Simultaneous Determination of Twelve Normal and Modified Nucleosides in mRNA of Cancer Cells by Ultra-high Performance Liquid Chromatography-Tandem Mass Spectrometry
2020-08-22LUOYangxuDAIKaijinDUJuanXIAOHuadiZHENGLingCHENXuncaiMAAndeLUOQizhi
LUO Yang-xu,DAI Kai-jin,DU Juan,XIAO Hua-di,ZHENG Ling,CHEN Xun-cai,MA An-de*,LUO Qi-zhi
(1.Hygiene Detection Center,School of Public Health,Southern Medical University,Guangzhou 510515,China;2.Guangdong Southern Medical University Technology Development Department CO.,Ltd.,Southern Medical University,Guangzhou 510515,China;3.Forensic Science Center,Southern Medical University,Guangzhou 510515,China;4.Department of Forensic Toxicology,School of Forensic Medicine,Southern Medical University,Guangzhou 510515,China)
Abstract:An ultra-high performance liquid chromatography-tandem mass spectrometric(UPLC-MS/MS) method was developed for the simultaneous determination of twelve normal and modified nucleosides in mRNA of cancer cells.Total RNA extraction,mRNA purification and mRNA hydrolyzation were performed using cultured cancer cell samples from commercial kits,with 8-bromoguanosine(8Br-G) as internal standard.Twelve normal and modified nucleosides were separated on an Agilent RRHD SB-C18(2.1 mm×50 mm,1.8 μm) column with 0.1% formic acid methanol solution and 0.1% formic acid solution as mobile phases,and determined by UPLC-MS/MS with positive electrospray ionization(ESI+) under multiple reaction monitoring(MRM) mode.The method could meet the requirements for method validation.There were good linear relationships for twelve normal and modified nucleosides in mRNA of cancer cells in the certain concentration ranges with their correlation coefficients(r) larger than 0.99.The limits of quantitation and limits of detection were in the ranges of 0.17-3.13 ng/mL and 0.08-1.56 ng/mL,respectively.The method was then applied to human nasopharyngeal carcinoma 5-8F cell line,human cervical cancer Hela cell line and human embryonic kidney epithelial 293T cell line,with successful determination of various modified nucleosides.The method is capable of simultaneous quantitation for twelve normal and modified nucleosides in mRNA with short analysis time(6 min per sample),low number of cells(2×106 per sample) and high sensitivity,which could provide a powerful tool for cancer research and clinical diagnosis.
Key words:nucleoside;RNA modification;mRNA;cancer cell;ultra-high performance liquid chromatography-tandem mass spectrometry(UPLC-MS/MS)
Although several modified nucleosides in mRNA have been reported,most of them are still unknown.Before understanding how these modified nucleosides act as effective targets for cancer diagnosis and therapy,and whether there is joint effect or subtractive effect between the modified nucleosides,the accurate qualitative and quantitative data about them is primarily necessary.However,it is limited by current separation and detection technique owning to the following two reasons:① The modified nucleosides usually have much similar chemical structure such as structural isomers,regioisomers and isotopic isomers,resulting in low selectivity;② Most of them are in a dynamic low level of concentrations in mRNA and the complexity of biological matrices challenges the detection strategies.Hence,it is necessary to develop a simultaneous quantitative analysis method with high selectivity and sensitivity for the detection of the modified nucleosides in mRNA[21].
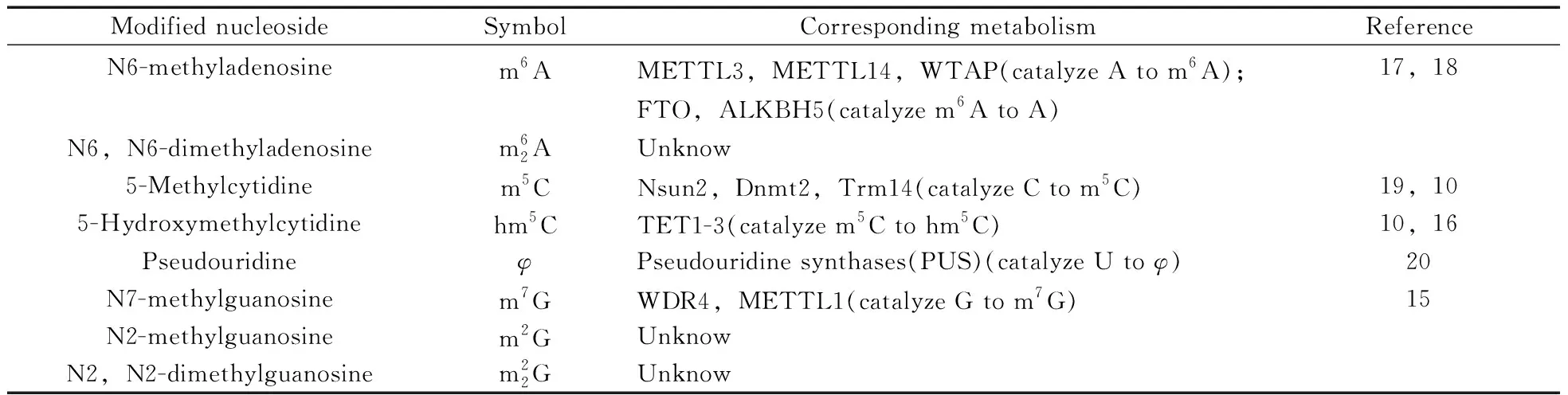
Table 1 Corresponding metabolism of the modified nucleosides discussed in this study
Traditionally,liquid chromatography(LC) coupled with UV(HPLC-UV),gas chromatography(GC),and capillary electrophoresis(CE) have been used for the determination of nucleosides in biological matrices.However,these techniques are associated with some disadvantages.HPLC-UV detection provides unsatisfactory sensitivity and selectivity because of endogenous interferences from the complex biological matrices.GC method requires a sample derivatization step which is laborious and time-consuming[22-26].In addition,genomic-based technologies,such as enzyme-linked immunosorbent assay(ELISA),immuno-northern blotting(INB)[27],microarrays and RNA sequence[28],have been developed for the nucleoside detection,but its widespread application has been limited.In case of ELISA and INB method,only a limited number of antibodies are available for certain nucleosides and some antibodies are of poor specificity[29].Microarrays and RNA sequence are designed to detect RNA modifications,which do provide advantages related to sensitivity,broad applicability to mRNA.However,it is laborious and time-consuming.Besides,it is difficult to simultaneously detect multiple nucleosides in the mRNA.At present,most of the detection methods are restricted to quantify the whole modified nucleosides in the clinical samples(such as blood and urine) but the modified nucleosides in specific mRNA,which could not elucidate the entire functions of modified nucleosides in cancer process.
Currently,ultra-high performance liquid chromatography-tandem mass spectrometric(UPLC-MS/MS) method has emerged as a powerful tool for identifying and quantifying RNA modifications due to its unique advantages,including small requirement of sample amount,superior sensitivity and selectivity,and the capacity of simultaneously quantitative analysis of multiple analytes in a single run within short time[30-34].In existing UPLC-MS/MS methods,the quantification of nucleosides from total RNAs or tRNAs have been studied,while there is no study demonstrating the simultaneously quantitative analysis of multiple modified nucleosides in mRNA,to the best of our knowledge[31-33,35].The modified nucleosides in mRNA are typically rare,which means a lower limit of detection is needed.In this work,an efficient UPLC-MS/MS method conducted with positive electrospray ionization(ESI+) in multiple reaction monitoring(MRM) mode was subsequently developed and validated for simultaneous quantitation of twelve normal and modified nucleosides in mRNA[36-37].
1 Experimental
1.1 Regents
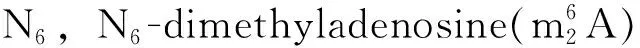
1.2 Apparatus
The UPLC-MS/MS analyses were performed on a Shimadzu UPLC system(Kyoto,Japan) consisting of two LC-20ADvp pumps,a CTO-20ACvp column heater,a SOL-20A autosampler and a CBM-20A/20Alite controller.An AB Sciex API 4000 triple quadrupole mass spectrometer(Foster City,USA) equipped with an ESI&Turb spray ionization source.All data was acquired and analyzed using Analyst software(Version 1.5.2,AB Sciex,Foster City,USA).
1.3 UPLC-MS/MS conditions
The mass spectrometric detection was operated in ESI+and MRM mode was applied to quantify the target analytes.MRM scans of the [M+H]+precursor ions for the twelve compounds and their IS,and two most predominant fragments of each target analyte were utilized.The MS conditions were presented as follows:ion source:ESI&Turb spray;ion polarity:positive;ionspray voltage:5 500 V;source temperature:600 ℃;gas:nitrogen;ion source gas 1:0.38 MPa(55 psi);ion source gas 2:0.47 MPa(65 psi);curtain gas:0.10 MPa(15 psi);collision gas:medium.The optimal parameters including MRM transitions,declustering potentials and collision energies were shown in Table 2.The structures of these compounds are presented in Fig.1.
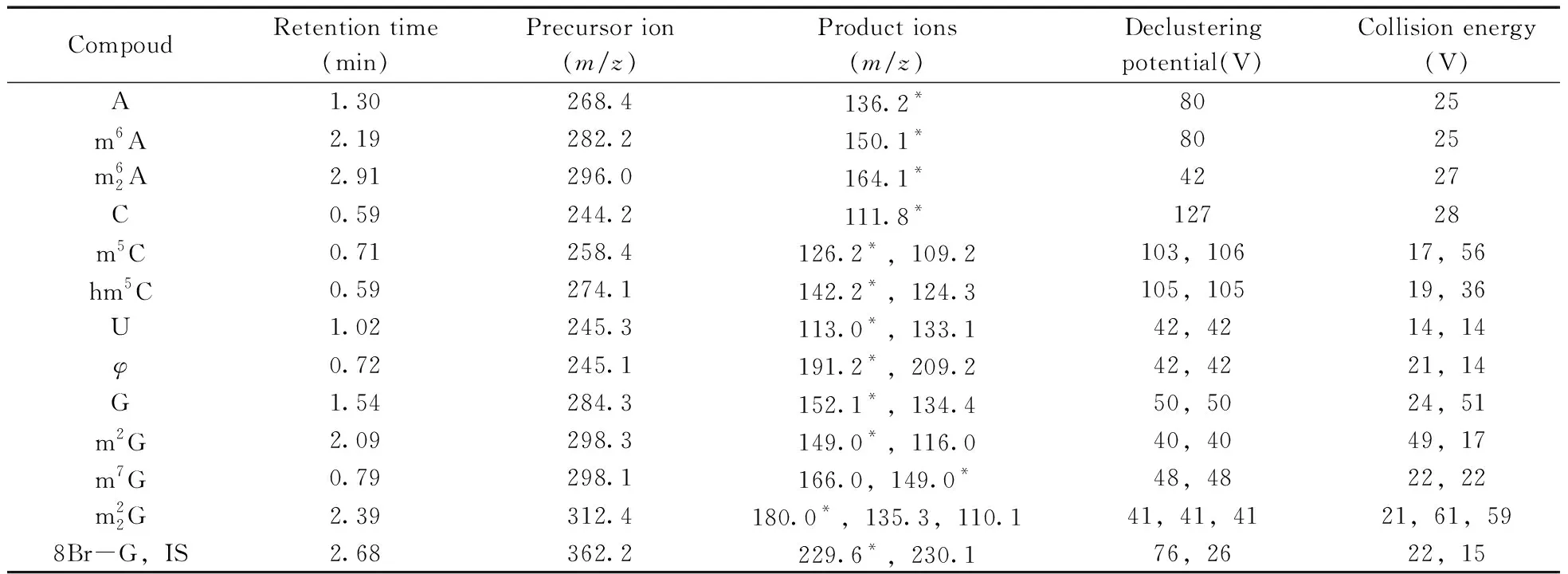
Table 2 Optimal MRM parameters for target analytes
LC separation was achieved using an Agilent RRHD SB-C18column(2.1 mm×50 mm,1.8 μm,Santa Clara,USA) with an Agilent precolumn(2.1 mm×5 mm,1.8 μm,Santa Clara,USA).The column temperature was maintained at 35 ℃ and the flow rate was 0.30 mL/min.The mobile phase consisted of 0.1%(by volume) formic acid methanol solution as the eluent A and 0.1% formic acid water solution as the eluent B.The linear gradient was shown as follows:95% B at 0 min,60% B at 2 min,95% B at 3.5 min,95% B at 6 min.The autosampler tray temperature was set at 15 ℃ and the injection volume was 10 μL.

Fig.1 Chemical structures of target analytes
1.4 Standard solutions,calibration standards and quality control samples
The standard stock solutions of target compounds were prepared by weighting and dissolving individual dry compounds in ultra-pure water with concentration of 0.1-10 mg/mL and stored at-80 ℃ in a glass vials for further sample preparation.The working solutions of analytes were freshly prepared by appropriate dilution of these standard stock solutions with ultra-pure water as required by each experiment.The calibration standards,samples at LOQ and LOD concentrations,and quality control(QC) samples at low(QCL),medium(QCM) and high(QCH) concentrations were prepared by spiking the blank matrix solution with a known quantity of analytes.8Br-G was used as IS and the working solution of IS was prepared by dissolving and diluting 8Br-G with ultra-pure water to obtain a final concentration of 500 ng/mL.
1.5 Cell culture
The protocol for cell culture was approved by Southern Medical University,Guangzhou,China.The human nasopharyngeal carcinoma 5-8F cell line(5-8F cell line),the human cervical cancer Hela cell line(Hela cell line) and the human enbryonic kidney epithelial 293T cell line(293T cell line) were purchased from American Type Culture Collection(ATCC)(Manassas,USA).These cells were cultured in 6 cm dishes(Corning Costar,Cambridge,USA) at a density of 2×106per dish,and were cultured in RPMI-1640,DMEM and DMEM medium supplemented with 10% fetal bovin serum and 1%(by volume) penicillin-streptomycin solution,respectively.These cells were maintained in a humidified incubator at 37 ℃ with 5% CO2.After 36 h,the cellular dish was digested by trypsin-EDTA solution,and then rinsed twice with pre-cold PBS.The cell suspension solution was immediately frozen in liquid nitrogen and stored at-80 ℃ for further sample preparation.
1.6 Sample and blank matrix preparation
Total RNA was extracted from cell suspension solutions using Simply P Total RNA Extraction Kit(Bioer Technology,Hangzhou,China) according to manufacturer's instructions.Then,mRNA was purified from total RNA extract using DynabeasRmRNA Purificaiton Kit(Ambion,Thermo Fisher Scientific,Waltham,USA) according to manufacturer's instructions.The mRNA hydrolyzation step was performed using Nuclease P1(N8630,Sigma-Aldrich,St.Louis,USA) and Cloned Alkaline Phosphatase(CIAP)(2250A,Takara,Kusatsu,Japan) successively as previously described[15,35,38].The details of the hydrolyzation step were as follow:20 μL mRNA extract was mixed with 0.5 μL of 1 U/μL muclease P1 solution and 2 μL of 100 mmol/L ammonium acetate solution and heated in a water bath at 42 ℃ for 6 h,and then the solution was added with 2.5 μL MES buffer and 0.5 μL of 1 U/μL CIAP solution and heated in a water bath at 37 ℃ for 6 h.All obtained samples were added with 10%(by volume) IS solution prior to UPLC-ESI-MS/MS analysis.The blank matrix solution was prepared by treating cell-free culture medium by the above method and dissolved in ultra-pure water for method validation.
1.7 Method validation
The analytical characters of the method,including selectivity,linearity and sensitivity,precision and accuracy,matrix effect and stability,were strictly validated according to the guidance of China Food and Drug Administration(CFDA) for Bioanalytical Method Validation Criteria[39].
The selectivity was tested by comparing blank matrix samples and QC samples(n=6).The responses of any interferences should be below 20% responses of target analytes and 5% response of IS.As tested nucleosides were endogenous compounds in mRNA of cells,the calibration standards at different concentrations were spiked in blank matrix solutions to yield a calibration curve(n=3).The calibration curves were constructed to confirm the linear relationship between the ratios of peak areas of each target analyte to the area of IS(y) and the nominal concentration of the analyte(x,ng/mL).The regression coefficient was calculated as regression parameter by weighted(1/x2) least square linear regression.The limit of quantitation(LOQ) and detection(LOD) for each analyte were measured by injecting working solutions serially diluted by blank matrix solutions(n=6) and calculated at a signal-to-noise ratio(S/N) of at least 10>∶1 and 3>∶1,respectively.The accuracy was determined by comparing nominal concentrations with measured concentrations of QC samples at low,medium and high concentrations and expressed as relative error(RE,%).The precision was determined by calculating relative standard deviation(RSD,%) of the replicate analyses.Intra- and inter-day precision and accuracy were investigated using QC samples at low,medium and high concentrations on the same day(n=6) and over three days(n=18).The accepted criteria were no more than 15% for precision RSD(%) and within±15% for accuracy RE(%).Matrix effect was investigated by comparing the measured concentrations of target analytes in blank matrix solutions spiked with low and high concentrations with corresponding neat standard working solutions in 3 duplicates.Stability validation was performed through measuring the QC samples at low and high concentrations in 3 duplicates under conditions including 24 h storage at 4 ℃ in the dark,1 month storage at-80 ℃ and freeze and thaw(3 cycles from-80 ℃ to 4 ℃) in 3 duplicates.
2 Results and discussion
2.1 Method development and optimization

2.1.1 Optimization of MS parametersThe optimization of MS parameters was carried out by injecting a constant stream of single standard solutions into the ESI source using a syringe infusion pump.The positive ESI mode was found to be more effective.The [M+H]+precursor ions were used for target analytes and their internal standard,because the molecular ions were the most dominant for each compound.The optimal ESI parameters were selected as described in “Section 1.3”.As far as we know,MRM mode with characteristic precursor and product ions is the golden standard for simultaneous quantification of multiple compounds in complex biological matrices.To minimize the effect of interferences from the biological samples and achieve the highest sensitivity and selectivity,the most intense transitions and fragmentation conditions were optimized as shown in Table 2.
2.1.2 Optimization of UPLC-ESI-MS/MS conditionsFurthermore,to develop a simple,fast and reliable UPLC-ESI-MS/MS method for simultaneous quantification of nucleosides in mRNA of cancer cells,an efficient liquid chromatographic separation of various nucleosides before MS detection is crucial.The optimization of LC conditions was primarily focused on the choice of column.Modified nucleosides are mostly polar and are usually analyzed by standard reversed-phase HPLC utilizing traditional columns with C18stationary phases.Several different commercial columns,including Agilent RRHD SB-C18column(2.1 mm×50 mm,1.8 μm,Santa Clara,USA),Agilent Poroshell 120 SB-C18column(4.6 mm×100 mm,2.7 μm,Santa Clara,USA) and Thermo Scientific BDS HYPERSIL C18column(4.6 mm×50 mm,2.4 μm,Waltham,USA) were tested.RRHD SB-C18column was found to be the most suitable one with satisfactory separation and retention time.The packing material consists of adimethyl-n-octadecylsilane monolayer bonded to a double-endcapped,porous silica support which reduce adsorption of basic and highly polar compounds.Meanwhile,the 1.8 μm particle diameter provide high-speed and higher resolution separations for most complex samples.To further improve the peak shape,resolution and reproducibility on RRHD SB-C18column,mobile phases,gradient profiles,column temperatures and volumes of the injected sample were tested and optimized.The optimal LC conditions were selected as described in “Section 1.3”.The combination of effective LC separation and MRM detection could help reduce the interferences and simultaneously quantify target nucleosides.The typical MRM chromatograms of quality control(QC) samples and blank matrix samples were presented in Fig.2.As a result,the optimized method is capable of simultaneously quantifying up to 12 nucleosides in mRNA of cancer cells within 6 min.

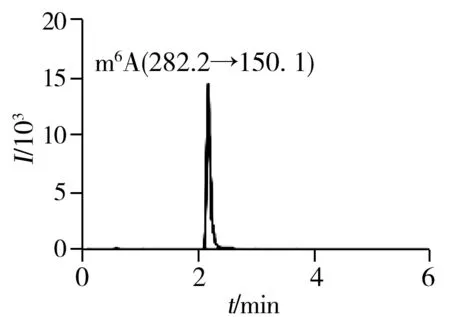








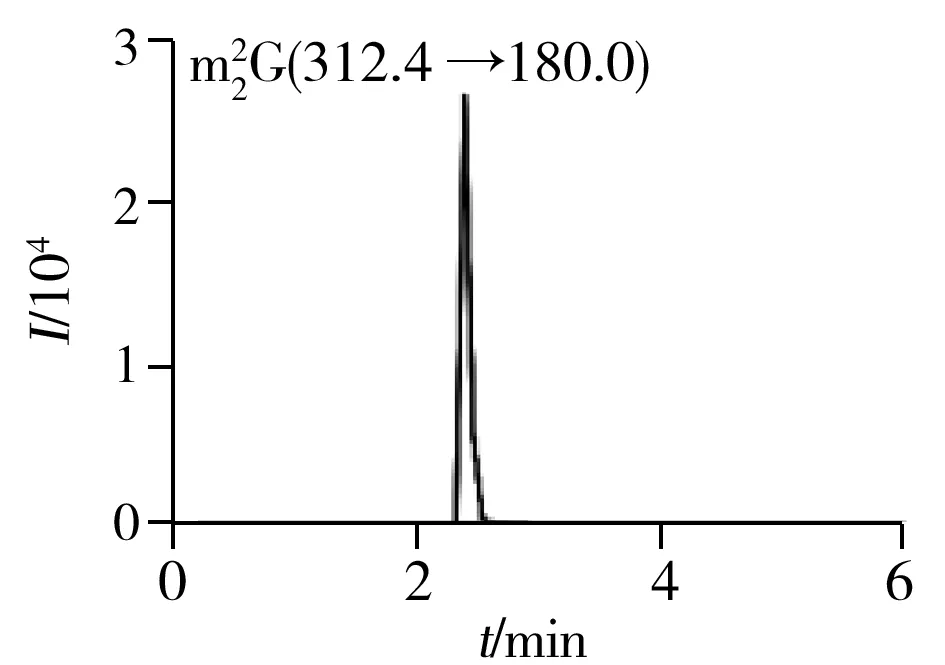
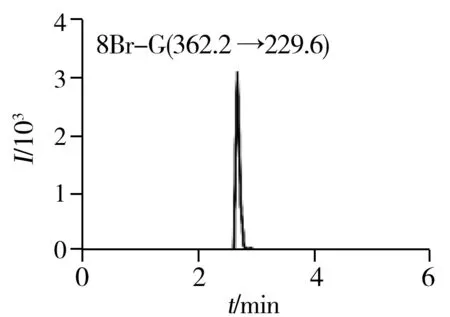
Fig.2 Typical MRM chromatograms of quality control(QC) samples
2.2 Method validation
2.2.1 SelectivityThe typical MRM chromatograms of QC samples were presented in Fig.2,which suggested that there were no significant endogenous interferences around the retention time for all the analytical targets and IS.The results indicated our developed method was capable of determining twelve normal and modifiednucleosides in mRNA with an acceptable selectivity.
2.2.2 Linearity and sensitivityThe calibration equations,correlation coefficients(r),linear ranges,limits of quantitation(LOQ) and detection(LOD) were summarized in Table 3.The calibration curves were established with more than 6 points and showed good linearities with correlation coefficients(r) more than 0.99.The LOQ and LOD for all the analytes were in the range of 0.17-3.13 ng/mL and 0.08-1.56 ng/mL,respectively.

Table 3 The calibration equations,correlation coefficient(r),linear ranges,limits of quantitation(LOQ) and detection(LOD) of twelve nucleosides
2.2.3 Precision and accuracyThe intra- and inter-day accuracy and precision of QC samples at low(QCL),medium(QCM) and high(QCH) concentration were summarized in Table 4.The relative standard deviations(RSD) were not more than 8.7% for intra-day precision and 12% for inter-day precision,respectively.The relative error(RE) values were in the range of-14%-7.6% for intra-day precision,and-15%-8.2% for inter-day precision.It is worth mentioning that an RSD(%) of 15% for precision and an RE(%) within±15% for accuracy are deemed acceptable.The results indicated that the developed method was accurate,stable and reproducible.
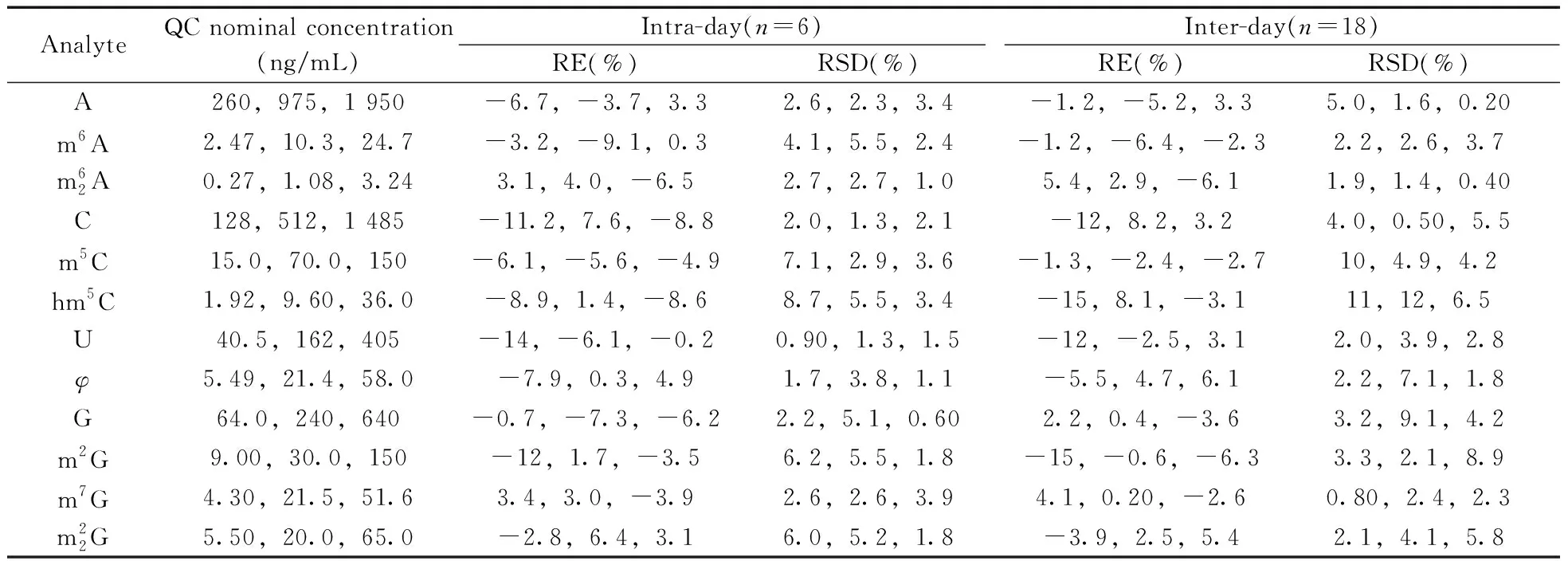
Table 4 Precision(RSD,%) and accuracy(RE,%) for the determination of twelve nucleosides
2.2.4 Matrix effectThe matrix effect was evaluated on QC samples at low and high concentration levels in 3 duplicates,respectively,and then presented as the measured concentration ratio(%) of spiked blank matrix samples(QC samples) to corresponding neat standard solutions.As shown in Table 5,the matrix effects(%) ranged from 92.0% to 106%.The results indicated that the matrix effect in this method was low enough for the reliable analysis to real application.

Table 5 Matrix effect and stability for the determination of twelve nucleosides
2.2.5 StabilityThe stability data of all analytes were calculated by comparing QC samples measured after storage at different conditions(see Table 5) to QC samples immediately measured after fresh preparation in 3 duplicates.As shown in Table 5,the results showed all analytes were stable under different conditions including 24 h storage at 4 ℃ in the dark(Condition 1),1 month storage at-80 ℃(Condition 2) and freeze and thaw(3 cycles from-80 ℃ to 4 ℃)(Condition 3).
2.3 Application in three cell lines


Table 6 mRNA modifications in 5-8F,293T and Hela cell lines






Fig.3 Typical MRM chromatograms of normal(upper) and modified(below,enlarged image) nucleosides in 5-8F cell line(a),Hela cell line(b) and 293T cell line(c)
3 Conclusions