Screening of aptamers and their potential application in targeted diagnosis and therapy of liver cancer
2019-07-24GuoQingZhangLiPingZhongNuoYangYongXiangZhao
Guo-Qing Zhang, Li-Ping Zhong, Nuo Yang, Yong-Xiang Zhao
Abstract Aptamers are a class of single oligonucleotide molecules (DNA or RNA) that are screened from random DNA or RNA oligonucleotide chain libraries by the systemic evolution of ligands by exponential enrichment technology. The selected aptamers are capable of specifically binding to different targeting molecules,which is achieved by the three-dimensional structure of aptamers. Aptamers are similar in function to monoclonal antibodies, and therefore, they are also referred to as "chemical antibodies". Due to their high affinity and specificity and low immunogenicity, aptamers are topics of intense interest in today's biological targeting research especially in tumor research. They not only have high potential for clinical advances in tumor targeting detection but also are highly promising as targeted tumor drug carriers for use in tumor therapy. Various experimental studies have shown that aptamer-based diagnostic and therapeutic methods for liver cancer have great potential for application. This paper summarizes the structure, characteristics, and screening methods of aptamers and reviews the recent research progress on nucleic acid aptamers in the targeted diagnosis and treatment of liver cancer.
Key words: Aptamer; Systemic evolution of ligands by exponential enrichment; Liver cancer; Outlook
INTRODUCTION
Tumors are one of the leading causes of human death worldwide. With the development of society, the population has increased, and the risk of tumors has also greatly increased. Tumor diagnosis and treatment have become an issue of intense concern. Liver cancer is a common malignant tumor, and its diagnosis and treatment are the research focus of many scholars.
Aptamer refers to a type of single oligonucleotide molecule (DNA or RNA) that is screened and enriched from a library of random single-stranded oligonucleotide sequences constructed in vitro, and aptamers can specifically bind to target molecules,such as small molecules, proteins, and even whole cells[1-3]. In 1990, Ellington et al[4]and Tuerk et al[5]screened for a random oligonucleotide that binds to T4 DNA polymerase with high affinity and specificity and confirmed the ability of this oligonucleotide sequence to bind to the target. The identified oligonucleotide was referred to as an aptamer, and the screening method was named systematic evolution of ligands by exponential enrichment (SELEX). The first aptamer drug, pegaptanib sodium (trade name: Macugen), was approved by the United States Food and Drug Administration in 2004 and is used to treat age-related macular degeneration[6]. Now,scientists have acquired a variety of aptamers screened using the SELEX technology,which have a special three-dimensional structure that can bind different target molecules[7,8]. The more detailed explanation for aptamers is given below.
APTAMERS
Structure and function of aptamers
An aptamer is a specific three-dimensional structure formed by the folding of a singlestranded nucleic acid molecule. The binding of an aptamer to the target molecule is achieved by its own special three-dimensional structure, with features such as hairpins, internal loops, bulges, pseudoknots, and G-tetramers[9]. The factors by which an aptamer binds to the target molecule are the complementary effects of the geometric shape, the interactions of aromatic ring overlap, the electrostatic interactions of the charged groups, the bases of the nucleic acid aptamer, the van der Waals forces, and the hydrogen bonds. Targets that can be bound include small molecules, proteins, cells, and even entire tissues. Proteins are the most numerous targets[10,11].
DNA and RNA aptamers
Both DNA and RNA aptamers can produce similar specificities and affinities, but each has its own distinct advantages. The primary difference between DNA and RNA lies in the sugar. The GH bond of deoxyribose in DNA has low reactivity and a stable structure, making it better able to resist the action of enzymes. The ribose in RNA is more reactive and less stable than deoxyribose, and double-stranded regions of RNA can stimulate the immune system via the Toll-like receptor. An RNA aptamer obtained from an RNA oligonucleotide chain library using the SELEX method needs to be modified for improved stability[12].
In the early stages, many researchers were more likely to screen for RNA aptamers because they have more diverse stereoscopic three-dimensional structures than DNA and higher affinity for targets[13]. However, in-depth studies of aptamers have presented increasing evidence that DNA aptamers are superior to RNA aptamers in application. For example, DNA aptamers have better structural stability and are easy to screen. Therefore, an increasing number of researchers are now more inclined to explore DNA aptamers, and in 2013, it was reported that more than 85% of aptamers were obtained from DNA oligonucleotide libraries[14].
SELEX METHOD FOR SCREENING APTAMERS
Basic principles of SELEX technology
The SELEX technology combines multiple fields, such as molecular biology,bioinformatics, and materials science, to develop aptamers for a variety of targets[15]. A large number (approximately 1014-15) of random DNA or RNA oligonucleotide chains are first synthesized, and then multiple screening and enrichment cycles are performed to select oligonucleotide chains from this library that can bind target molecules with high specificity and high affinity. New base sequences will appear during the screening process, and as the cycle progresses, the specificity and affinity with which the selected oligonucleotide strand binds the target molecule will gradually increase. The final oligonucleotide chain has the highest specificity and affinity for the target molecule and is referred to as the aptamer of the specific target molecule[16,17].
Establishment of a random oligonucleotide chain library
As a raw material, the oligonucleotide chain for aptamer screening is fixed at both ends and random in the middle. The random sequences can specifically bind to the target molecule, and the fixed sequences can be used for amplification and screening.Screening of DNA or RNA aptamers starts with synthesis of random DNA or RNA oligonucleotide chain libraries.
The first step in building a library is to design fixed sequences at both ends of the oligonucleotide chain, which are usually 20-30 nucleotides in length. The fixed sequences generally contain primer binding sites and restriction endonuclease binding sites required for PCR amplification, while binding sites containing RNA polymerase are needed when preparing RNA aptamers. The transcribed RNA sequences must also be able to bind to reverse transcriptase[18]. In designing the intermediate sequences, attention should be paid to length. Short chains are easier to design and fabricate and less expensive, but if the sequence is too short, it cannot form secondary structures that specifically and stably bind to target molecules, or there are not enough secondary structures in the library for screening. As the length of the intermediate sequence increases, the capacity of the library increases, but increasing the length of the intermediate sequence beyond a certain threshold will not significantly increase the number of unique stereo structures available for screening.Generally, the length of the intermediate sequence is 30-50 nucleotides, which can theoretically form approximately 1014-1024random oligonucleotide chains. The stereo structure formed by this number of nucleotides is sufficient to bind a target molecule[19]. After the sequence design is completed, a large number of DNA oligonucleotide chains with random sequences in the middle can be synthesized with a DNA synthesizer, and a DNA oligonucleotide chain library can be established.
Selection and enrichment of aptamers
After establishment of a DNA oligonucleotide chain library, it is necessary to screen for and enrich oligonucleotide chains that can bind to target molecules with high specificity and affinity. Mature screening techniques include magnetic bead separation[20], high-speed centrifugation, affinity chromatography[21], capillary electrophoresis[22], and nitrocellulose membrane filtration[23]. For example, studies have shown that a target aptamer can be obtained in only four cycles of capillary electrophoresis SELEX[24].
On the basis of traditional screening methods, many screening methods have been developed, such as counter SELEX[25], subtractive SELEX[26], photo SELEX[27], and negative SELEX[28], which can improve the selectivity of aptamers. Blended SELEX[29],complex targeting SELEX[30], expression cassette SELEX[31], and toggle SELEX[32]can improve the universality of aptamers. In addition, automated SELEX[33], non-SELEX[34],and FluMag SELEX[35]can shorten the screening cycle. Methods for combining different pools, such as tailored SELEX[36]and genomic SELEX[37], can increase the likelihood of screening for aptamers of interest. At present, cell-based screening (cell-SELEX) is widely used[38](Figure 1).
To shorten the screening process, the following screening methods have been developed. Multiple GO-SELEX employs graphene oxide (GO) for screening. This method is simple, fast, and high throughput, and thus reduces the screening cost and shortens the screening time[39]. Single-walled carbon nanotube-assisted cell-SELEX(SWCNT-assisted cell-SELEX) requires only six rounds of screening compared with the 15 rounds necessary in standard cell-SELEX and has a high screening efficiency[40].On-chip cell-SELEX, which incorporates microfluidic chips into the cell-SELEX process, requires only five rounds of screening, greatly shortening the screening process[41]. Sequence-constructive SELEX modifies the sequence of aptamers to improve the affinity and specificity for the targets[42]. Furthermore, Hi-Fi SELEX is a protocol for rapid and efficient DNA aptamer selection[43].
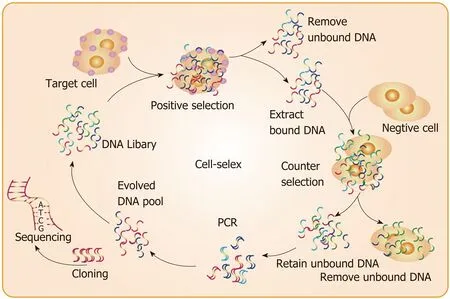
Figure 1 Schematic diagram of cell systemic evolution of ligands by exponential enrichment strategy. The target cells are used to screen and enrich the selection pools, and clonal sequencing is performed to obtain the required aptamers. SELEX: Systemic evolution of ligands by exponential enrichment.
The general procedures for screening and enrichment of DNA aptamers are as follows: in the first round of screening, target molecules are used to screen DNA oligonucleotide chain libraries and the oligonucleotide chain DNA is amplified via PCR; in the second round of screening, the amplified products are rescreened and the DNA is amplified via PCR; after 8-16 repeated rounds of screening, flow cytometry and clonal sequencing are performed to obtain the required high specificity and high affinity aptamers[15]. However, depending on the SELEX method used, the number of screening and enrichment rounds will vary. In most cases, too many rounds will waste consumable reagents without significantly improving the screening rate.However, too few rounds may not identify the best aptamers or impurities. The appropriate number of screening rounds is generally the number at which the affinity does not continue to increase. Because of the low fidelity of the polymerase used in SELEX, each screening enrichment cycle may produce variants, and thus, the screening enrichment cycles actually increase the capacity of oligonucleotide chain libraries. In addition to the effect of screening enrichment, the number of cycles needed for screening enrichment is one of the criteria for evaluating the advantages and disadvantages of screening enrichment methods. Many of the methods mentioned above are optimized in terms of the number of cycles.
Base sequencing, characterization, and molecular structure modification of aptamers
Screening of enriched aptamers requires base sequencing to obtain well-defined base sequences. Then, structural analysis and characterization of these screened aptamers need to be performed, to assess factors such as specificity, affinity, and stability.
The chemical synthesis of aptamers is more stable than antibodies. However,despite their high stability and tolerance to temperature, pH range, and some organic solvents, aptamers can be readily degraded or removed by the kidneys in vivo.Because of the limited interaction between natural base pairs and drugs, in vivo use requires prolonging the survival time of aptamers under physiological conditions,that is, optimizing the structure of aptamers. Conventional optimization methods include the following: (1) Substituting a natural nucleotide for a nucleotide modified with a group to avoid degradation of the nucleic acid[44]; (2) Chemically synthesizing a mirror image sequence of the selected aptamers with high plasma and serum stability to enhance the stability of aptamers to make them have significant therapeutic potential[45]; (3) Modifying aptamers with locked nucleic acids to enhance their ribozyme resistance. Methylene-linked 2'-oxygen and 4'-carbon in carbohydrate residues have strong ability to hybridize with nucleic acid molecules and are not easily degraded by enzymes[46]; and (4) Covalently attaching PEG or cholesterol to aptamers, prolonging their residence time in the human body and reducing renal filtration rate[47,48]. The half-life of survival of aptamers in human physiological environment can be extended from a few minutes to several hours by one or more of the above methods, and aptamers with modifiable properties are superior to antibodies or polypeptides in the diagnosis and treatment of tumor diseases.
CHARACTERISTICS OF APTAMERS AND COMPARISON WITH MONOCLONAL ANTIBODIES
Characteristics of aptamers
Aptamers have many advantages, such as high affinity and specificity, a wide range of action, small size, good stability, low cost, and easy synthesis and screening compared with traditional immunological and chemical recognition molecules, and aptamers have shown broad application prospects in the diagnosis and treatment of diseases[49].
Comparison of aptamers and monoclonal antibodies
There are some similarities between aptamers and monoclonal antibodies (mAbs). In terms of function, both aptamers and mAbs can bind target molecules with high specificity and affinity. In terms of application, both aptamers and mAbs can be used in medical research and in clinical diagnosis and treatment; therefore, aptamers are known as "chemical antibodies". Compared with mAbs, aptamers do not exhibit immunogenicity or antigenicity and do not cause a related immune response or immune rejection in the human body. This property makes aptamers reusable for the same patient in the diagnosis and treatment of clinical diseases[50].
APPLICATION OF APTAMERS IN THE TARGETED DIAGNOSIS OF LIVER CANCER
The early stage of tumorigenesis may present no symptoms, leading to tumors in the middle and late stages before symptoms are identified, which hinders the treatment of tumors. However, tumor markers already exist in the early stage of tumorigenesis,which is of great significance for the early diagnosis and treatment of tumors. The main difficulty in the early diagnosis of tumors is the identification of the characteristics of tumors and tumor cells. Tumor markers are important molecules in the diagnosis and treatment of tumors. They can target whole tumors or tumor cells.Fluorescence imaging of tumors and cells can diagnose tumors and monitor their treatment. Due to the high specificity and affinity of aptamers, a goal currently attracting intense interest is to screen for aptamers as tumor markers and apply them to the targeted diagnosis of tumors. Aptamers have become potential probe molecules for targeted diagnosis of liver cancer, and their application in vitro and in vivo of liver cancer can improve detection specificity and sensitivity[51](Figure 2).
Aptamers for in vitro detection of liver cancer
In recent years, with the in-depth study of aptamers, increasing numbers of liver cancer detection methods based on aptamers have been developed to provide different in vitro detection methods for the diagnosis of liver cancer (Table 1).
The aptamer-nanoparticle system (Apt-NP) formed by the conjugation of liver cancer-related aptamers and nanoparticles, can be used for the diagnosis of liver cancer. Recently, Hu et al[52]labeled HepG2 cells with biotin-conjugated TLS11a aptamer (Bio-TLS11a)., and then co-incubated with streptavidin-conjugated fluorescent silica nanoparticles (SA-FSNPs). HepG2 cells were detected in vitro by the interaction between biotin and streptavidin. Hu et al[52]pointed out that these SAFSNPs can specifically and sensitively detect aptamer-labeled HepG2 cells and show no obvious toxic effects, showing the good application prospect of this method for liver cancer cells and other types of cancer cells.
With the deeper research on nano-molecules, more and more nano-molecules have been discovered and used together with aptamers for targeted detection of liver cancer cells. Wang et al[53]established a new method for detecting circulating tumor cells (CTCs) in hepatocellular carcinoma (HCC). They combined the aptamer of the carbohydrate sialic acid Lewis X with a hydroxyapatite/chitosan (HA/CTS) nanofilm to form the CTC-BioTChip platform, which effectively captures and identifies HCC CTCs. A small amount of aflatoxin B1 can cause liver cancer to occur. Joo et al[54]developed a rapid method for the determination of aflatoxin B1(AFB1) based on GO and fluorescein imide modified AFB1-specific aptamer. When the aptamer binds to AFB1, the conformation of the aptamer changes and interacts with GO such that the fluorescence intensity is reduced or even quenched.
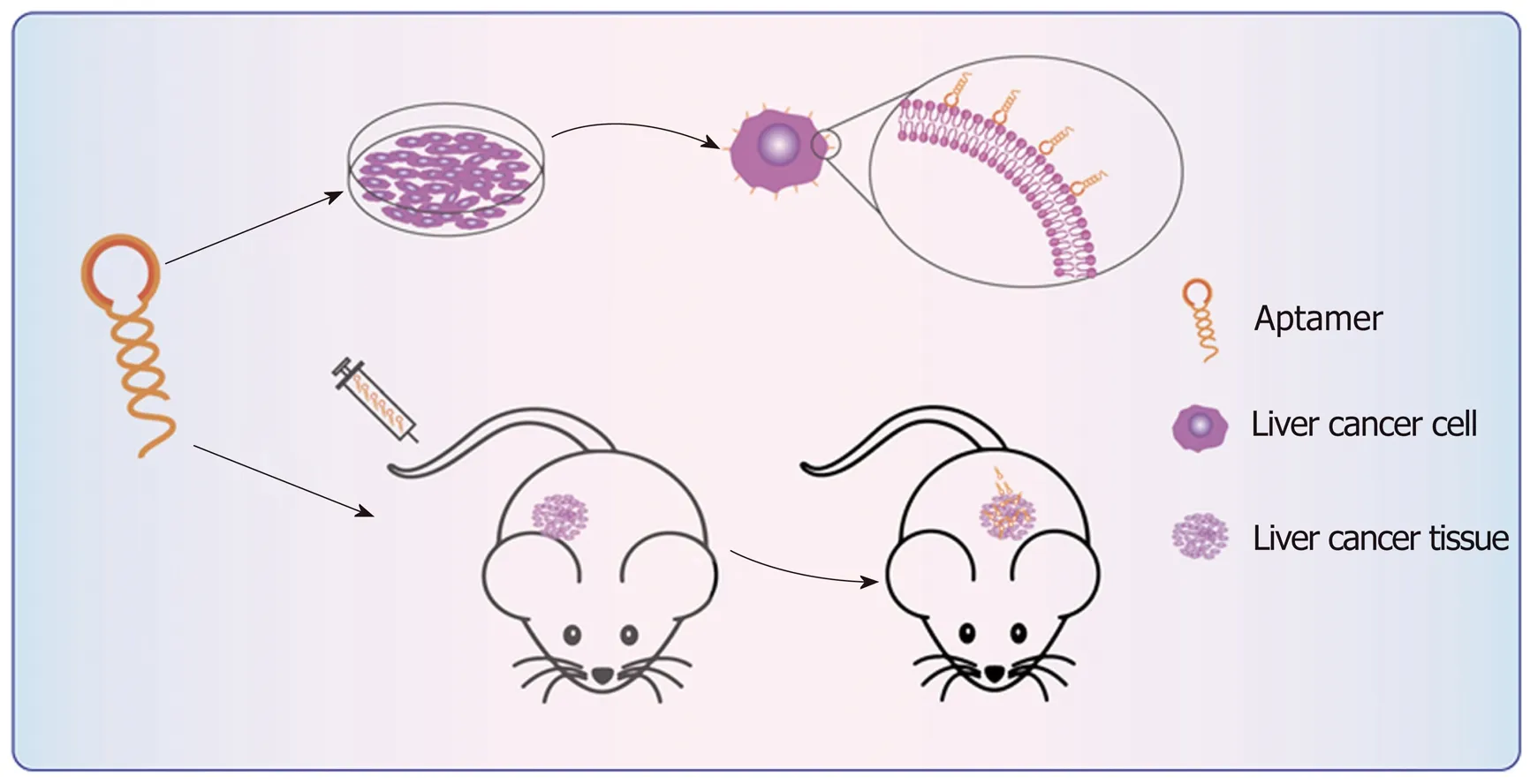
Figure 2 Schematic diagram of aptamer targeting to liver cancer. The aptamers bind to the surface of the liver cancer cells with high specificity and high affinity after incubation with the liver cancer cells. After the aptamers are injected into the mouse model through the tail vein, they specifically bind to the liver cancer tissue site.
In the past few years, the potential role of quantum dots in fluorescence imaging has gradually attracted the attention and research of more scholars. QD-labeled aptamers are also a new strategy for detecting liver cancer cells. It has been found that the SL2-B aptamer labeled with QDs can specifically recognize HepG2 cells and be taken up by HepG2 cells, and the results can be detected by fluorescence microscopy.Moreover, as the incubation time of HepG2 cells with the conjugate increased, it also showed potential antiproliferative activity against HepG2 cells[55]. This technique of QDs labeling aptamers provides a new strategy for in vitro detection and research of liver cancer.
Dickkopf-1 (DKK1) has potential application value for the early diagnosis of HCC.Zhou et al[56]obtained two kinds of aptamers that recognize DKK1, one is slow-off aptamer, and the experimental results show that it can be used for aptamer-based ELISA assay, while another fast-off aptamer is more suitable for flow cytometry and spot-blot. In addition, the combination of molecular beacons[57], which have stem-ring structures and contain fluorescent and quenching agents, and fluorescent dyes[58]with aptamers targeting tumors has also been proven to be useful for detecting or imaging tumors. This provides more potential methods for in vitro detection of liver cancer.
Aptamers for in vivo imaging of liver cancer
In vivo imaging is still a common method for the clinical diagnosis of tumors.Techniques include magnetic resonance imaging (MRI), ultrasound molecular imaging, computed tomography (CT), and fluorescence molecular tomography,which have been verified in small rodents[59]. Nowadays, due to the extremely high incidence of liver cancer and the increasingly intensive research on aptamer, the application of various aptamer-based contrast agents, probes, and fluorescent groups to in vivo oncology imaging has become the research topic of numerous scholars(Table 2).
MRI is a non-invasive tumor diagnosis technique. To improve the diagnostic efficacy of MRI, Yan et al[60]obtained an aptamer capable of binding to endoglin(CD105) with high specificity by SELEX. This aptamer is conjugated to paramagnetic and fluorophore on the G5 dendrimer to form a targeted nanoprobe that can be used to detect tiny HCCs. Zhong et al[61]developed a nanoprobe targeting CD105 for MRI by the use of aptamer-modified magnetic carboxymethyl chitosan, which can improve the early diagnosis of HCC. In addition, Zhao et al[62]prepared a targeted MRI molecular probe based on ultrasmall superparamagnetic iron oxide (USPIO) mediated by phosphatidylinositol-3 (GPC3) aptamer. The probe was verified in xenograft mice,and the experimental results confirmed the targeted imaging effect of the probe on HCC expressing GPC3.
Various studies have shown that aptamers have great potential advantages in the targeted diagnosis of liver cancer, but this application is still in its infancy. For some malignant tumors with a low expression rate of tumor markers, further exploration is still needed[63].
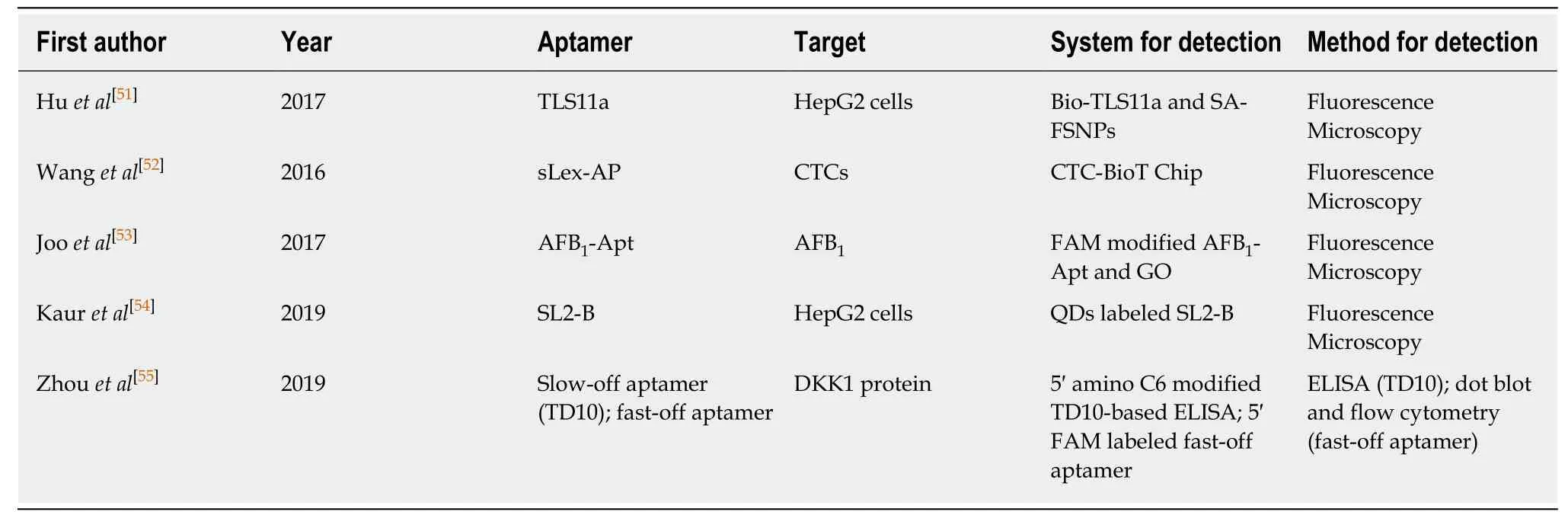
Table 1 Current literature summary of aptamers for in vitro detection of liver cancer
APPLICATION OF APTAMERS IN TARGETED THERAPY OF LIVER CANCER
In addition to acting as therapeutic agents, aptamers can also be used as targeting agents to conjugate with antitumor drugs, oligonucleotides, or nanocarriers for targeted tumor therapy. A drug delivery system based on aptamers has many advantages. It can not only significantly improve the pharmacokinetic properties of drugs in vivo, but also can reduce toxic and side effects to some extent certain due targeting properties. Because aptamers are small (nanosized) molecules, the whole drug delivery system can effectively penetrate all levels of blood vessels into the tumor site[64].
The traditional methods of liver cancer therapy have poor targeting, inefficient drug delivery, poor efficacy, and severe side effects, which together present great challenges in the therapy of liver cancer. The targeted therapy of liver cancer is now the focus of scientists' research. Currently, aptamers have been widely studied and applied in various fields because of their characteristics. Modified aptamers have been applied to the treatment of liver cancer, improving the efficiency of treatment and greatly reducing the side effects of drug treatment[65]. Aptamers can be used as therapeutic agents to intervene in the treatment of liver cancer. They can also be conjugated with antineoplastic drugs, oligonucleotides, or nanocarriers for the treatment of tumors, such as chemotherapy, gene therapy, immune therapy,radiotherapy, and phototherapy[66](Table 3).
Hu et al[67]constructed a bispecific system using the hepatoma cells-specific aptamer TLS11a and anti-CD3, which mediates T cell activation, targets cytotoxic T cells to the liver cancer site, and thus enhances the anti-tumor effect. The experimental results show that TLS11a/CD3 can connect hepatoma cells and T cells, avoiding the occurrence of immune escape and mediating the lysis of hepatoma cells and the release of cytokines, and the released cytokines can continue to enhance anti-tumor effects. This aptamer-based bispecific system provides a new technology platform for targeted immunotherapy of liver cancer.
The recombinant adenovirus carrying the PTEN gene (Ad5-PTEN) is an effective anti-tumor agent, but its application is greatly limited due to its poor stability and toxic effects to normal tissues. To overcome the application limitations of Ad5-PTEN,Xiao et al[68]formed a novel gene delivery system, EPAP, by linking EpDT3, an aptamer that specifically binds to epithelial cell adhesion molecule (EpCAM), to Ad-PTEN. The results show that EPAP is non-toxic to normal cells and tissues, can target and inhibit EpCAM-positive hepatoma cells, enhance anti-tumor activity, and improve the gene therapy efficiency of liver cancer.
Today, mesoporous silica nanoparticles (MSNs) are a potential therapeutic platform for cancer therapy. MSNs are a good nano-drug delivery platform because of its good stability, high load, high biocompatibility, adjustable pore size, and protection against inactivation or degradation of the incorporated drug[69]. Babaei et al[70]developed a PEGylated MSN system capable of targeting EpCAM and controlling the release of 5-fluorouracil. First, the authors loaded 5-fluorouracil into mesoporous silica, hybridized with gold nanoparticles to its outer surface, PEGylated, and then conjugated with EpCAM aptamer, and the resulting complex can be used for HCC treatment. This system has potential application prospects in the targeted therapy of early liver cancer. In addition, it can also be used for CT imaging.
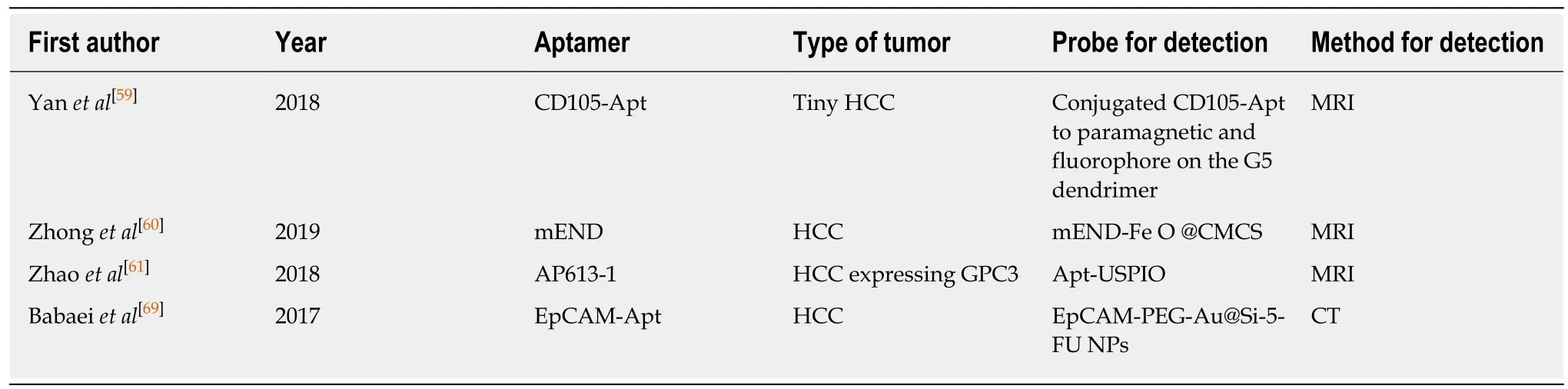
Table 2 Current literature summary of aptamers for in vivo imaging of liver cancer
Black Phosphorus Quantum Dots (BPQDs) have a unique photocatalytic activity and have potential applications in photodynamic therapy, but the application of BPQD is limited due to the instability of the tumor microenvironment. To solve this problem, Lan et al[71]constructed a nanostructure based on the TLS11a aptamer. Lan et al[71]synthesized BPQDs/Pt hybrid mesoporous silica (BMSF@Pt), and then conjugated the TLS11a aptamer to BMSF@Pt to form Apt-MBMSF@Pt nanostructure.This structure can specifically target to HCC cells, killing HCC cells by selfcompensation of oxygen, enhancing photodynamic efficacy, and providing a new method for targeted therapy of HCC.
MiRNA-195 (miR195) is a potent gene drug that inhibits vascular endothelial growth factor (VEGF), and some studies have demonstrated the inhibitory effect of miR195 in HCC[72]. Liu et al[73]conjugated histidine-modified disulfide cross-linked stearyl polyarginine peptide (H3R5) to cell penetrating peptide-modified aptamer,ST21, which is capable of specifically binding to HCC cells. Then, under the action of PEG, miR195 and Fasudil were loaded to the conjugation. Finally, a nano-system,FasudilST21-H3R5-PEGmiR195, based on ST21-H3R5-PEG was constructed. The results showed thatFasudilST21-H3R5-PEGmiR195has a strong ROCK2 blocking effect and VEGF silencing effect, showing a higher anti-tumor effect, and has great application prospects for targeted therapy of HCC.
Liposomes are complex structures composed of cholesterol and phospholipids that can contain hydrophilic and lipophilic drugs and reduce the cytotoxicity of drugs.Liposomes are very effective drug nanocarriers. Cabazitaxel is a broad-spectrum antineoplastic drug, but its clinical adverse reactions hinder its use. Cheng et al[74]modified capsaicin liposomes with a TLS1c aptamer targeting MEAR cells and demonstrated the highly specific targeting ability and strong antitumor effect of this complex on MEAR cells. This drug delivery system is also an effective potential therapeutic strategy. Nanoparticles, polymer micelles, and biomimetic nanocarriers can also be used as drug carriers to construct drug delivery systems, providing potential for the further development of targeted therapy for liver cancer[75].
CONCLUSION
In conclusion, the diagnosis and treatment of liver cancer developed on the basis of aptamer solve the problems of low diagnostic sensitivity, large side effects of anticancer drugs, and difficulty in tumor monitoring. Various experimental results have confirmed the great potential of these strategies in the clinical application to liver cancer, and provide good scientific evidence for the clinical application of aptamers in liver cancer.
As “chemical antibodies”, aptamers have been considered an important research direction in the targeted diagnosis and treatment of tumors. With the continuous development of aptamers with different targeting sites, the application of aptamers with different targeting sites has become increasingly extensive. Aptamers have been modified and applied to various aspects of targeting research for liver cancer. With the development of different detection methods and treatment methods based on aptamers, aptamers will continue to develop in the clinical application of liver cancer,and finally realize early diagnosis and treatment of liver cancer.
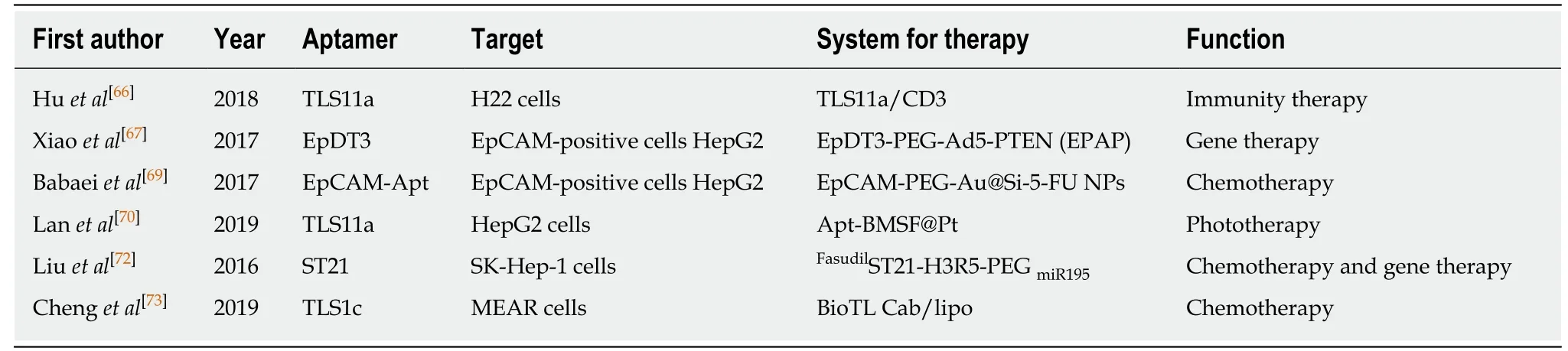
Table 3 Current literature summary on the application of aptamers in targeted therapy for liver cancer
杂志排行
World Journal of Gastroenterology的其它文章
- Diuretic window hypothesis in cirrhosis: Changing the point of view
- Fluoroquinolones for the treatment of latent Mycobacterium tuberculosis infection in liver transplantation
- Reactivation of hepatitis B virus infection in patients with hemolymphoproliferative diseases, and its prevention
- Current status of endoscopic retrograde cholangiopancreatography in patients with surgically altered anatomy
- Choledochal cysts: Similarities and differences between Asian and Western countries
- Gastro-duodenal disease in Africa: Literature review and clinical data from Accra, Ghana
