Assessment of biological functions for C3A cells interacting with adverse environments of liver failure plasma
2020-05-10ZuHongLiZhongYangXieXiaoXiOuyangKaiZhouHuangXiaoPengYuYaLeiZhaoYanHongZhangDanHuaZhuJiongYuLanJuanLi
Zu-Hong Li , Zhong-Yang Xie , Xiao-Xi Ouyang , Kai-Zhou Huang, Xiao-Peng Yu,Ya-Lei Zhao, Yan-Hong Zhang, Dan-Hua Zhu, Jiong Yu, Lan-Juan Li
State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases,the First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310003, China
ABSTRACT
Keywords:
C3A cells
Acute liver failure
Biological function
CYP450 enzymes
Introduction
Liver failure is a severe clinical syndrome with high mortality rates [1] . Immune dysregulation is now recognized to be central to the pathogenesis of acute liver failure (ALF), and it is likely that the clinical features and outcomes are related to innate immune response to liver injury in patients [2] . It is postulated that immune dysfunction mediated the initial inflammatory insult to the subsequent multiple organ dysfunction syndrome [3 , 4] .Local tissue injury triggers local mediators of innate immunity to release an array of pro-inflammatory mediators through activation of macrophages, polymorphonuclear phagocytes, endothelial cells and complement system. Systemic inflammation occurs following intravascular augmentation of local inflammatory mediators.Pro-inflammatory cytokines recruit circulating neutrophils, B and T cells and increase vascular permeability. A rise in circulating levels of anti-inflammatory cytokines and mediators in response to systemic pro-inflammatory cytokine release, termed compensatory anti-inflammatory response syndrome, develop to prevent overwhelming inflammation. This counter-regulatory homeostatic mechanism is defined by persistently elevated circulating levels of anti-inflammatory cytokines, and by impairment in cellular immune function. Finally, the phase of immunological dissonance represents the persistence of an “unbalanced” inflammatory response that results in refractory multiple organ failure and increased risk of death.
Orthotopic liver transplantation is often the only treatment available to improve the patient survival [5] . However, liver transplantation is largely limited by the shortage of donor organs worldwide. The bioartificial liver (BAL) has been developed to be a promising alternative for patients with liver failure as a bridge to liver transplantation or to regeneration of liver functions [6 , 7] . To date, the choice of biocomponent for BAL is still under debate. Ideally, primary human hepatocytes should be preferred cell resource for BAL application. Unfortunately, these hepatocytes are not easily available due to the same shortage of organs. Hence, human hepatoma-derived cell lines are used as surrogates for primary hepatocytes as they maintain some hepatocyte-specific characteristics [8] .
The C3A cell line, a clonal derivative of hepatoblastoma-based HepG2 cell line, has been utilized in BAL for its better differentiated hepatocyte phenotype. Previous studies [9 , 10] have shown that the synthetic function of albumin for C3A-BAL is comparable to that of primary hepatocytes based BAL. It was also reported that C3A-BAL has the capacity for gluconeogenesis. But, in other studies [11 , 12] , C3A-BAL consumes glucose and produces lactate.Moreover, ammonia elimination and ureagenesis were reported in different C3A-BALs [11 , 13] . Further, despite the lack of convincinginvitrodata, C3A cell-based BALs have been evaluated in patients with ALF [14] .
Along with the importance of cell resource in BAL, the interaction between adverse environments of liver failure plasma and hepatocytes also plays a significant role to achieve optimal performance for BAL system. There are some concerns that the accumulation of toxic substances in plasma, including cytokines deriving from deranged immune response, lipopolysaccharides and metabolic products, such as ammonia, bile acids and bilirubin, potentially exerts detrimental effects on biological functions of hepatocytes [15] . It has been reported that culture with liver failure plasma could induce cytotoxicity with diminished DNA and protein synthesis in HHY41 cells [16] . Similarly, fulminant liver failure serum inhibits growth rate and synthesis of RNA, DNA and protein in cultured HepG2 cells [17] . In another study, however, the overall cellular functions were not impaired for in vitro human hepatocytes exposed to ALF serum [18] . Up to now, there are only a few studies working at the metabolic alternations of C3A cells under the culture environment with liver failure plasma, mainly concerning on carbohydrate metabolism, total protein synthesis and ureagenesis [19] , barely on the modulations of detoxification functions, such as CYP450 enzymes. The aim of this study was to investigate the effects of plasma from patients of ALF on the growth and biological functions of C3A cells in vitro, especially on the alterations of molecular levels and functions for CYP450 enzymes.
Materials and methods
Materials
The C3A cell line was obtained from the American Type Culture Collection (ATCC). Cell Counting Kit-8 (CCK-8), lactate dehydrogenase (LDH) Assay Kit and RIPA lysis buffer were purchased from Beyotime Biotechnology (Shanghai, China). TRIzol reagent was from Invitrogen Corporation (Carlsbad, CA, USA). PrimeScriptRT reagent Kit and SYBR R ○Premix Ex TaqTM II (Tli RNaseH Plus) for quantitative PCR were bought from Takara Bio Inc. (Tokyo, Japan). Mini-PROTEAN TGX precast protein gels was from Bio-Rad (Hercules,CA, USA). Ammonia Assay Kit was from Sigma-Aldrich (St. Louis,MO, USA). Albumin Human ELISA Kit and antibodies for Western blotting were bought from Abcam (Cambridge, MA, USA). The reagent of P450-GloTM Assays was purchased from Promega (Madison, WI, USA). BCA Kit was purchased from Thermo Fisher Scientific (Waltham, MA, USA).
Plasma samples collection
ALF plasma was obtained from 5 patients, referred to ALF diagnosis criteria of EASL 2017 [20] , at the onset of artificial liver system treatment mainly based on plasmapheresis. All patients signed a consent form approved by the ethics committee. Normal plasma was collected from fresh frozen plasma supplied for clinical treatment of plasmapheresis. The plasma samples were stored at -80 °C until used. The procedure was approved by the ethical committee of the First Affiliated Hospital, Zhejiang University School of Medicine and in accordance with theDeclarationofHelsinkiof 1975.
Cell viability and LDH assay
Cell viability was measured by CCK-8 assay [21-23] . Briefly,each group of cells was seeded at a density of 3 ×104cells per well in 96-well plates. After 12 h, the culture medium was replaced by fresh DMEM medium containing 10% FBS (control group), fresh DMEM medium containing 10% normal plasma (normal plasma group) and ALF plasma (ALF plasma group), respectively. After 24 h incubation, the cells were washed with PBS and tested according to the manufacturer’s instructions.
To determine the LDH release, each group of cells was seeded at a density of 3 ×104cells per well in 96-well plates. After 12 h, the culture medium was replaced by fresh DMEM medium containing 10% FBS, fresh DMEM medium containing 10% normal plasma and ALF plasma, respectively. After 24 h incubation, the cells were washed with PBS and incubated with DMEM medium without serum or plasma for another 8 h. Then, the incubated medium of each group was collected and tested following the procedure of LDH Assay Kit [21-23] .
Quantitative RT-PCR
Each group of cells was seeded at a density of 5 ×105cells per well in 12-well plates. After 24 h, the culture medium was replaced by fresh DMEM medium containing 10% FBS, fresh DMEM medium containing 10% normal plasma and ALF plasma, respectively. After 12 h incubation, the cells were washed with cold PBS and used to extract total RNA with TRIzol reagent. Then, total mRNA was reverse-transcribed into cDNA using PrimeScriptRT reagent Kit according to the manufacturer’s protocol. SYBR Green quantitative PCR was applied to quantify the PCR amplification. The relative mRNA expression levels of target genes were normalized to those of GAPDH or actin.
Western blotting analysis
For Western blotting analysis, each group of cells was seeded at a density of 1 ×106cells per well in 6-well plates. After 12 h, the culture medium was replaced by fresh DMEM medium containing 10% FBS, fresh DMEM medium containing 10% normal plasma and ALF plasma, respectively. After 24 h incubation, the cells were harvested and lysed in RIPA lysis buffer containing protease inhibitor cocktail. The protein concentration was determined by BCA protein assay. Targeted proteins in lysates were separated by SDS-PAGE electrophoresis using 4%-15% Mini-PROTEAN TGX precast protein gels, and were immuno-blotted with their specific antibodies.
Ammonia assay
To measure the ammonia concentration in the supernatant,each group of cells was seeded at a density of 3 ×104cells per well in 96-well plates. After 12 h, the culture medium was replaced by fresh DMEM medium containing 10% FBS, fresh DMEM medium containing 10% normal plasma and ALF plasma, respectively. After 24 h incubation, the cells were washed by PBS and continuously incubated with fresh DMEM medium containing 2 μg/mL of ammonia without addition of FBS and phenol red for about 12 h. Then, the culture supernatants were collected and detected using Ammonia Assay Kit according to the manufacturer’ protocol.

Table 1Clinical characteristics of patients with ALF who received artificial liver system treatment.

Table 2Blood biochemical parameters in normal and ALF plasma samples.
Albumin secretion
Each group of cells was seeded at a density of 3 ×104cells per well in 96-well plates. After 12 h, the culture medium was replaced by fresh DMEM medium containing 10% FBS, fresh DMEM medium containing 10% normal plasma and ALF plasma, respectively. After 24 h incubation, the cells were washed by warm PBS and continuously incubated with fresh DMEM medium containing 10% FBS for 24 h. Then, the culture medium were centrifuged to remove debris and collected for further examination. Albumin concentration in samples was tested using Albumin Human ELISA Kit [21-23] .
Cytochrome P450 activity assay
The measurement of CYP450 activity was performed with P450-GloTM Assays [21-23] . Each group of cells was seeded at a density of 3 ×104cells per well in 96-well plates. After 24 h, the culture medium was replaced by fresh DMEM medium containing 10%FBS, fresh DMEM medium containing 10% normal plasma and ALF plasma, respectively. After 24 h incubation, the cells were washed by PBS and treated with test compounds under the guideline of P450-GloTM Assays.
Statistical analysis
Experiments were duplicated at least three times. Data were expressed as mean ±standard deviation (SD). Difference between two experimental groups was analyzed using Student’st-test. AP<0.05 was considered statistically significant.
Results
The clinical details of five patients with ALF who received artifi cial liver system treatment are given in Table 1 . Blood biochemical parameters, including albumin, alkaline phosphatase (ALP),aspartate aminotransferase (AST), alanine aminotransferase (ALT),gamma-glutamyltransferase (GGT), complement 3 (C3) and 4 (C4),in ALF and normal plasma samples are listed in Table 2 .
Effects of ALF plasma on cell viability and cell damage
The cell viabilities were decreased 15% in ALF plasma group than that in control group, while cell viabilities of C3A cells in normal plasma group were similar to that in control group( Fig. 1 (A)). LDH assay was further carried out to examine the cell damage under different culture conditions. The LDH leakage of C3A cells in ALF plasma group had 1.3-fold elevation compared with that in control group ( Fig. 1 (B)). There was a minor increase of LDH leakage for C3A cells in normal plasma group.
Evaluation of hepatic gene transcription
Gene transcription for CYP450 enzymes showed different transcript level changes under the culture of ALF plasma. Transcript levels of CYP1A2 and CYP2B6 were increased about 3-fold and 1.5-fold in ALF plasma group compared with those in control group ( Fig. 2 (A) and (B)). In contrast, the mRNA levels of CYP2C8,CYP2D6, CYP2E1, CYP3A4 and CYP3A7 were decreased 3-, 1.3-, 1.4-,6.4- and 5.2-fold in ALF plasma group, respectively ( Fig. 2 (C)-(G)).CYP3A5 transcript level had no obvious decrease from that in control group ( Fig. 2 (H)). In normal plasma group, all of the CYP450 enzyme genes obtained higher transcript levels than those in control group.
The transcription of UDP-glucuronosyl transferase 1A1(UGT1A1) in ALF plasma group was increased 2.5-fold relative to the control group (P<0.01, Fig. 3 (A)). Transcript levels of UGT2B4 and human coagulation factor X (HCF-X) in ALF plasma group were approximately the same as those in control group( Fig. 3 (B) and (E)). After the culture under ALF plasma exposure,transcript levels of UGT2B7 (P<0.01), glutathione S-transferases(GST) and albumin (P<0.01) were decreased 2.2-, 1.7- and 1.7-fold compared with those in control group, respectively ( Fig. 3 (C), (D),and (F)). As the same profiles of mRNA levels for CYP450 enzymes,transcription levels of C3A cells cultured with normal plasma were higher than those in control group.
Effects of ALF plasma on protein expression
The protein expression of CYP1A2, CYP2D6, CYP3A4 and hepatocyte nuclear factor 4 alpha (HNF-4α) were detected by Western blotting analysis. For CYP1A2, CYP2D6 and CYP3A4, protein levels in C3A cells cultured with ALF plasma did not significantly differ from those in normal plasma group and control group ( Fig. 4 ).Likewise, different culture conditions had no obvious effects on protein levels of HNF-4αin three groups ( Fig. 4 ).

Fig. 1. The effects of ALF plasma on cell viability and cell damage as measured by CCK-8 assay (A) and LDH leakage assay (B). Values are expressed as mean ±SD, n = 3.**P < 0.01, versus control group.
Effects of ALF plasma on albumin secretion and ammonia metabolism
The concentration of secreted albumin was 3.3 μg/mL for C3A cells in control group, while it was decreased to 1.7 μg/mL for C3A cells in ALF plasma group (P<0.01). Secreted albumin of C3A cells was 5.4 μg/mL after cultured with normal plasma (P>0.05,Fig. 5 (A)). To assess the ammonia metabolism after different treatments, culture medium containing 2 μg/mL of ammonia was utilized to determine the final ammonia concentration. The results indicated that C3A cells in ALF plasma group can produce as much as 8.9 μg/mL of ammonia in 12 h incubation, while C3A cells in control group and normal plasma group produced 6.3 μg/mL and 7.4 μg/mL ammonia in 12 h incubation, respectively ( Fig. 5 (B)).
Assessment of CYP450 enzyme activity
For CYP1A1 enzyme, there was 25% activity reduction for C3A cells in ALF plasma group compared with that in control group( Fig. 6 (A)). In contrast, the enzyme activity of CYP1A2 had no obvious alternation in ALF plasma group relative to control group( Fig. 6 (B)). As shown, the CYP3A4 activity of C3A cells was only 23% in ALF plasma group in comparison with that in control group(P<0.01, Fig. 6 (D)). As the same, CYP2B6 activity also reduced to 59% for C3A cells in ALF plasma group (P<0.01, Fig. 6 (C)). In normal plasma group, the enzyme activities of CYP1A1, CYP1A2 and CYP3A4 had 10%, 50% and 40% reduction, respectively, while the CYP2B6 activity kept identical with that in control group.
Discussion
The occurrence of liver failure always accompanies with accumulation of a wide range of potential cytotoxic substances in blood, originating from either necrotic debris of hepatocytes or metabolites that are normally eliminated in the hepatocytes. Some studies pointed out that hydrophobic bile acids can induce hepatocyte apoptosis in vivo or in vitro, which involved different mechanisms including ligand-independent CD95 activation, JNK, oxidative stress and CD95 membrane targeting [24 , 25] . In addition, it was shown that serum from patients with fulminant liver failure can cause hepatocyte detachment and apoptosis by aβ1-integrin pathway [26] .
In this study, we demonstrated that culture condition with ALF plasma mildly inhibited cell viability and increased LDH leakage in C3A cells after 24 h incubation. Previous experimental studies [16 , 17] have exhibited the inhibitory effects of ALF plasma/serum from patients on growth and macromolecular synthesis in different cell lines. Shi and coworkers found that growth rate of HepG2 cell was significantly inhibited in 10% ALF serum during the 4 day culture [17] . McCloskey et al. reported that ALF plasma induced significantly increased51Cr release, and decreased total protein synthesis in HHY41 cells following 16 h incubation[16] . Complement system cascade can be very effective in inducing cellular apoptosis events, which are present in normal plasma or ALF plasma. Components of complement 3 and 4 (C3 and C4)in normal and ALF plasma samples were determined and listed in Table 2 , indicating that levels of both C3 and C4 in normal plasma were much higher than that in ALF plasma. However, as shown in Fig. 1 , cell viabilities and LDH in normal plasma group were similar to that in control group, while ALF plasma group exhibited decreased cell viabilities and elevated LDH leakage.Thus, it was unlikely that the inhibited cell viability was caused by complement system cascade in ALF plasma group. In a recent study, Melgaço et al. explored the role of C3 in ALF caused by viral hepatitis, showing low levels of C3a in ALF plasma samples,and increased apoptotic events in HepG2 cells exposed under ALF plasma [27] . Viral load is another possible factor that affects cell viability. HBV exhibits a very narrow host range and a strong tropism for liver parenchymal cells, assuming that susceptibility to HBV infection is restricted to differentiated cells. Although several human hepatoma-derived cell lines, including HepG2 cells,support HBV replication after HBV DNA transfection, none of them are susceptible to HBV infection [27 , 28] . There is no evidence that HepG2-derived C3A cells are susceptible to HBV infection, and thus causes negligible adverse effects on cell viability.
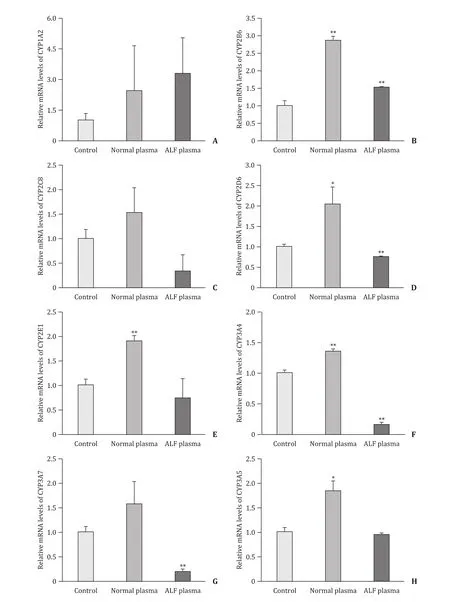
Fig. 2. The effect of ALF plasma on transcription levels of CYP450 genes including CYP1A2 ( A ), CYP2B6 ( B ), CYP2C8 ( C ), CYP2D6 ( D), CYP2E1 ( E), CYP3A4 ( F), CYP3A7 ( G)and CYP3A5 ( H). Values are expressed as mean ± SD, n = 3. *P < 0.05, **P < 0.01, versus control group.
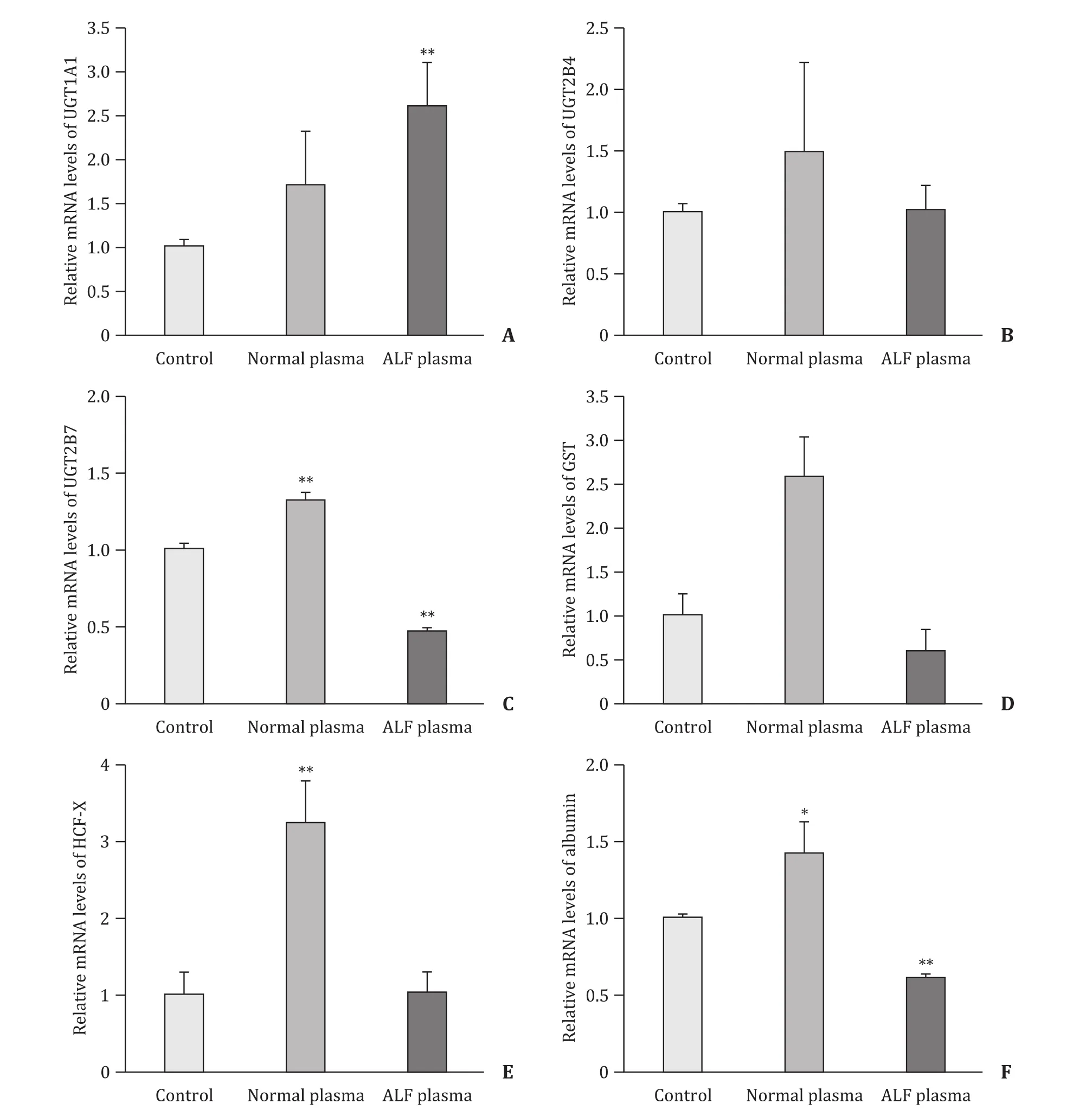
Fig. 3. The effect of ALF plasma on transcription levels of hepatic genes including UGT1A1 ( A), UGT2B4 ( B), UGT2B7 ( C), GST ( D), HCF-X ( E) and albumin ( F). Values are expressed as mean ± SD, n = 3. *P < 0.05, **P < 0.01, versus control group.

Fig. 4. The protein expression of CYP1A2, CYP2D6, CYP3A4 and HNF-4 αassessed by Western blotting analysis.

Fig. 5. The effect of ALF plasma on albumin secretion ( A) and ammonia production ( B). Values are expressed as mean ± SD, n = 3. **P < 0.01, versus control group.

Fig. 6. The effect of ALF plasma on CYP450 enzyme activity: CYP1A1 ( A), CYP1A2 ( B), CYP2B6 ( C), CYP3A4 ( D ). Values are expressed as mean ± SD, n = 3. **P < 0.01, versus control group.
The CYP450 constitute the major enzymes family capable of catalyzing the oxidative biotransformation of most drugs and other lipophilic xenobiotics [29 , 30] . CYP1A1 and CYP1A2 are the two members of CYP1 family. In humans, CYP1A2 is constitutively expressed at higher levels only in liver, while CYP1A1 is found in liver at relatively low levels [31] . In comparison with control group,CYP1A2 gene transcription enhanced in ALF group, but the protein level and enzyme activity had almost no changes in 24 h culture.The enzyme activity of CYP1A1 displayed only a modest decrease in ALF group. CYP1A2 gene is highly inducible by numerous xenobiotics such as methylcholanthrene and other polycyclic aromatic hydrocarbons [32] . It is quite likely that the metabolites in patient’s plasma such as bile acid, possessing similar chemical structure with polycyclic aromatic hydrocarbons, cause the up-regulation of CYP1A2 mRNA level. However, the protein level did not keep pace with the elevation of CYP1A2 mRNA, possibly because of the nonlinear relationship between mRNA abundance and protein expression. CYP2B6, CYP2C8, CYP2D6 and CYP2E1 all are the members of CYP2 family. In humans, all of them belong to the minor hepatic CYP450s. The culture of C3A cells under ALF plasma increased the CYP2B6 gene transcription, but interestingly enzyme activity of CYP2B6 reduced about 40% compared with control group. It is speculated that some components in plasma enhance the activity of CYP2B6 promoter, while several small molecular metabolites become potent inhibitors for CYP2B6 enzyme. Although the mRNA level of CYP2D6 had mild reduction, there is no significant difference between control group and ALF group on protein expressions. CYP3A4, CYP3A5 and CYP3A7 are the main members of CYP3 family. CYP3A4 is in the majority of individuals abundantly expressed in liver, while expressions of the minor isoforms, CYP3A5 and CYP3A7, are generally lower compared with CYP3A4 [33] . The effects of ALF plasma on C3A cells resulted in a significant downregulation of CYP3A4 gene transcription and enzyme activity, but the protein expression almost kept identical with that of control group. A previous study documented that cytokine can mediate the down-regulation of CYP3A4 via JAK/STAT pathway [34] , thus the cytokine accumulated in plasma perhaps is one of the important reasons to down-regulate the CYP3A4 gene transcription.Since there was little change on protein level, the obvious reduction of enzyme activity is possibly due to the inhibiting factors in plasma or post-translational modification of CYP3A4 enzyme under culture conditions.
UGT predominantly catalyze endo- and xenobiotics to form hydrophilic conjugates via a reaction referred to as glucuronidation,which involves the covalent linkage of glucuronic acid to a substrate bearing a suitable functional group [35] . Among all of the UGT isoforms known to date, UGT1A1 is the only enzyme responsible for elimination of bilirubin. It was reported that the UGT1A1 gene can be activated by the arylhydrocarbon receptor in response to their ligands [36] . In this study, the up-regulation of UGT1A1 gene transcription after ALF plasma exposure was considered to be related to the accumulated bilirubin in ALF plasma, by which some transcription factors were activated to initialize the elevation of UGT1A1 mRNA expression. HNF-4α, highly expressed in liver,is a member of the nuclear receptor superfamily. HNF-4αregulates constitutive expression of a large number of target genes,including several gene encoding human CYP450 enzymes, UDP-glucuronosyltransferases and sulfotransferases [37] . Although there were deleterious factors in ALF plasma, it was seemed that the protein level of HNF-4αwas not affected in C3A cells under the culture conditions with ALF plasma.
Albumin secretion plays an important role in biological functions of hepatocytes in liver. Culture conditions with ALF plasma down-regulated the albumin mRNA level, and subsequently decreased albumin concentration to approximately half amount of that in control group. Ammonia metabolism is a key function due to the involvement of dysfunction on ammonia detoxification in disturbing cerebral function in liver failure. Mavri-Damelin et al. [38] have reported that because the absence of ornithine transcarbamylase and arginase I mRNA and protein for urea cycle,ammonia cannot be detoxified via urea cycle in C3A cell line.Arginase II, which produces urea independently of the urea cycle and crucially is not involved in ammonia detoxification, is the source of detectable amount of urea in this cell line. Another study also documented continuous accumulation of ammonia over time in cultured C3A cells [39] . Our data is inconsistent with the above studies; ammonia was accumulated in any culture environment.Furthermore, ALF plasma enhances ammonia accumulation, which is also inconsistent with clinical scenarios.
It is obvious that the C3A cells show deficiencies in their biological functionality, particularly in comparison with primary hepatocytes. This is probably related to the mutual exclusivity of proliferative capacity and differentiation as found in some cell sources [40 , 41] . To improve hepatic functionality in C3A cell line,genetic modification maybe one alternative method to augment their performance in BAL systems. The beneficial effects of overexpression of ARG1 and PXR genes in HepG2 cells, and of HNF-4αgene in OUMS-29 cells have been illustrated [42] . Moreover, BALs containing GS and CYP3A4 genes overexpressed HepG2 cells have proven efficacies in an animal model of ALF [43] .
In conclusion, these findings revealed that culture conditions with ALF plasma caused inhibitory effects on cellular growth and viability in C3A cells. Moreover, upon ALF plasma exposure,CYP450 enzymes exhibited various alterations on mRNA expression, protein levels and enzyme activities. Ammonia accumulation and decreased albumin secretion were also found to be the response to adverse environments. Although this study implies that some strategies are probably required to improve the performance of C3A cells against the detrimental effects of ALF plasma from patients, we consider that the most important issue is to define the nature of cytotoxic substances in ALF plasma to optimize the BAL system.
CRediT authorship contribution statement
Zu-Hong Li:Conceptualization, Data curation, Methodology,Writing - original draft.Zhong-Yang Xie:Data curation, Formal analysis, Methodology, Writing - original draft.Xiao-Xi Ouyang:Conceptualization, Data curation, Methodology, Writing - review &editing.Kai-Zhou Huang:Methodology, Resources.Xiao-Peng Yu:Resources.Ya-Lei Zhao:Methodology.Yan-Hong Zhang:Software.Dan-Hua Zhu:Resources.Jiong Yu:Resources.Lan-Juan Li:Conceptualization, Project administration, Funding acquisition, Supervision, Writing - review & editing.
Funding
This study was supported by grants from the Independent Project Fund of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, the National Key Research and Development Program of China (2016YFC1101304/3), the Key Program of the National Natural Science Foundation of China ( 81330011 ) and Science Fund for Creative Research Groups of the National Natural Science Foundation of China (81721091).
Ethical approval
This study was approved by the Ethics Committee of the First Affiliated Hospital of Zhejiang University School of Medicine(20170226).
Competing interest
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Neoadjuvant chemoradiotherapy before resection of perihilar cholangiocarcinoma: A systematic review
- Hepatobiliary&Pancreatic Diseases International
- Human microbiome is a diagnostic biomarker in hepatocellular carcinoma
- Current practice of anticoagulant in the treatment of splanchnic vein thrombosis secondary to acute pancreatitis
- Enhanced recovery after surgery program in the patients undergoing hepatectomy for benign liver lesions
- Transarterial chemoembolization versus percutaneous microwave coagulation therapy for recurrent unresectable intrahepatic cholangiocarcinoma: Development of a prognostic nomogram
