Uncovering the potential targets of Viburnum odoratissimum for the treatment of related diseases
2020-01-01XiobinXueGuodongYo
Xiobin Xue,Guodong Yo,b
a.Key Laboratory of Computational Chemistry-Based Natural Antitumor Drug Research &Development,Liaoning Province,School of Traditional Chinese Materia Medica,Shenyang Pharmaceutical University,Shenyang 110016,China;
b.Jiangsu Kanion Pharmaceutical Company Ltd.,Lianyungang 222001,China
Abstract Viburnum odoratissimum,a landscape plant,is rich in vibsane-type diterpenes,triterpenes,monoterpenoids,sesquiterpenoids,coumarins,and lignans and so on.In the present work,network analysis was carried out to predict the potential targets of V.odoratissimum and the treatment of related diseases.13 main target proteins were predicted,as well as the diseases which might interact with the compounds from V.odoratissimum. The results showed that vibsane-type diterpenes and triterpenes had the potential to treat neoplasm-related and inflammation-related diseases,and the key target proteins were MDM2,NOS,AR,DNMT1,NR3C1,RARA and PTPN1.Among them,AR,NOS,DNMT1,NR3C1,DNMT1 and PTPN1 were also the crucial targets for triterpenes in the treatment of diabetes mellitus.Besides,ALOX15,PIPN1,TLR9,MIT,DNMT1,RAC1,RTGS2,RARA and ACHE were the potential targets for coumarins and lignan in the treatment of neoplasm-related diseases.Furthermore,ESR1,AR,NOS2,NR3C1 were the primary targets for monoterpenoids and sesquiterpenoids because of their potential neuroprotective effects.It is noteworthy that the study of the potential targets and related diseases can provide new insights for further development of V.odoratissimum.
Keywords:Viburnum odoratissimum; target proteins; network; diseases
1 Introduction
Natural products have long been regarded as an excellent source for drug discovery due to their structural diversity and a wide range of variety of biological activities such as anti-tumor,neuroprotective,anti-inflammatory,ichthyism and anti-oxidant activities.Viburnum odoratissimumis an evergreen shrub mainly distributed in Hunan,Fujian,Guangdong,Taiwan and Zhejiang provinces in China.It is also distributed in Japan and North Korea [1].
Studies on the chemical components ofV.odoratissimumin recent years have indicated that it contains vibsane-type diterpenes,triterpenes,monoterpenoids,sesquiterpenoids,coumarins,and lignans.Modern pharmacological studies have proved that these compounds possess various pharmacological effects,such as anti-tumor,neuroprotective,anti-inflammatory,ichthyism and anti-oxidant activities.Its branches and leaves are used as traditional Chinese medicines for clearing heat and damp,activating the channels and collaterals and detoxicating.However,the potential targets of these compounds and the treatment of related diseases have not yet been reported.This study aimed to reveal the potential targets ofV.odoratissimumand the treatment of related diseases through network pharmacology analysis.
Target protein identification of natural products is an important aspect of drug development and design [2].Hence,we used a number of databases to identify potential targets for compounds fromV.odoratissimumand the treatment of related diseases.This work could provide new insights for further research onV.odoratissimum.
2 Material and methods
2.1 Compound preparations
A total of 152 compounds inV.odoratissimumwere obtained through literature survey.The chemical structures of each component were mapped using ChemDraw 14.0 software.The structural optimization model was then constructed via PubChem (https://pubchem.ncbi.nlm.nih.gov/) and saved as a SMILES file.
2.2 Predicting target proteins
TargetNet (http://targetnet.scbdd.com/home/index/) has 623 target models that could be used for netting or predicting the binding of multiple targets for any given molecule [3].Our study used AUC ≥ 75% and possibility ≥ 0.5 as the cutoff values for potentially active target proteins.The target proteins were obtained and converted to corresponding gene names using UniProt (http://www.uniprot.org).
2.3 Predicting target-related diseases
Gene-Cloud of Biotechnology Information(GCBI) (https://www.gcbi.com.cn/gclib/html/index)includes more than 50,000 human genes and provides gene information by integrating and combing different databases.Gene radar is used to predict which genes are associated with diseases based on the literature [4].Hence,we screened the related diseases with the multigene radar for further analysis.
2.4 Construction of networks
2.4.1 Construction of compound-target network
Based on the target prediction results of different types of compounds ofV.odoratissimum,the compound-target correspondence relationship was established in the excel table and submitted to Cytoscape 3.7.2 software to construct compoundtarget network.In the network,nodes represented compounds and targets,node size represented the degree,and edges represented the interaction of compounds and target proteins.
2.4.2 Construction of target-disease network
The first 25 potential target proteins of each compound were screened according to the degree.The first 20 diseases related to these genes were predicted and the corresponding documents were poured into Cytoscape 3.7.2 software to construct target-disease network.In the network,nodes represented targets and diseases,node size represented degree value,and edges represented the interaction of the target proteins and diseases.
2.4.3 Construction of compound-target-disease network
Based on the predictions,compound-targetdisease network was constructed using Cytoscape 3.7.2 software.
3 Results and discussion
There were 152 compounds inV.odoratissimum,such as vibsane-type diterpenes,triterpenes,monoterpenoids,sesquiterpenoids,coumarins,and lignans.Several databases were used to predict the potential targets for these compounds and the treatment of related diseases.In these networks,compounds,target proteins and diseases were represented by hexagon,octagon and diamond nodes,respectively.Node size represented the degree of compound-target-disease combination.Edges represented compound-target-disease interaction.Results were obtained by screening the target protein (network degree of node ≥ 19) and treatment of related diseases (network degree of node ≥ 17).
3.1 Construction of vibsane-type diterpenes networks
3.1.1 The compound-target network of vibsane-type diterpenes
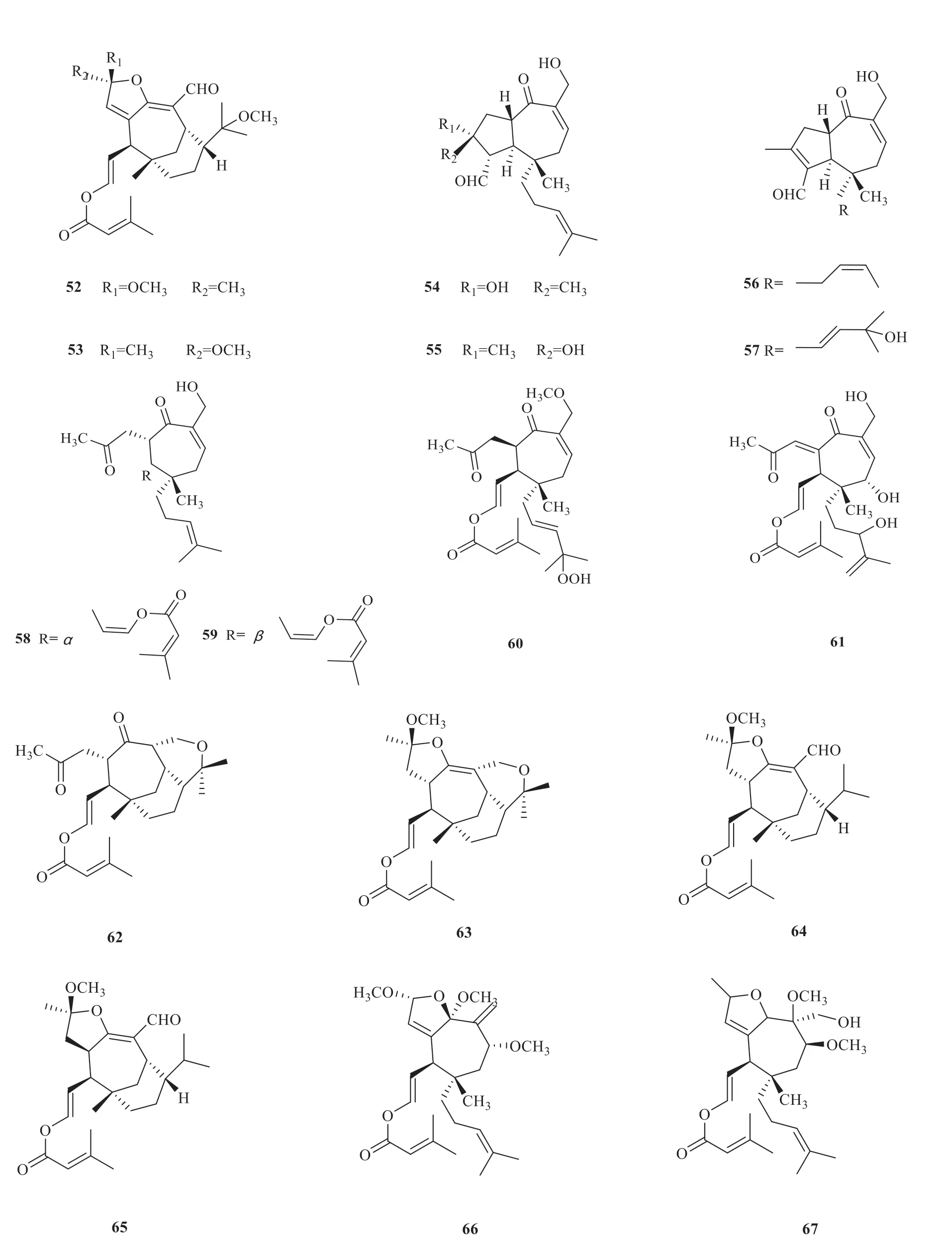
Vibsane-type diterpenes are mainly present inV.odoratissimumwith strong biological activity.According to the carbon skeletons,vibsanetype diterpenes have been classified into three subtypes,including 11-membered ring (1-26) [5-10],7-membered ring (27-69) [5-7,9,11-19]and rearranged type (70-87) [20-27](Fig.1).
(to be continued)
(to be continued)

Continued fig.1
(to be continued)
Continued fig.1
(to be continued)
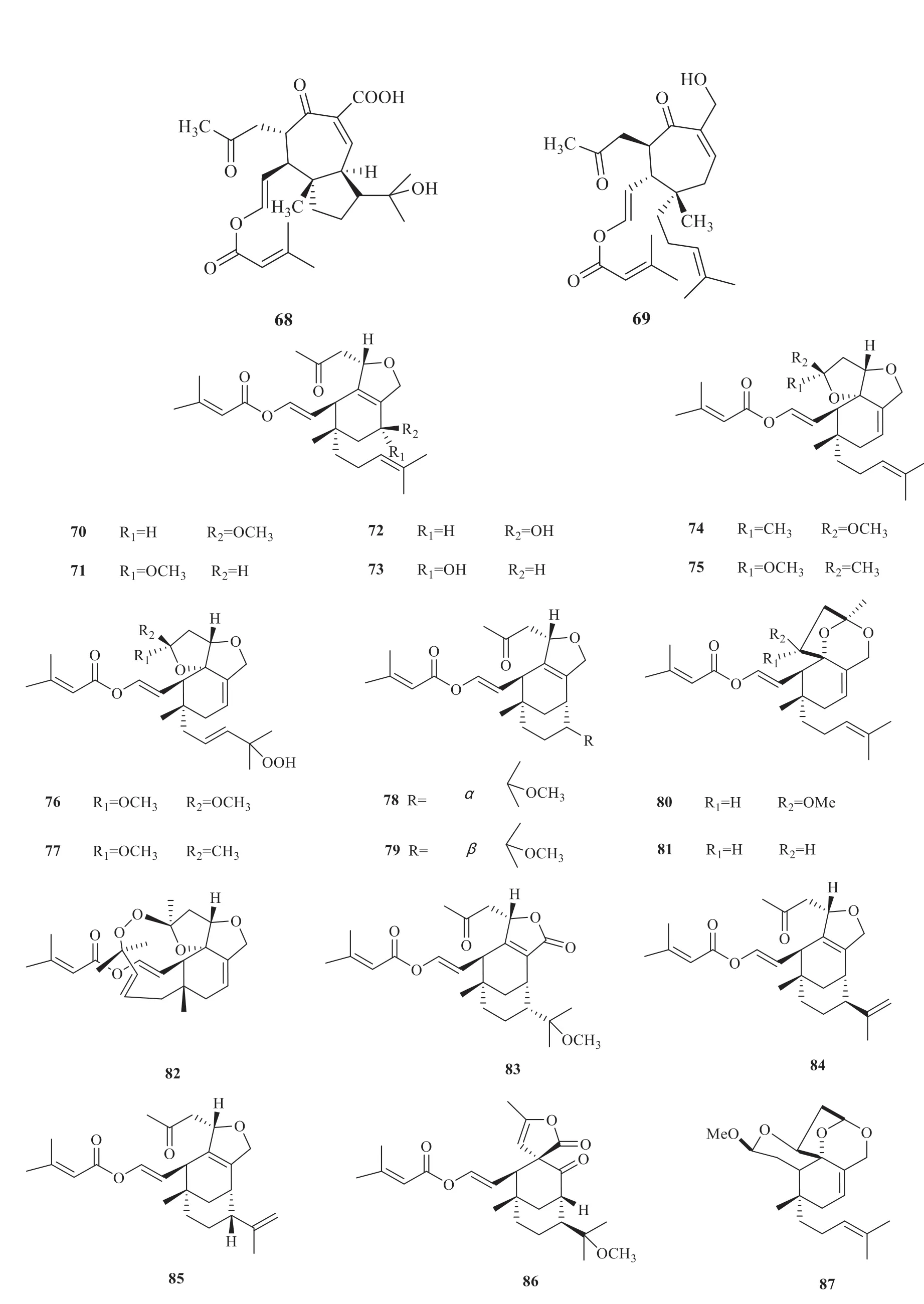
Continued fig.1
(to be continued)
This network was composed of 161 nodes and 995 edges (Fig.2).The network showed that some compounds could act on multiple target proteins.In addition,the inverted triangles represented the classification of compounds.Target protein screening results showed that 7-membered ring compounds had the strongest correlation with the target proteins,and DNMT1,AR and ACHE were most closely related to vibsane-type diterpenes.In conclusion,the results provideed a basis for further research on those compounds.
3.1.2 The target-disease network of vibsane-type diterpenes
To reveal the interaction of target proteins with diseases,a target-disease network based on potential target proteins was constructed using the screening results obtained from the GCBI databases.The results of the target-disease network construction and analysis were shown in Fig.3 and Table 1.Network analysis based on the target-disease interaction data revealed that MDM2,NOS,AR,DNMT1,NR3C1 and RARA might have an effect on neoplasm-related diseases,genetic diseases and mental disorders.
Vibsane-type diterpenes have been reported to possess excellent anti-tumor activities [28].In addition,vibsanin B (5),a vibsane-type diterpene,has been reported to target Hsp90,providing a promising drug lead for treating inflammationrelated diseases [29].Our analysis also indicated that vibsane-type diterpenes were associated with the neoplasm-related and inflammation-related diseases.
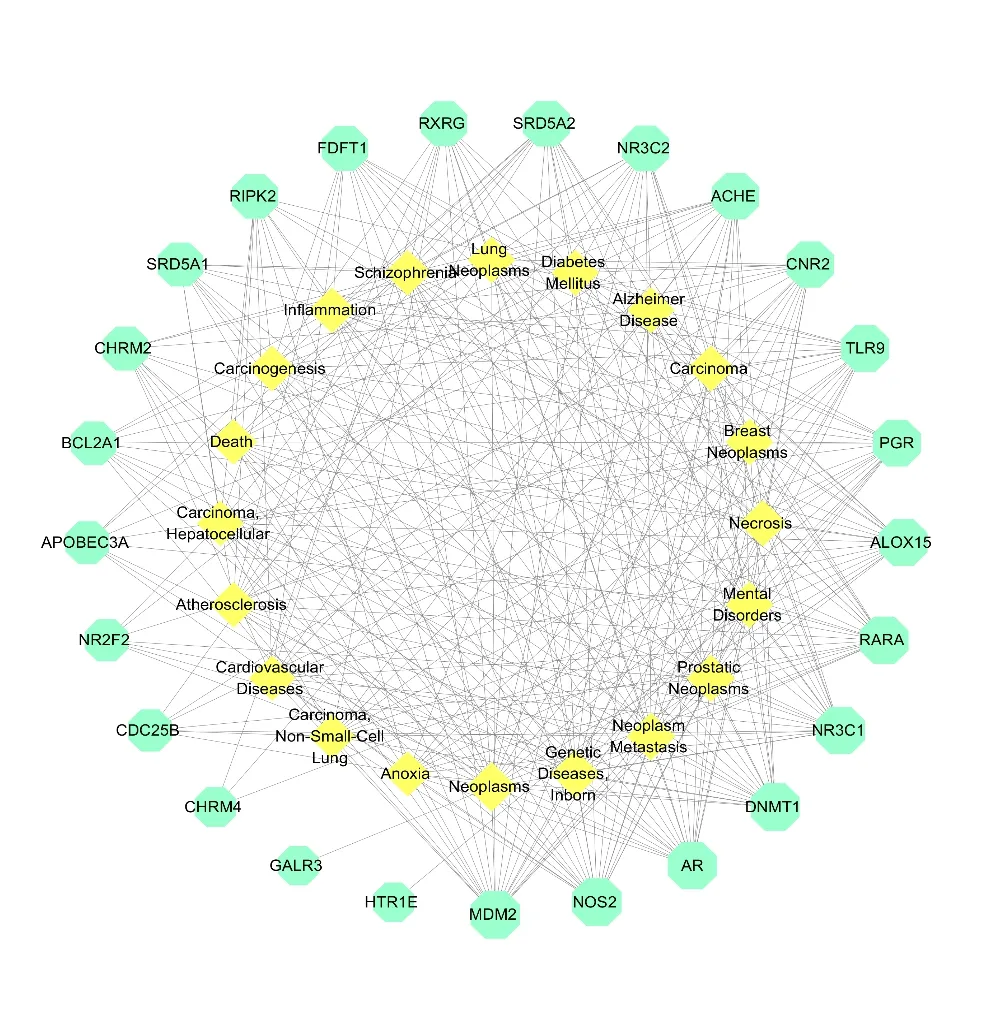
Fig.3 The target-disease network of vibsane-type diterpenes.Nodes on the outer ring represent the target proteins and nodes on the inner ring represent the diseases.
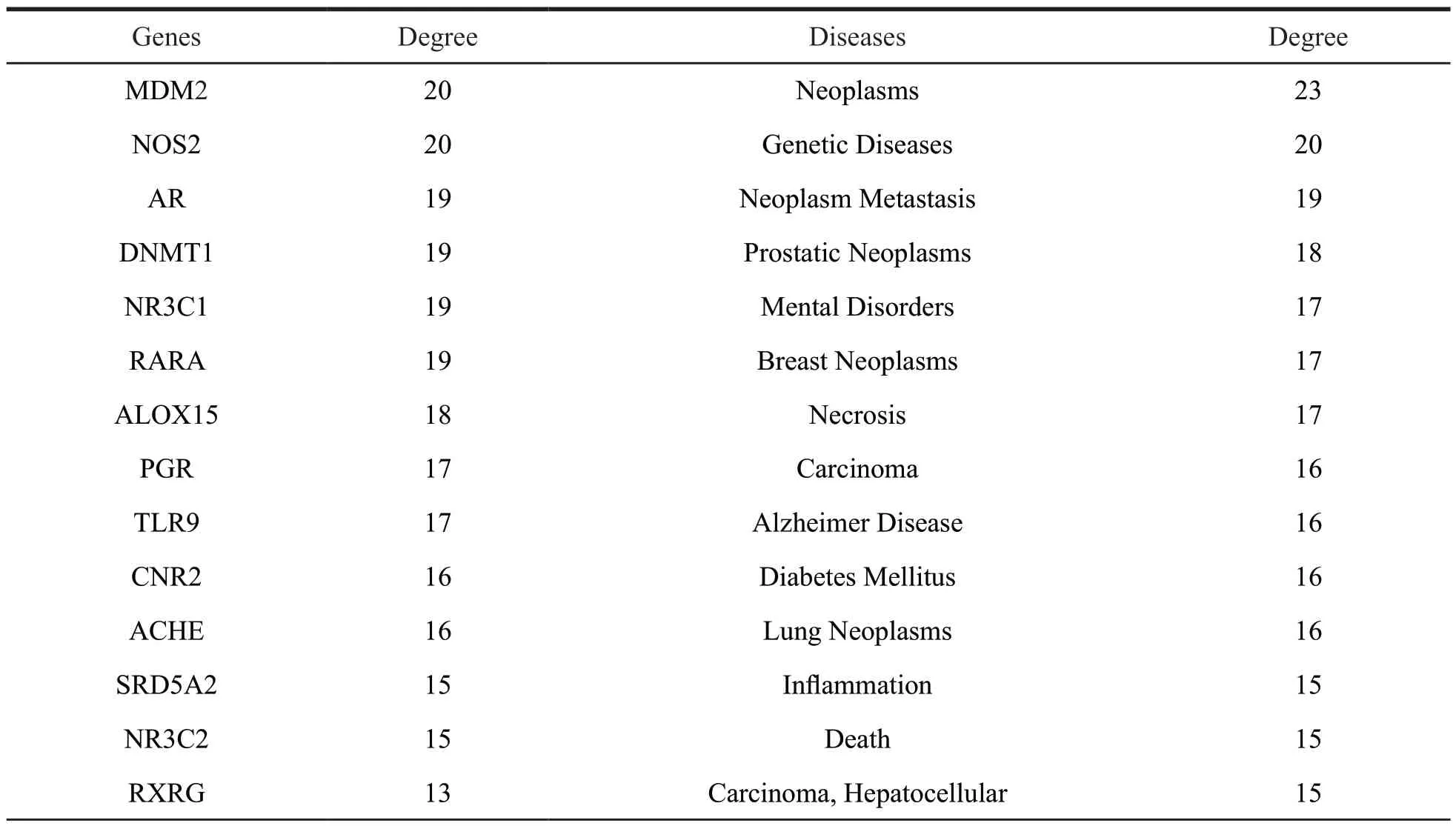
Table 1 The detailed information of targets and diseases for vibsane-type diterpenes
(to be continued)
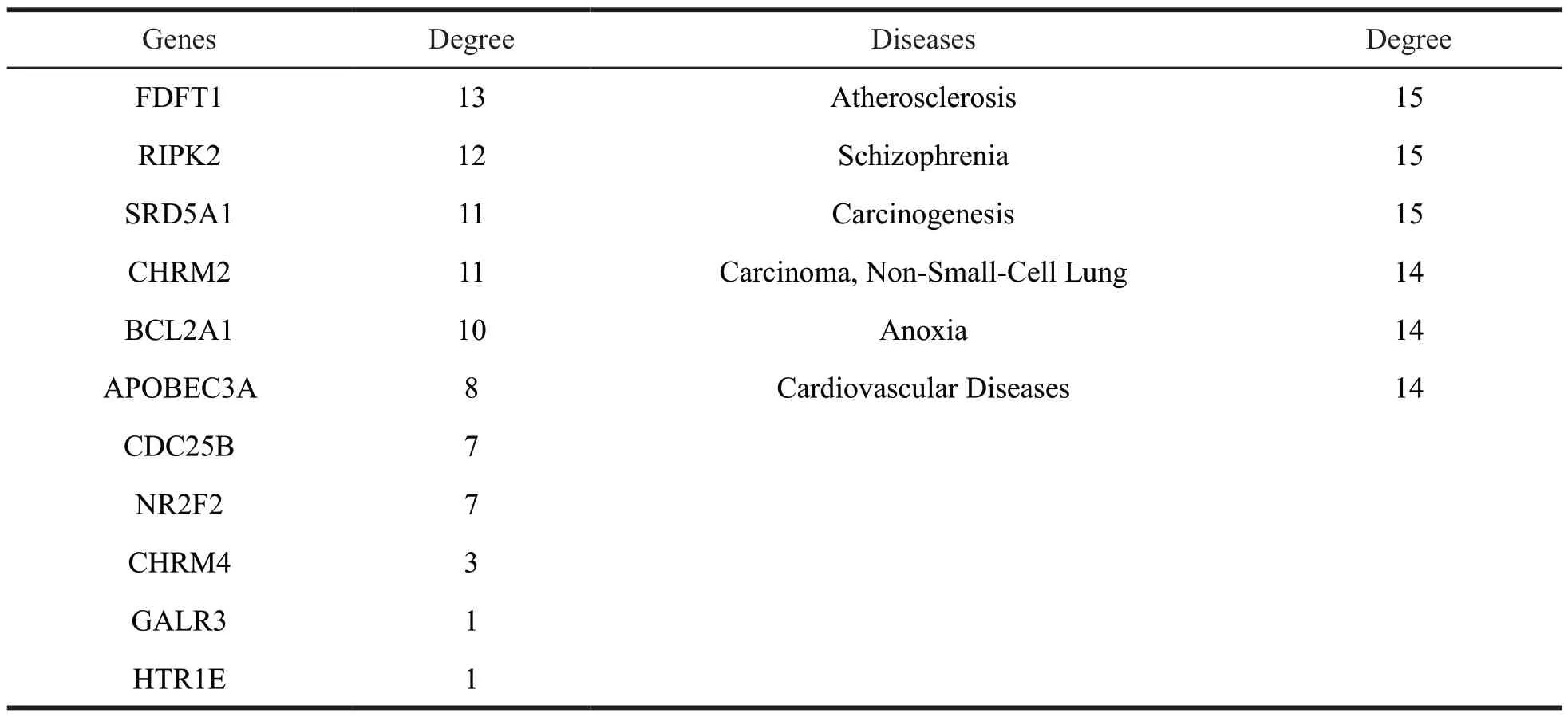
Continued table 1
3.1.3 The compound-target-disease network of vibsane-type diterpenes
As shown in Fig.4,in order to better observe the relationship between compounds,targets,and related diseases,a compound-target-disease network was constructed.Based on the literature reports and prediction results,the vibsane-type diterpenes were found to have the potential to treat neoplasm-related and inflammation-related diseases.
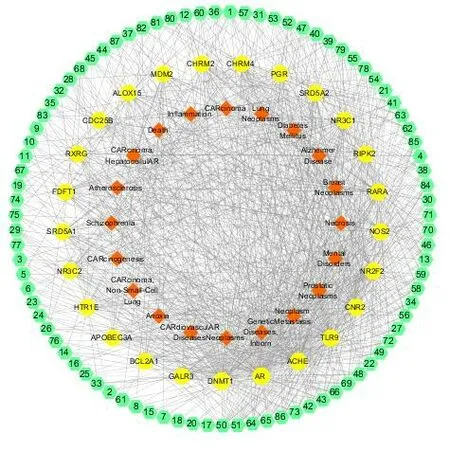
Fig.4 The compound-target-disease network of vibsane-type diterpenes.The three rings from the outside to the inside represent the ring of compounds,the target proteins and the diseases,respectively.
3.2 Construction of triterpenes networks
3.2.1 The compound-target network of triterpenes
To date,31 triterpenes compounds (88-118)have been isolated fromV.odoratissimum[8,25,30-33](Fig.5).The compound-target network analysis showed 84 nodes and 476 edges in total (Fig.6).In the network,the degree of a node depended on the number of routes connected to the node.The larger the degree,the stronger the interaction.According to the topological property of the network,target proteins such as AR,NOS2,NR1H4 and RIPK2 might play a pivotal role in the whole network of triterpenes.
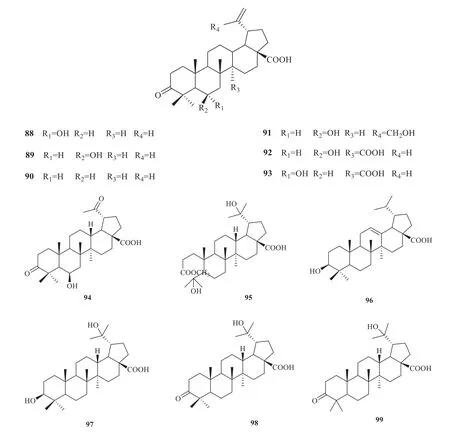
Fig.5 Triterpenes from V.odoratissimum
(to be continued)
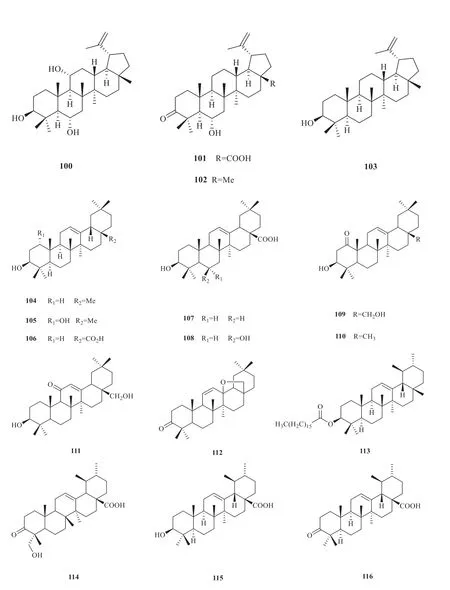
Continued fig.5
(to be continued)

Continued fig.5
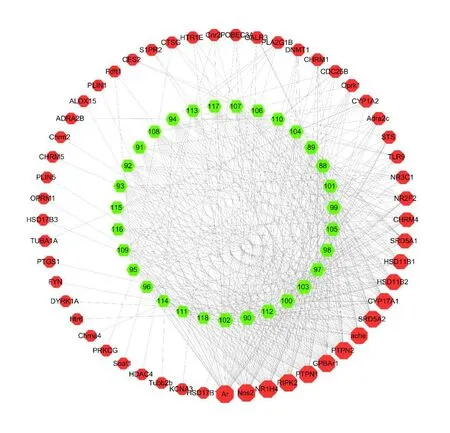
Fig.6 The compound-target network of triterpenes.Nodes on the outer ring represent the target proteins and nodes on the inner ring represent the compounds.
3.2.2 The target-disease network of triterpenes
Based on the compound-target network which illustrated the relationship between compounds and target proteins,25 most promising target proteins and the treatment of related diseases were screened out.The results of the target-disease network construction and analysis in Fig.7 and Table 2 demonstrated that the neoplasm-related diseases,genetic diseases,liver diseases and inflammation-associated diseases were the major disease categories.Our results also indicated that some potential targets such as AR,NOS2,NR3C1,DNMT1 and PTPN1 were closely related to diabetes mellitus.
Besides,modern pharmacological surveys have found some triterpenes showed cytotoxic activity against several tumor cell linesin vitro[25].It was also found that compound 98 could stimulate the absorption of glucose in insulin resistant HepG2 cells without affecting cell viability [32].The network also helped to demonstrate the application of triterpenes as leading compounds in the treatment of neoplasm-related diseases and diabetes mellitus.In summary,network pharmacology identified the primary target proteins for these triterpenes and the treatment of related diseases.
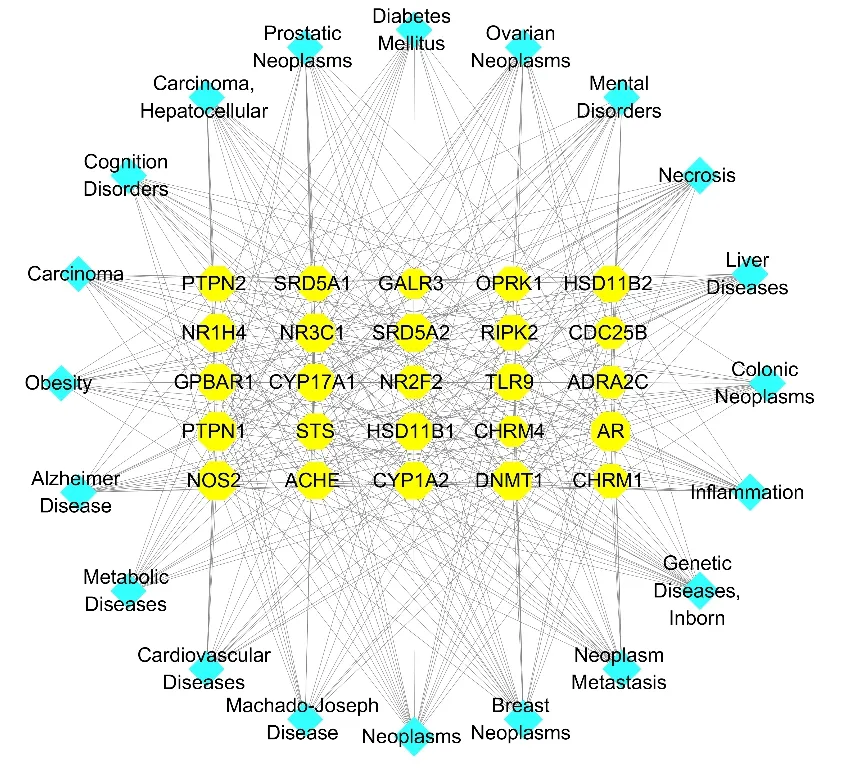
Fig.7 The target-disease network of triterpenes.Nodes on the outer ring represent the diseases and nodes in the inner rectangle represent the target proteins.
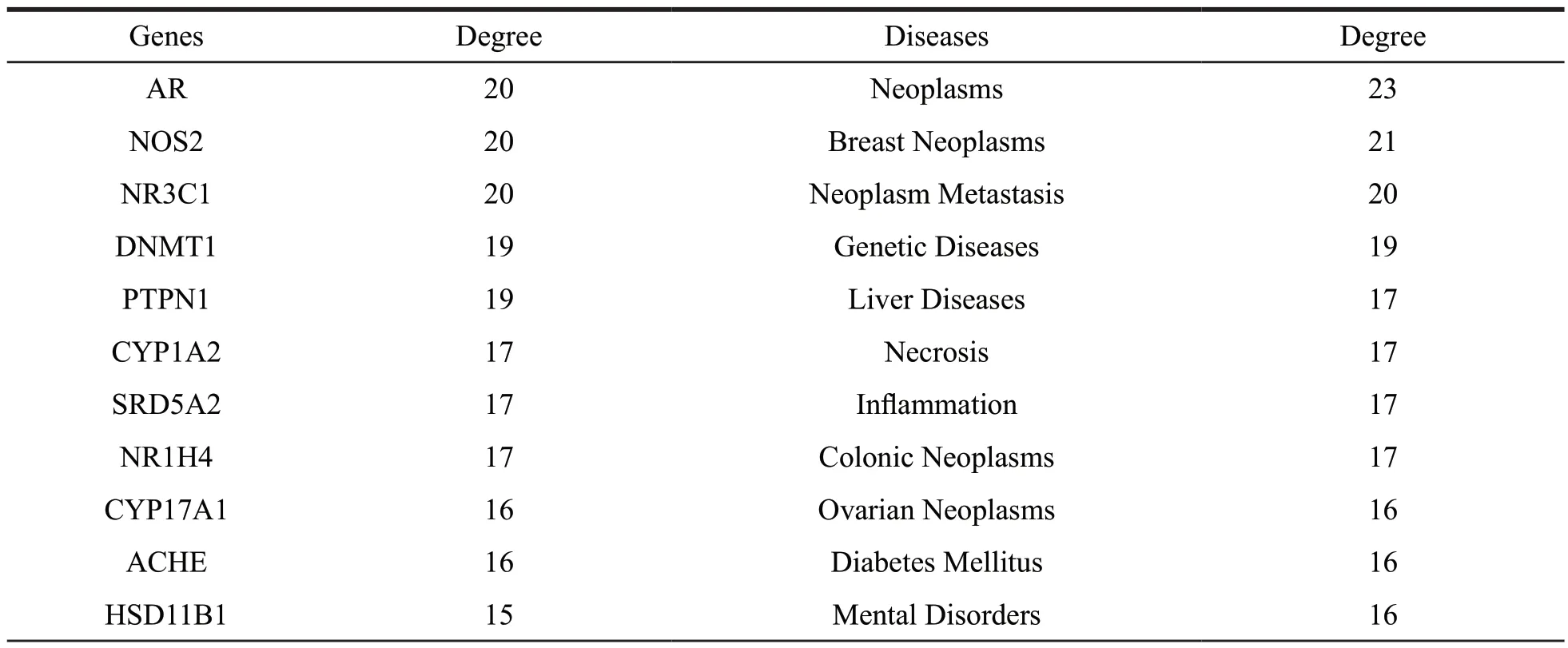
Table 2 The detailed information of the targets and diseases for triterpenes
(to be continued)
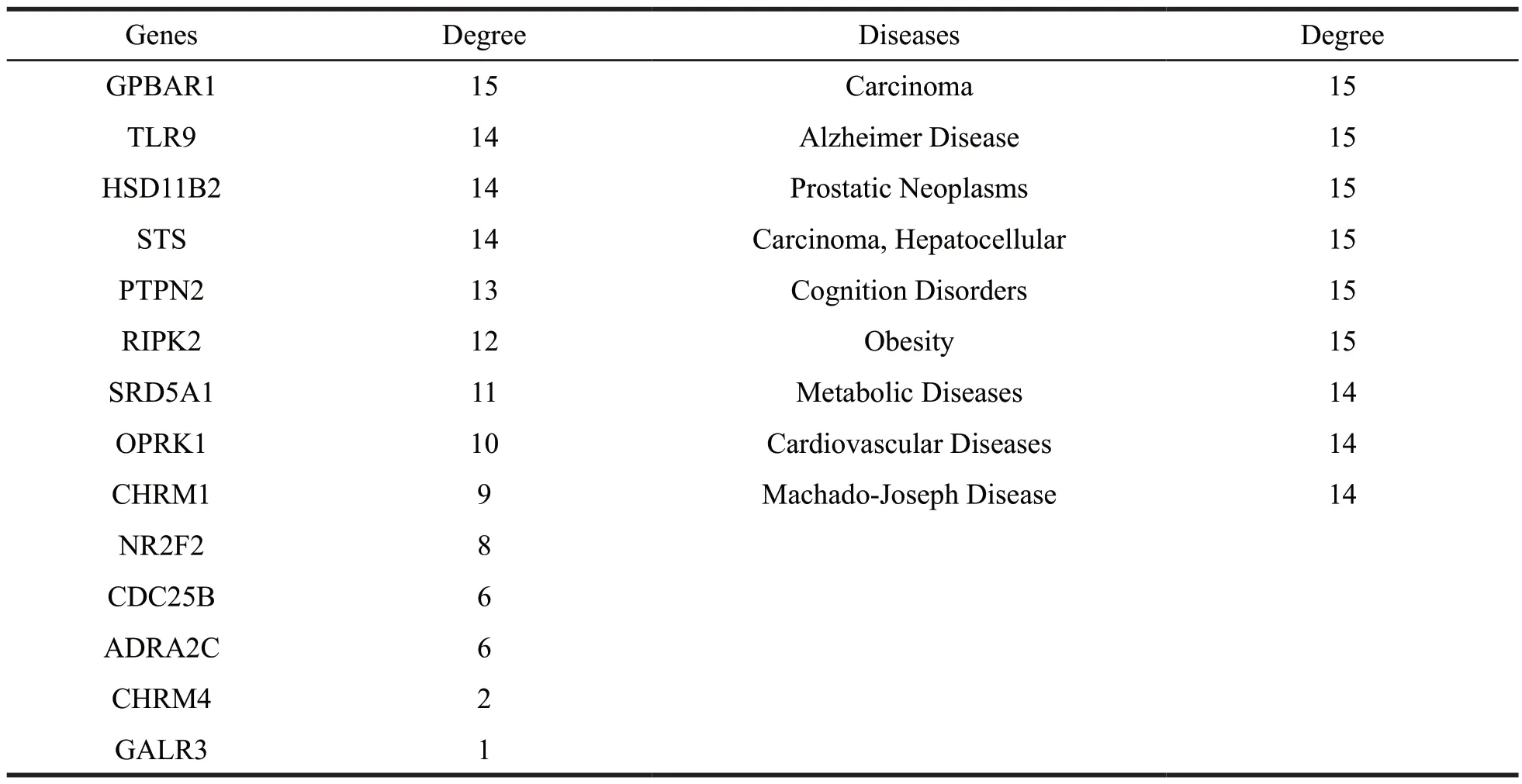
Continued table 2
3.2.3 The compound-target-disease network of triterpenes
The compound-target-disease network was constructed by screening the first 25 targets that were closely related to the compounds (Fig.8).The network showed that triterpenes had a potential in treating neoplasm-related diseases and diabetes mellitus.

Fig.8 The compound-target-disease network of triterpenes.Rings from the outside to the inside represent the ring of the compounds,the target proteins and the diseases,respectively.
3.3 The construction of monoterpenoids and sesquiterpenoids networks
3.3.1 The compound-target network of monoterpenoids and sesquiterpenoids
Based on the network,5 monoterpenoids(119-123) (Fig.9) and 5 sesquiterpenoids (124-128) (Fig.10) were identified as the potential targets [34].The compound-target network comprised 87 nodes and 176 edges in total(Fig.11).The network indicated that each target was associated with multiple compounds.Besides,the target proteins such as AR,PPOR and ACHE,were more closely related to monoterpenoids and sesquiterpenoids.
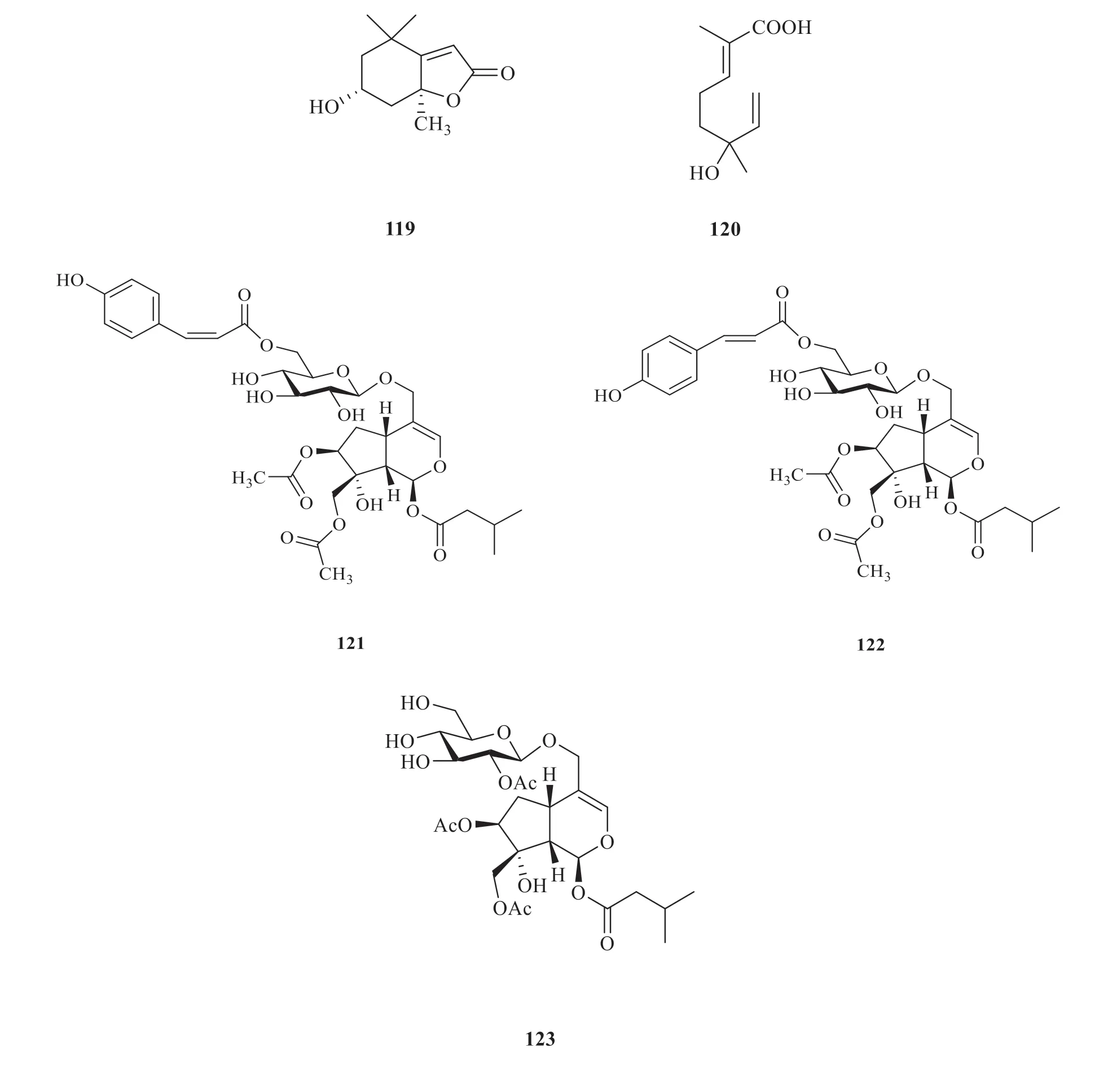
Fig.9 Monoterpenoids from V.odoratissimum
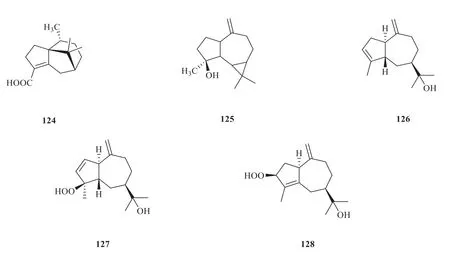
Fig.10 Sesquiterpenoids from V.odoratissimum
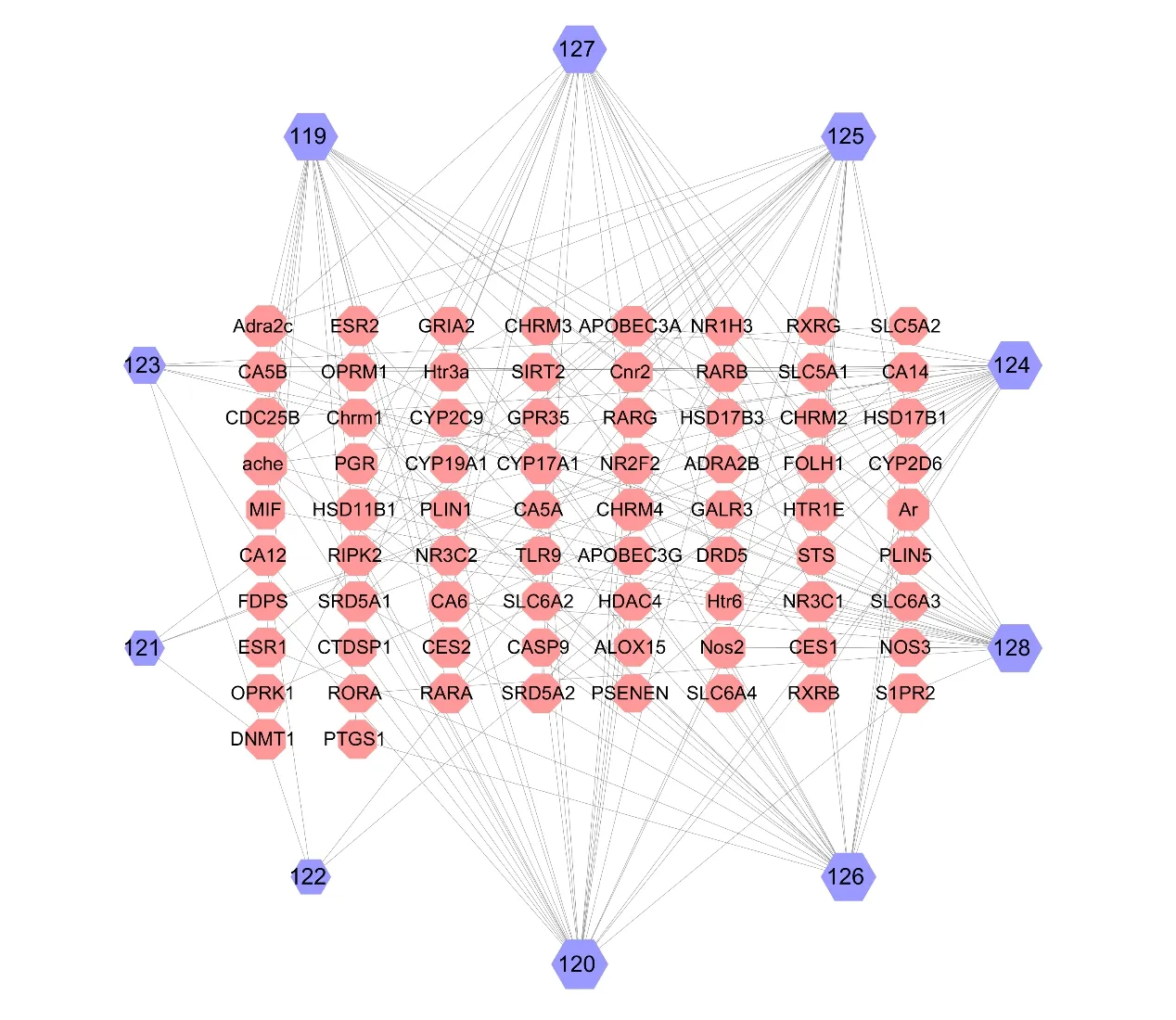
Fig.11 The compound-target network of monoterpenoids and sesquiterpenoids.Nodes on the outer ring represent the compounds,and nodes in the inner rectangle represent the target proteins.
3.3.2 The target-disease network of monoterpenoids and sesquiterpenoids
To further clarify the relationship between target proteins and diseases,a target-disease network was constructed.The results of the targetdisease network construction and analysis were shown in Fig.12 and Table 3.Our results indicated that four target proteins,ESR1,AR,NOS2 and NR3C1,located at the core position,were mainly involved in the neoplasm-related diseases,mental diseases and genetic diseases.It had been reported that compounds 121-124 exhibited significant neuroprotective effects [35].Therefore,the results which indicated that the monoterpenoids and sesquiterpenoids might have neuroprotective effect,demonstrated the reliability of the network analysis.
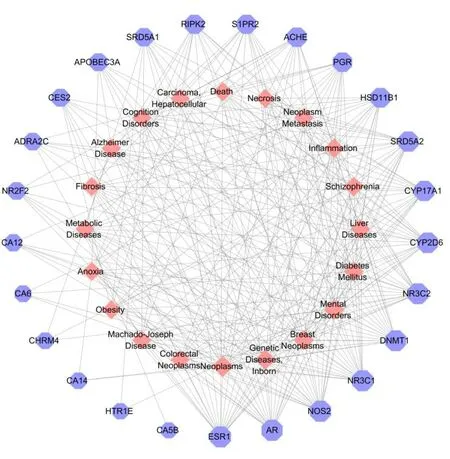
Fig.12 The target-disease network of monoterpenoids and sesquiterpenoids.Nodes on the outer ring represent the diseases and nodes on the inner ring represent the target proteins.
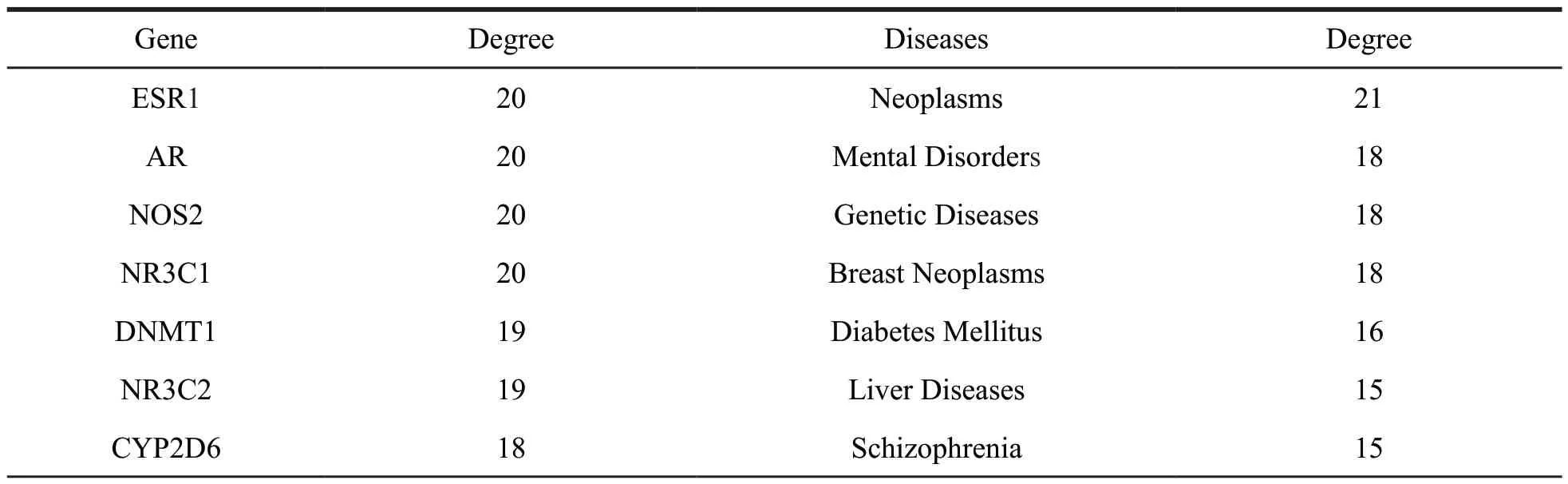
Table 3 The detailed information of the targets and diseases for monoterpenoids and sesquiterpenoids
(to be continued)
Continued table 3
3.3.3 The compound-target-disease network of monoterpenoids and sesquiterpenoids
By screening the first 25 targets that were closely related to the compounds,the compoundtarget-disease network was constructed.The network showed that monoterpenoids and sesquiterpenoids might play a role in neoplasm-related and mental diseases (Fig.13).
3.4 The construction of coumarins networks
3.4.1 The compound-target network of coumarins
It was reported that four coumarin compounds(129-132) were isolated fromV.oddratissimum(Fig.14) [25].As shown in Fig.15,the compoundtarget network analysis showed 38 nodes and 98 edges in total.This network results indicated the degree of binding of these compounds to the target proteins was similar.
3.4.2 The target-disease network of coumarins
To further understand the interaction between the compounds and their corresponding target proteins,we constructed the compound-target network,as shown in Fig.16.Diseases were predicted based on 25 target proteins screened.22 target proteins had the greatest interaction with the diseases in the top 20 GCBI databases (Table 4).The degree of each node depended on other important features in the network.The higher the degree of the target protein in the network was,the more likely it was to play an important role in the development of the disease.Our results showed that five target proteins,including DNMT1,MIT,TLR9,PIPNT and ALOX15,were primarily associated with the neoplasm-related diseases and genetic diseases,but experimental studies on these diseases have not been reported yet.
Fig.13 The compound-target-disease network of monoterpenoids and sesquiterpenoids.Rings from the outside to the inside represent the ring of the target proteins,the diseases and the compounds,respectively.
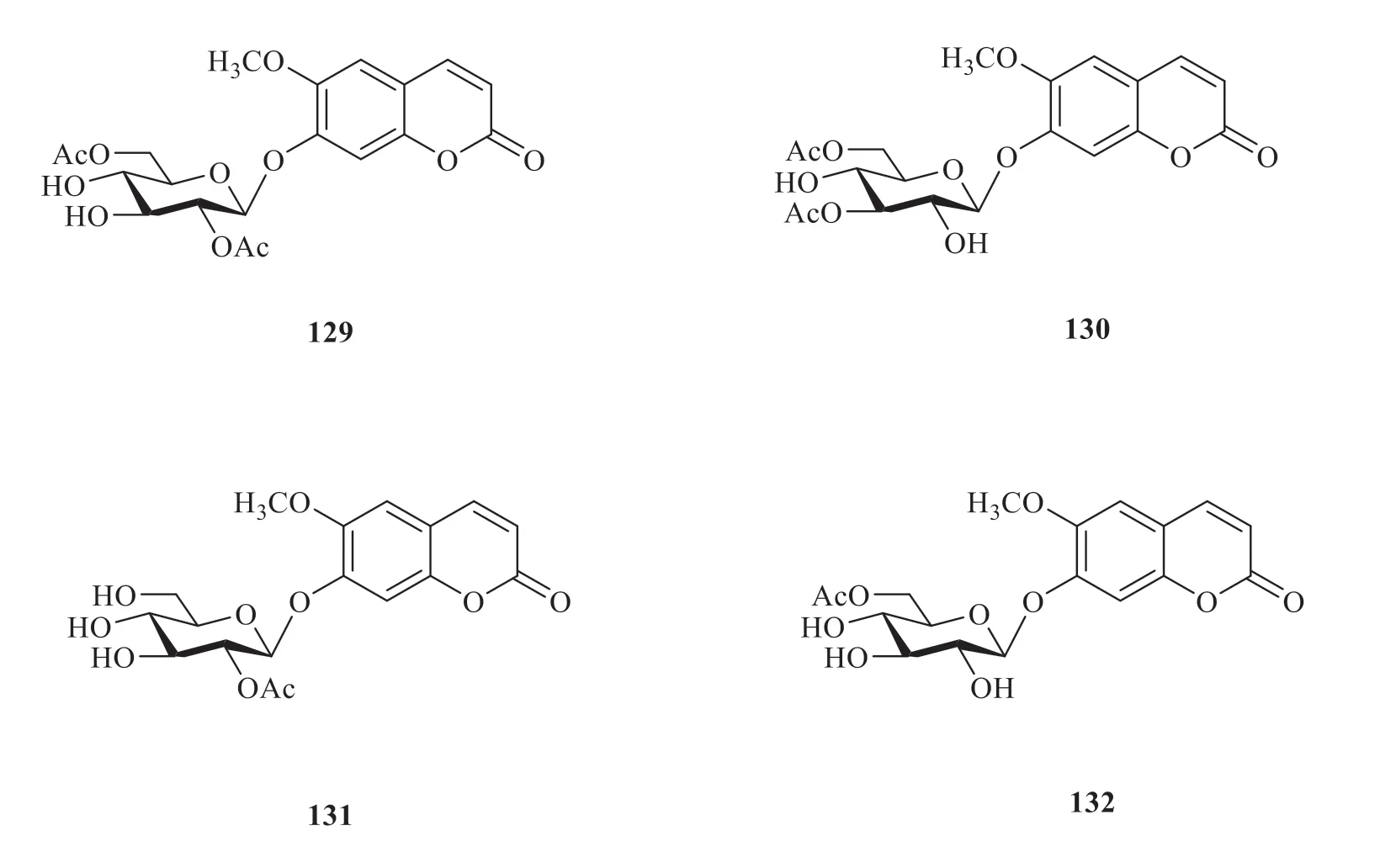
Fig.14 Coumarins from V.odoratissimum
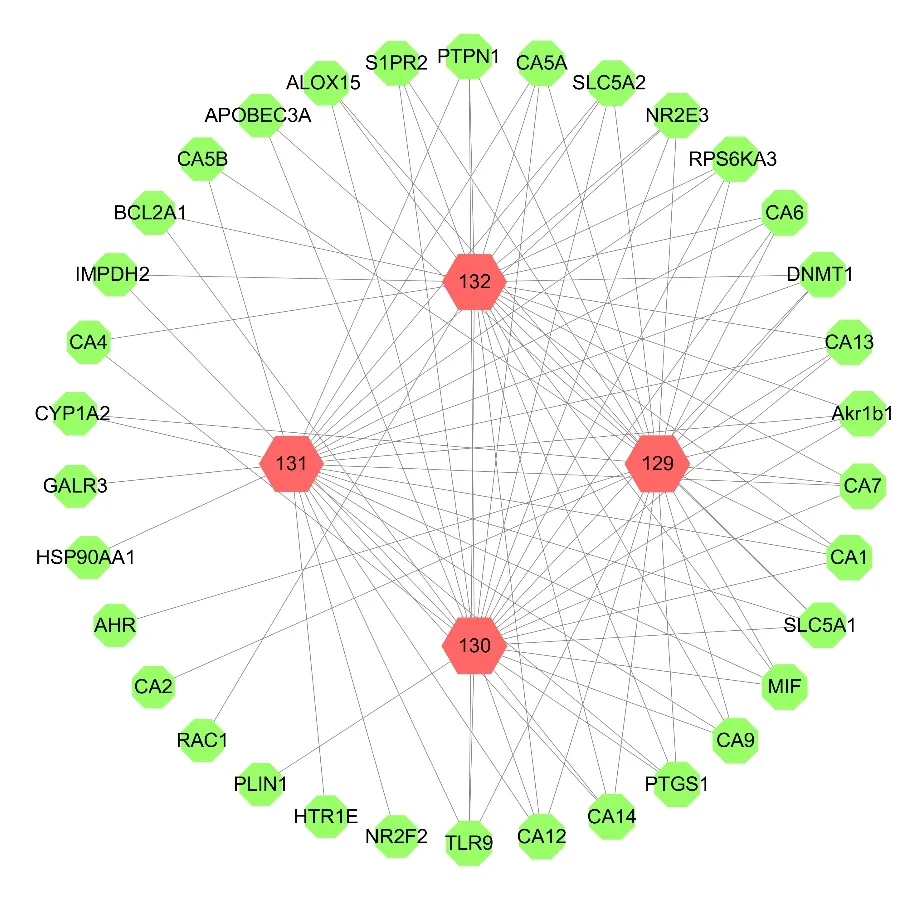
Fig.15 The compound-target network of coumarins.Nodes on the outer ring represent the targets and nodes on the inner ring represents the compounds.
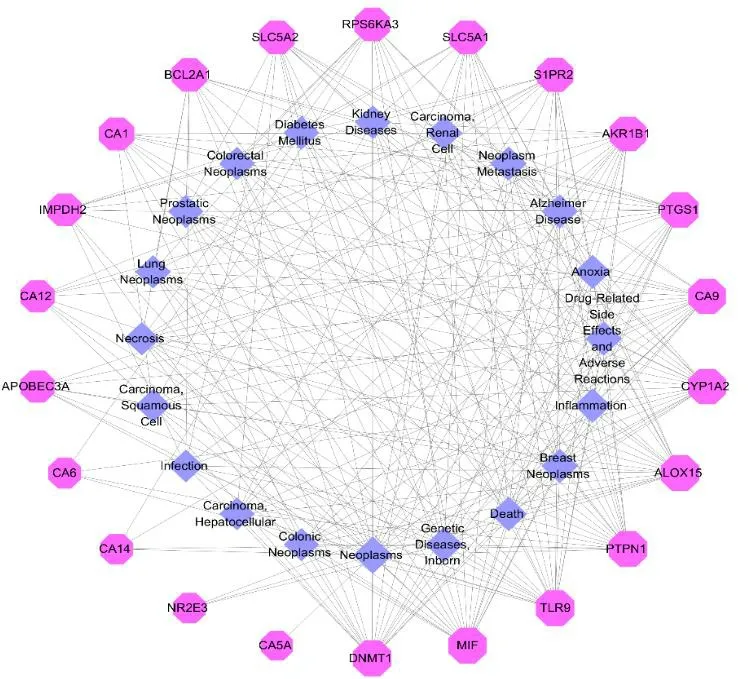
Fig.16 The target-disease network of coumarins.Nodes on the outer ring represent the diseases and nodes on the inner-ring represent the target proteins.
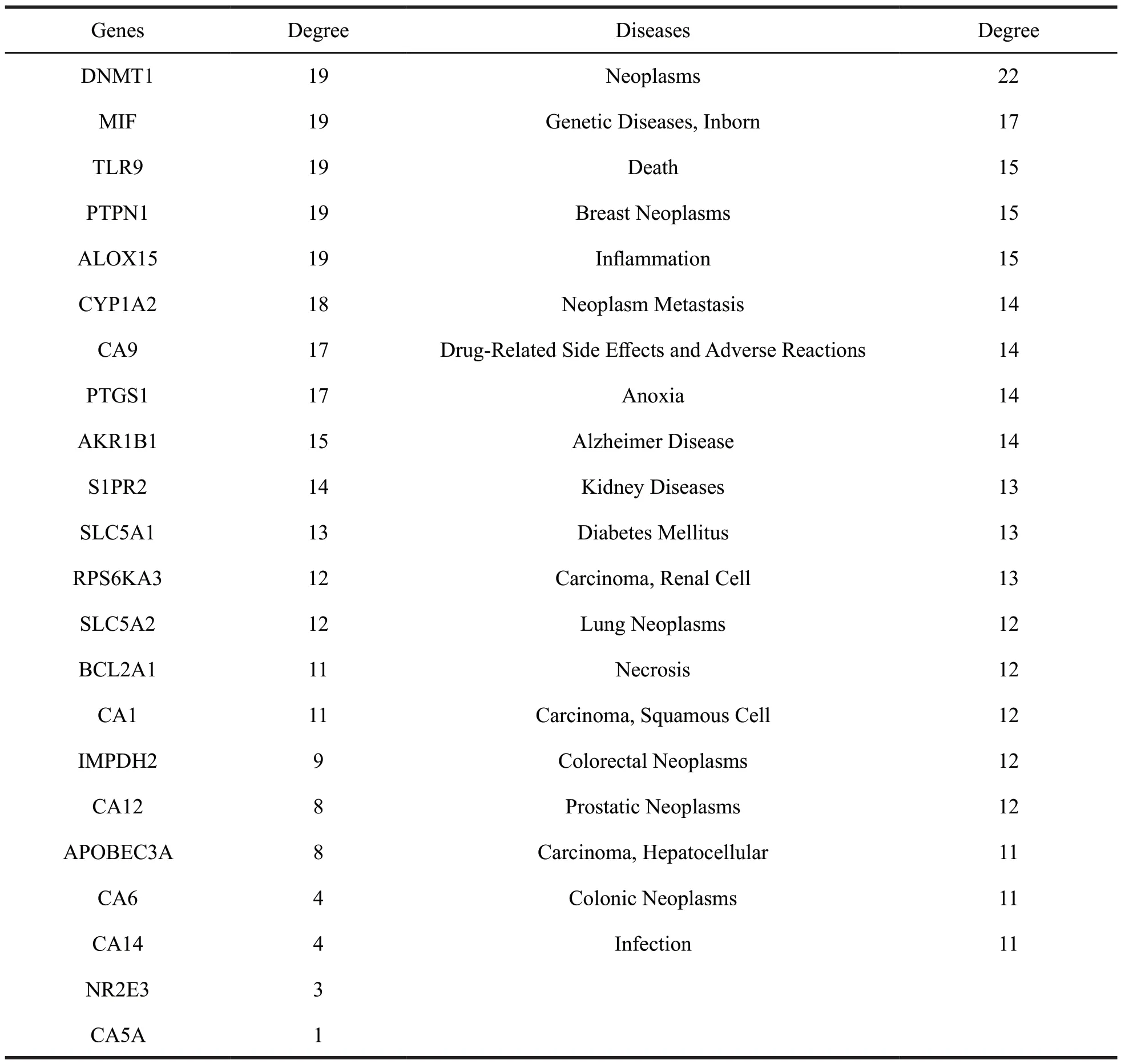
Table 4 The detailed information of the targets and diseases for coumarins
3.4.3 The compound-target-disease network of coumarins
To further understand the interaction between compounds and their corresponding targets and diseases,we constructed the compound-targetdisease network (Fig.17).The results indicated that coumarins were primarily related to neoplasm-related diseases and genetic diseases.These findings laid a foundation for further study of such compounds in the future.
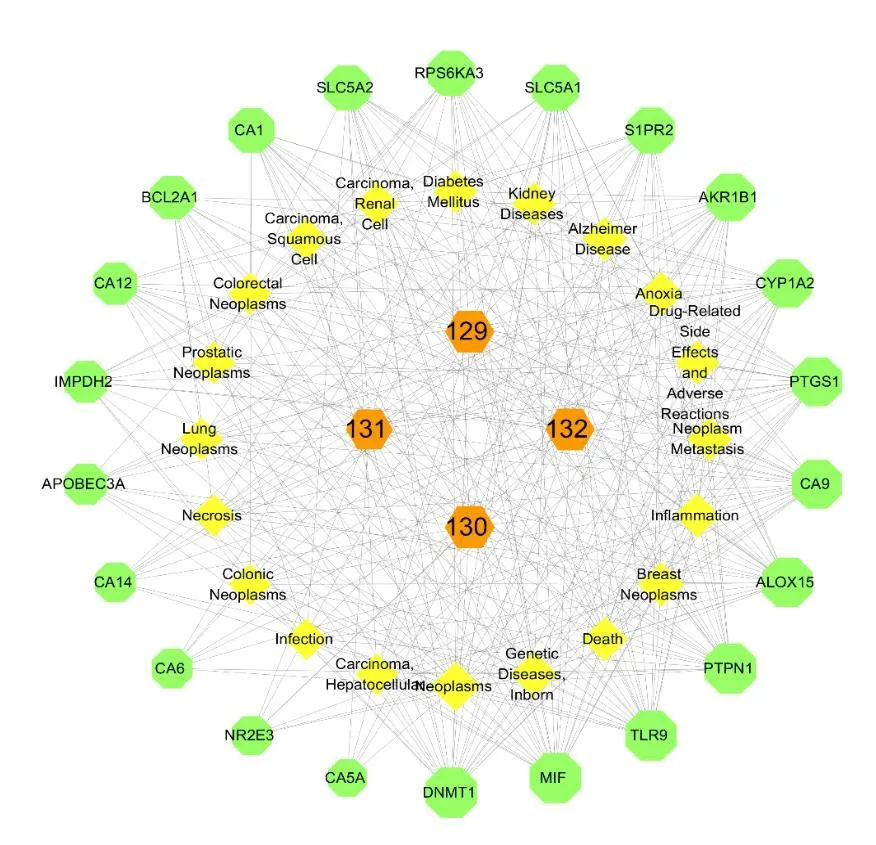
Fig.17 The compound-target-disease network of coumarins.Nodes from the outer ring to the inner ring represent the target proteins,the diseases and the compounds,respectively.
3.5 Construction of lignans networks
3.5.1 The compound-target network of lignans
12 lignans (133-144) have been isolated fromV.oddratissimum[36,37](Fig.18).To further identify the relationship between compounds and the target proteins,a compound-target network was constructed(Fig.19).A total of 48 nodes and 539 edges between the compounds and the targets were acquired.By comparing the size of the hexagons,it was not difficult to find that few target proteins interacted with compounds 133,134 and 137.In addition,the target proteins TUBB2B and CA6 were more closely related to the compounds and interacted more frequently with the compounds than other target proteins.
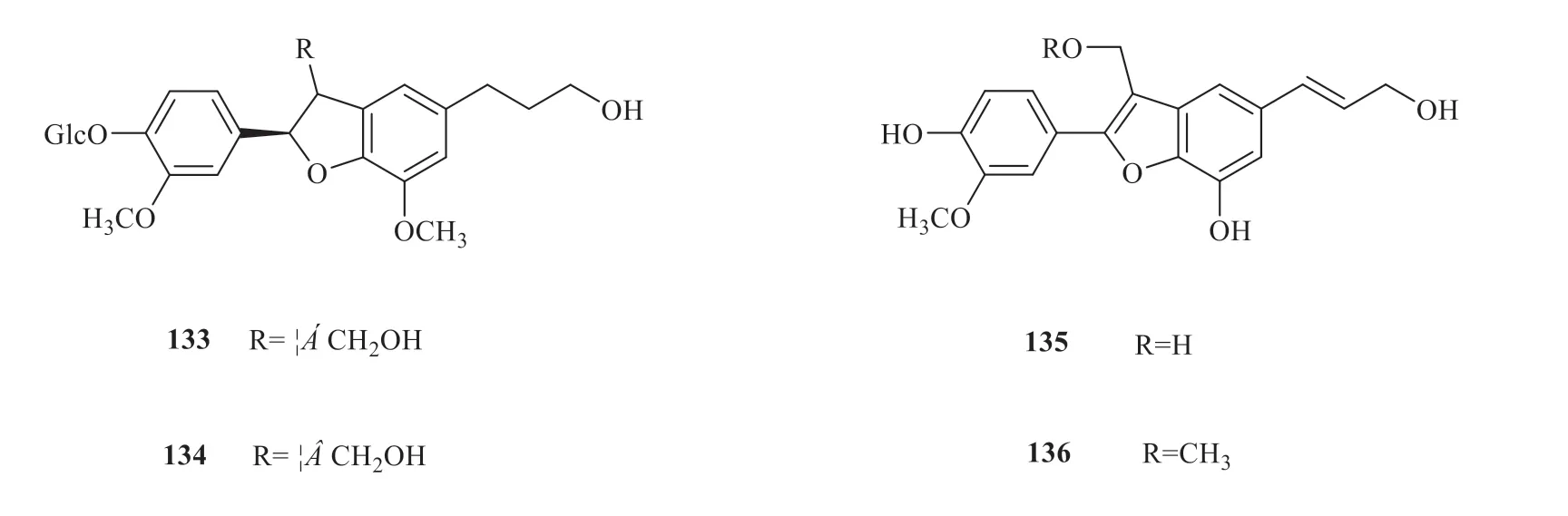
Fig.18 Lignans from V.odoratissimum
(to be continued)
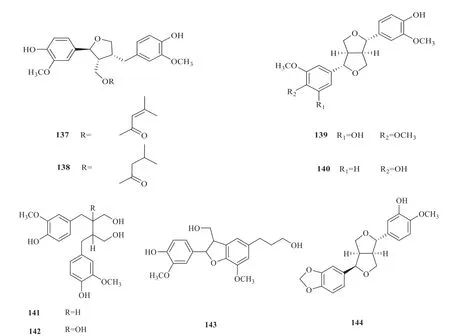
Continued fig.18
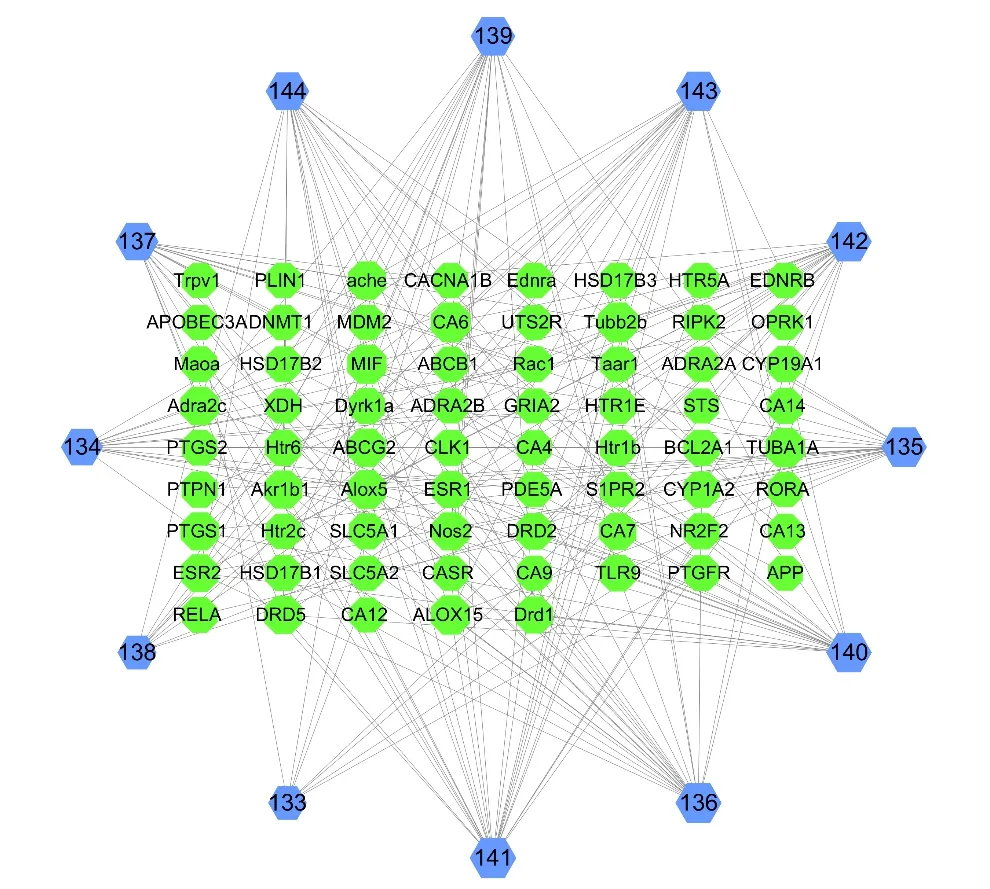
Fig.19 The compound-target network of lignans.Nodes on the outer ring represent the compounds and nodes in the inner rectangle represent the target proteins
3.5.2 The target-disease network of lignans
To further clarify the interaction between potential target proteins and diseases,a targetdisease interaction network was constructed as shown in Fig.20 and Table 5.Our result indicated ALOX15,PTPN1,TLR9,MIF and DNMT1 were mostly related to the diseases,including neoplasm-related diseases and genetic diseases.
In addition,(+)-9’-O-senecioyllariciresinol(137),a lignan,had moderate inhibitory effect on the human tumor cell lines A431 and T47D [10].These results indicated that ALOX15,PTPN1,TLR9,MIF and DNMT1 had certain effect on the treatment of various diseases.

Fig.20 The target-disease network of lignans.Nodes on the outer ring represent the diseases and nodes in the inner rectangle represent the target proteins.
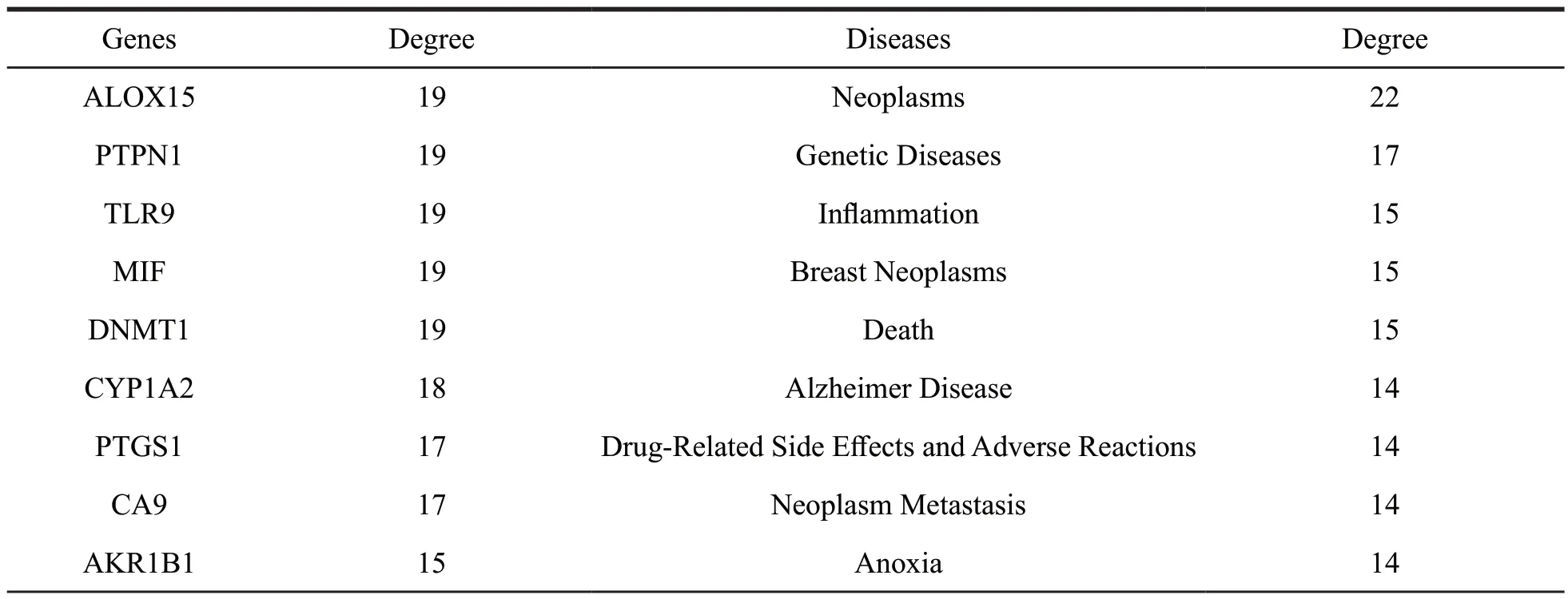
Table 5 The detailed information of the targets and diseases for lignans
(to be continued)
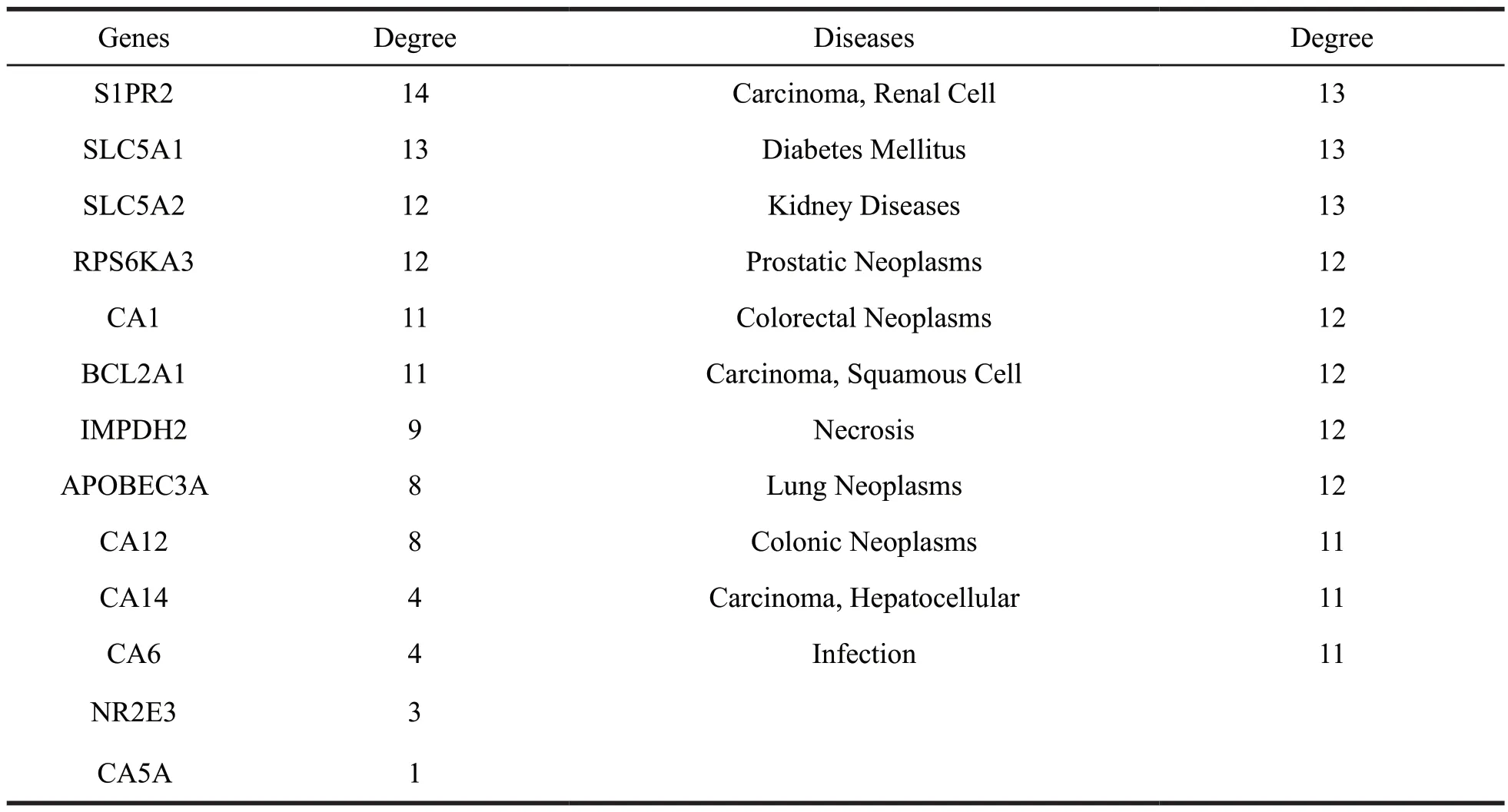
Continued table 5
3.5.3 The compound-target-disease network of lignans
A compound-target-disease network was constructed to identify potential targets of different diseases that different compounds act upon (Fig.21).The results indicated that lignans might play a role in the treatment of neoplasm-related diseases and genetic diseases.
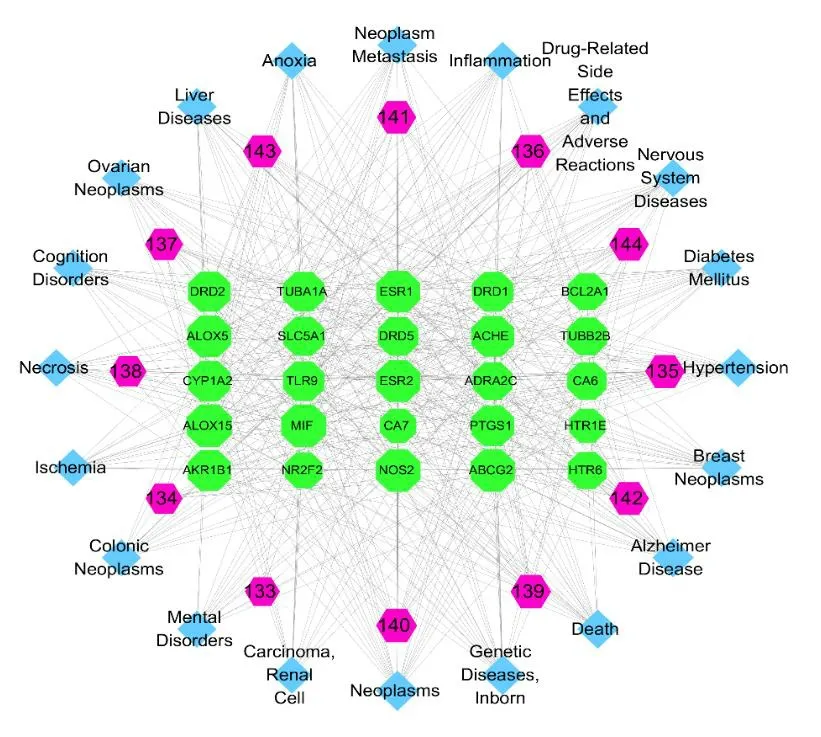
Fig.21 The compound-target-disease network of lignans.Nodes from the outer ring to the inner ring represent the diseases and the compounds,respectively.Nodes in the inner rectangle represent the target proteins.
3.6 Construction of other compounds networks
3.6.1 The compound-target network of other compounds
V.odoratissimumhas other compounds,for example,sterol,organic acids and flavones (145-152)(Fig.22) [37,38].As shown in Fig.23,there were 85 nodes and 176 edges in this network.Network analysis showed that each compound was associated with multiple target proteins.
3.6.2 The target-disease network of othercompounds
To elucidate the complex interaction between target proteins and their corresponding diseases,we constructed a target-disease network based on potential target proteins.The results of the construction and analysis of the target-disease network were shown in Fig.24.The results showed that RAC1,PTGS2,RARA,MIF and ACHE were the protein targets that acted as‘hubs’ in the network and these target proteins were the main targets in the treatment of neoplasm-related diseases.Therefore,the network analysis will help to screen active target proteins and diseases.
Fig.22 Sterol,organic acids and flavones from V.odoratissimum
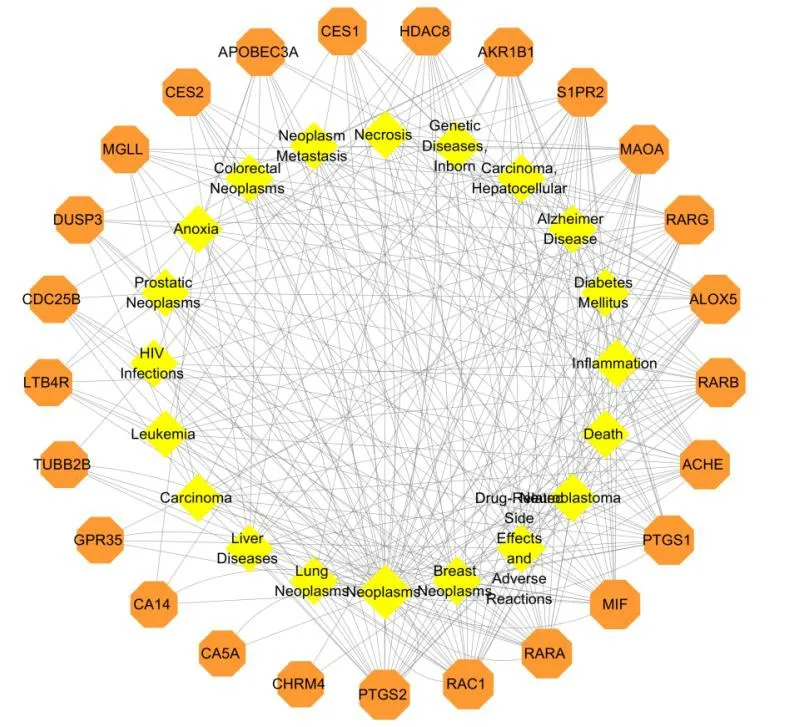
Fig.24 The target-disease network of other compounds.Nodes on the outer ring represent the diseases and nodes on the inner ring represent the target proteins.
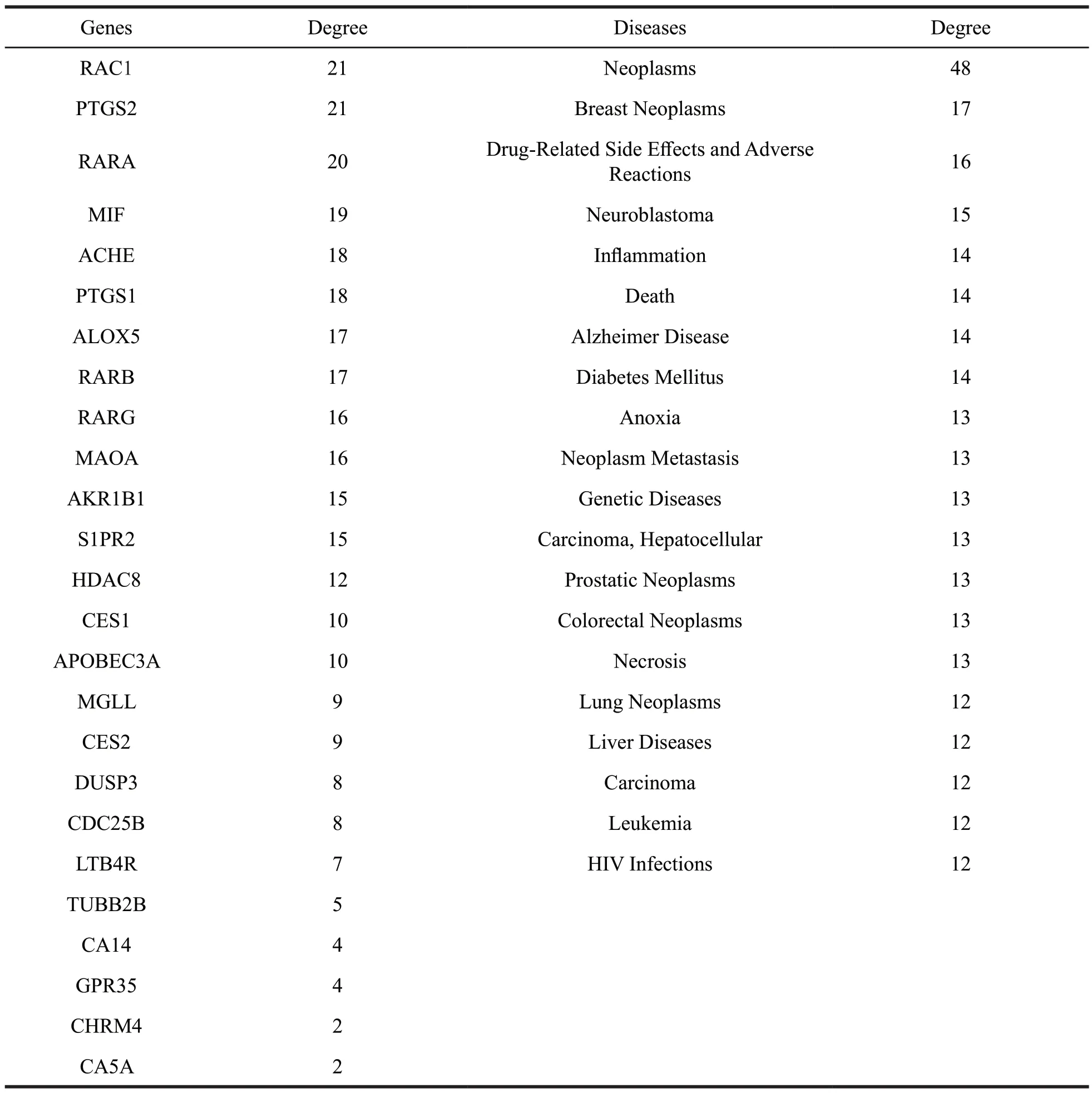
Table 6 The detailed information of the targets and diseases for other compounds
3.6.3 The compound-target-disease network of other compounds
Predicting compound-target-disease interaction can help decipher the underlying biological mechanisms.Therefore,a compoundtarget-disease network was constructed as shown in Fig.25.We hypothesized that these compounds might have the effect in the treatment of neoplasmrelated diseases.
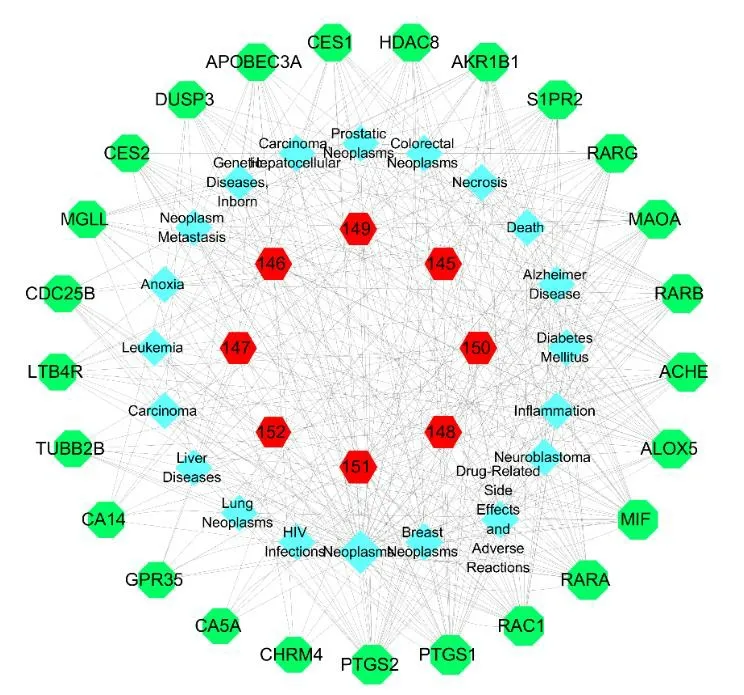
Fig.25 The compound-target-disease network of other compounds.Nodes from the outer ring to the inner ring represent the target proteins,the diseases and the compounds,respectively.
4 Conclusion
In summary,the results successfully predicted,illuminated and confirmed compound-targetdisease interaction.V.odoratissimumhas significant advantages in the treatment of various diseases,which is consistent with previous studies.
According to the results of potential targets screening for these compounds,the target proteins,MDM2,NOS2,AR,NR3C1,ESR1,RAC,PTGS2,RARA,DNMT1,MIT,TLR9,PIPNT,ALOX15,were the key target proteins for these compounds.In addition,vibsane-type diterpenes and triterpenes were associated with inflammation-related diseases,while monoterpenoids and sesquiterpenoids might have neuroprotective activity.Triterpenes were the potential therapeutic agents for diabetes mellitus.Our results provide strong evidence for further studies on the potential targets ofV.odoratissimumand the treatment of related diseases.
These results not only provide clues to the exploration of compound-target-disease interaction,but also demonstrate a feasible method to discover the potential target proteins for other plants and the treatment of related diseases.
Conflict of interest
The authors have declared that there is no conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China [Grant No.81703385].
杂志排行
Asian Journal of Traditional Medicines的其它文章
- Contribution Regulations for Asian Journal of Traditional Medicines
- Research progress on chemical constituents and pharmacological activities of Viburnum tinus L.
- Studies on the chemical components and biological activities of the genus of Juncus
- Research progress on chemical constituents in Periploca forrestii and their pharmacological activities
- Studies on chemical constituents of Ailanthus altissima (Mill.)Swingle and their antioxidant activity
- Physicochemical properties and antioxidant activities of polysaccharides extracted from Panax ginseng C.A.Meyer with graded percipitation method
