture called Med-BDA based on the latest zeta paradigm, provide a list of success strategies to ensure a successful execution of Med-BDA, identify all potential benefits of BDA and finally, compare our work with related literature reviews of healthcare analytics to prove that our aforementioned contributions are unique.
Our work endeavors to assist healthcare organizations in planning an actionable roadmap comprising big data technical experts, BDA experts, and technology and process enhancements to lead a comprehensive valuation of their current BDA abilities and capabilities, value drivers and prioritize their BDA-related goals. By applying our roadmap, organizations can develop a cost-effective and timely strategy for their BDA initiatives that leads to incremental benefits. A limitation of our roadmap is that our proposed architecture has not been currently implemented practically; the first prototype implementation is scheduled to start at Agha Khan University Hospital in July 2020, a well-known international chain of hospitals(https://hospitals.aku.edu/pakistan/Pages/default.aspx). The future work primarily involves experimenting with Med-BDA in the light of our proposed success strategies. In proposing Med-BDA, we have concretely solved the BDA problem for the healthcare sector because the implementation team now has a solution methodology and tool for every possible challenge. We will be proposing extensions to Med-BDA (if needed) in case of any abnormal changes in BDA technology stack. Finally, it is important to mention that variants of Med-BDA can be developed for other important big data applications, e.g., analysis of IoT-based production data [156],learning compressed data representations through latent factor models [157], and analysis of mobile data streams [158]. To develop the architecture, the particular requirements of a given domain needs to be initially analyzed and then the technology stack can be selected based on these requirements by big data domain experts.
杂志排行
IEEE/CAA Journal of Automatica Sinica的其它文章
- A Multi-Layered Gravitational Search Algorithm for Function Optimization and Real-World Problems
- Dynamic Hand Gesture Recognition Based on Short-Term Sampling Neural Networks
- Dust Distribution Study at the Blast Furnace Top Based on k-Sε-up Model
- A Sensorless State Estimation for A Safety-Oriented Cyber-Physical System in Urban Driving: Deep Learning Approach
- Theoretical and Experimental Investigation of Driver Noncooperative-Game Steering Control Behavior
- An Overview of Calibration Technology of Industrial Robots
