Proteomics of the mediodorsal thalamic nucleus of rats with stressinduced gastric ulcer
2019-07-10ShengNanGongJianPingZhuYingJieMaDongQinZhao
Sheng-Nan Gong, Jian-Ping Zhu, Ying-Jie Ma, Dong-Qin Zhao
Abstract BACKGROUND Stress-induced gastric ulcer (SGU) is one of the most common visceral complications after trauma. Restraint water-immersion stress (RWIS) can cause serious gastrointestinal dysfunction and has been widely used to study the pathogenesis of SGU to identify medications that can cure the disease. The mediodorsal thalamic nucleus (MD) is the centre integrating visceral and physical activity and contributes to SGU induced by RWIS. Hence, the role of the MD during RWIS needs to be studied.AIM To screen for differentially expressed proteins in the MD of the RWIS rats to further elucidate molecular mechanisms of SGU.METHODS Male Wistar rats were selected randomly and divided into two groups, namely, a control group and an RWIS group. Gastric mucosal lesions of the sacrificed rats were measured using the erosion index and the proteomic profiles of the MD were generated through isobaric tags for relative and absolute quantitation(iTRAQ) coupled with two-dimensional liquid chromatography and tandem mass spectrometry. Additionally, iTRAQ results were verified by Western blot analysis.RESULTS A total of 2853 proteins were identified, and these included 65 dysregulated (31 upregulated and 34 downregulated) proteins (fold change ratio ≥ 1.2). Gene Ontology (GO) analysis showed that most of the upregulated proteins are primarily related to cell division, whereas most of the downregulated proteins are related to neuron morphogenesis and neurotransmitter regulation. Ingenuity Pathway Analysis revealed that the dysregulated proteins are mainly involved in the neurological disease signalling pathways. Furthermore, our results indicated that glycogen synthase kinase-3 beta might be related to the central mechanism Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0)license, which permits others to distribute, remix, adapt, build upon this work non-commercially,and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:http://creativecommons.org/licen ses/by-nc/4.0/Manuscript source: Unsolicited manuscript Received: January 27, 2019 Peer-review started: January 27,2019 First decision: March 20, 2019 Revised: May 8, 2019 Accepted: May 18, 2019 Article in press: May 18, 2019 Published online: June 21, 2019 P-Reviewer: Ierardi E, Liu HK S-Editor: Cui LJ L-Editor: Wang TQ E-Editor: Ma YJ through which RWIS gives rise to SGU.CONCLUSION Quantitative proteomic analysis elucidated the molecular targets associated with the production of SGU and provides insights into the role of the MD. The underlying molecular mechanisms need to be further dissected.
Key words: Mediodorsal thalamic nucleus; Proteome; Restraint water-immersion stress;Stress-induced gastric ulcer; Glycogen synthase kinase-3 beta
INTRODUCTION
Stress can cause behaviour, immune, and neural responses, and intense and persistent stress responses could lead to a wide range of physiological and psychological dysfunctions in the body[1-3]. Restraint water-immersion stress (RWIS) can cause serious gastrointestinal dysfunction and has been widely used to study the underlying molecular mechanisms of stress-induced gastric ulcer (SGU) and to screen potential curative drugs[4,5]. The primary central mechanism through which brainstem circuits regulate gastric function under normal physiological conditions has been fundamentally clarified[6-8], and the peripheral regulatory mechanism of gastric ulcers induced by RWIS has also drawn much attention[9-12]. Previous studies have reported that neuronal hyperactivity of vagal parasympathetic efferents, which mostly originate in the primary gastrointestinal control centre of the central medulla (mainly in the dorsal motor nucleus of the vagus and partly in the nucleus ambiguous) and are further mediated by the higher central-anterior hypothalamus (mainly the paraventricular nucleus and supraoptic nucleus), leads to gastric dysfunction[13-15]. In addition, we previously found that catecholaminergic, vasopressinergic, and oxytocinergic neurons might be associated with gastric ulcers induced by RWIS[16-18].The brain nuclei and neurotransmitters or hormones involved in RWIS are shown in Table 1[10,11,13-15,19,20]. However, we have not yet retrieved relevant papers that discuss the effects of proteomic changes on the brain of rats during exposure to RWIS.
The mediodorsal thalamic nucleus (MD) has complex connections with the prefrontal cortex and subcortical structures[21-26], and previous studies have revealed that the MD is the integration centre of visceral and physical activity and is involved in the modulation of emotion[27-31], cognition, and memory[32-36]. We previously found that the expression of Fos, which is used as a marker of neuronal activity, is significantly increased in the MD of rats after 2 h of RWIS[37]. Hence, the role of the MD during RWIS needs to be studied. A comparative proteomic analysis of RWISexposed and control rats might not only shed light on the role of the MD in gastrointestinal dysfunction induced by RWIS but also contribute to the detection of proteomic differences and the identification of targets for more specific therapies.
The combination of isobaric tags for relative and absolute quantitation (iTRAQ)with multidimensional liquid chromatography and tandem mass spectrometry(LC-MS/MS) analysis is one of the most sensitive proteomic technologies for detecting and quantitatively analysing low-abundance proteins, and this approach has been widely used to discover disease-specific targets and biomarkers for a variety of diseases[12,38-41]. To the best of our knowledge, an iTRAQ-based approach has been applied to study the central mechanism of gastrointestinal dysfunction induced by RWIS. To identify dysregulated proteins in the MD, which might help to further explore the molecular mechanisms and pathogenesis of SGU induced by RWIS, we quantitatively compared the proteomes of control and RWIS-exposed rats using the multiplex capability of the iTRAQ approach. The present study provided a novel understanding of the role of the MD, and the newly identified molecular differences might provide fundamental information for further study.

Table 1 Brain nuclei and neurotransmitters/hormones involved in restraint water-immersion stress
MATERIALS AND METHODS
Preparation of animals
Adult male Wistar rats, weiging 250-300 g and provided by the Experimental Animal Center of Shandong University, Jinan, China, were housed two per cage at 22 °C ± 2°C under a 12 h light-12 h dark cycle with free access to food and water before the initiation of RWIS. The sxperiments were initiated at least 7 days after the animals arrived at the facility. Prior to the experiments, rats were fasted for 24 h but allowed access to tap water ad libitum.
Stress protocols
Twenty-four male rats, numbered from 01 to 24, were used in the stress experiments.Twelve rats, which were selected using the random number table method, were subjected to RWIS for 4 h (RWIS group), and the other rats were not subjected to RWIS (control group). RWIS was performed as previously described[16]. Experiments were performed between 8:00 morning and 12:00 afternoon. All procedures were performed according to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of Shandong Normal University (Jinan, People’s Republic of China;No. AEECSDNU2018003). In addition, all efforts were made to minimize the animals’suffering and to reduce the number of animals used.
Evaluation of gastric mucosal damage
At the end of the procedure, rats were killed through a pentobarbital-Na+injection(100 mg/kg BW, i.p.). Stomachs were removed, placed in 1% formaldehyde solution for 30 min, and incised along the greater curvature. Gastric mucosal lesions were measured using the erosion index (EI)[42].
Sample collection
After brain dissection, the MD (coordinates: 2.04-3.84 mm rostral to the obex, depth of 5.0-6.0 mm below the surface of the obex, and 1.0 mm lateral to the midline) was harvested for protein extraction (WPI, Sarasota, United States)[43]. To minimize the individual differences of rats, every three individual samples were mixed for protein extraction, and protein extracts were prepared in duplicate for each group.
Protein extraction and iTRAQ labelling
The sample was resuspended in approximately eight times volume of lysis buffer (4%SDS and 100 mM HEPES (pH = 7.6) containing protease inhibitor cocktail and PMSF).The homogenate was sonicated for 30 min on ice. After centrifugation at 25 000 g for 30 min at 4 °C, the supernatant was collected and stored at -80 °C. The total protein concentration was measured using the BCA Kit (Trans Gen Biotech, Beijing, China).
Equal amounts of the extracted protein were mixed according to groups and then precipitated with acetone overnight. After the protein was resuspended in triethylammonium bicarbonate (TEAB) buffer, protein quantification was done using the BCA Kit. The proteins were reduced by 5 mmol/L DTT at 56 °C for 30 min and alkylated by 10 mM IAA at room temperature for 30 min. The sample was then diluted with 50 mM ammonium bicarbonate until the concentration of urea was lower than 1 M. Trypsin (Sigma) was added to the sample at a mass ratio of 1:50(enzyme:protein) for 12 h. iTRAQ-8plex labelling reagents (AB Sciex) were added to the peptide samples, which were then incubated at room temperature for 120 min.The reaction was stopped by the addition of water, followed by concentration of the product using SpeedVac and desalts. The purified peptides were collected and stored at -80 °C until use.
High-pH reverse-phase fractionation and LC-MS/MS detection
The peptides were fractionated on a Waters UPLC using a C 18 column (Waters BEH,Milford, United States, c18 2.1×50 mm, 1.7 µm). Peptides were eluted at a flow rate of 600 μL/min with a linear gradient of 5%-35% solvent B (acetonitrile) over 10 min,with solvent A composed of 20 mM ammonium formate with pH adjusted to 10. The absorbance at 214 nm was monitored, and a total of ten fractions were collected.
The fractions were separated by nano-HPLC (Eksigent Technologies) on a secondary reversed-phase analytical column (Eksigent, C18, 3 µm, 150 mm × 75 µm).Peptides were subsequently eluted using the following gradient conditions with phase B (98% ACN with 0.1% formic acid) from 5% to 40% B (5-65 min), 40% to 80% B(66-71 min), and 80% to 5% B (72-90 min), and total flow rate was maintained at 300 nL/min. Electrospray voltage of 2.5 kV versus the inlet of the mass spectrometer was used. The QTOF 4600 mass spectrometer was operated in information-dependent data acquisition mode to shift automatically between MS and MS/MS acquisition. MS spectra were acquired across the mass range of 350-1250 m/z. The 25 most intense precursors were selected for fragmentation per cycle with a dynamic exclusion time of 25 s.
Data processing
After product separation, all MS/MS spectra were analysed using Mascot (Matrix Science, London, United Kingdom; version 2.3.0). Mascot was set up to search the UniProt_rat database and was searched with a fragment ion mass tolerance of 0.1 Da and a parent ion tolerance of 25.0 ppm. Additionally, carbamidomethylation of cysteine and the iTRAQ 8PLEX conjugation of lysine and the N-terminus were specified in Mascot as fixed modifications. Oxidation of methionine and iTRAQ 8PLEX conjugation of tyrosine were specified in Mascot as variable modifications.
Scaffold (version Scaffold_4.4.1.1, Proteome Software Inc., Portland, OR, United States) was used to validate the MS/MS-based peptide and protein identifications.Peptide identifications were accepted at a false discovery rate (FDR) of less than 1.0%using the Scaffold Local FDR algorithm. Protein probabilities were assigned using the Protein Proph et al[44]gorithm. Only those proteins with greater than 90% probabilities were accepted.
Quantitative data analysis
Scaffold Q+ (version Scaffold_4.4.5, Proteome Software Inc., Portland, OR, United States) was used to quantify the iTRAQ peptide and for protein identifications. To determine quantitative changes, a ± 1.2-fold change in the MD of rats between the control and RWIS groups with P < 0.05, as calculated by the Student’s t-test, was used as the threshold for categorizing upregulated and downregulated proteins.
Bioinformatic analysis
Gene Ontology (GO) annotation and pathway enrichment analysis of all the identified and differentially expressed proteins were implemented using the online tool DAVID(http://david.abcc.ncifcrf.gov/). GO annotation includes biological processes (BP),cellular components (CC), and molecular functions (MF). Ingenuity Pathway Analysis(IPA) software (version 7.5, Ingenuity System Inc., Redwood City, CA, United States;www.ingenuity.com) was used to identify the biological functions and signalling pathways of the annotated differentially expressed proteins. The GO annotations involving signalling pathways and networks were ranked based on the enrichment of the differentially expressed proteins.
Western blot analysis
The results of the proteomic experiment were verified by Western blot analysis.Protein extracts (30 μg) were separated by SDS-PAGE followed by electrotransfer onto PVDF membranes (PVDF, Millipore, Bedford, MA, United States). The membranes were blocked with 5% non-fat dry milk in 0.01 M PBS containing 0.1% Tween-20 for 2 h at room temperature and then incubated at 4 °C overnight with the following primary antibodies: Anti-glycogen synthase kinase-3 beta (GSK3B) (mouse monoclonal, 47 kDa; 1:2000 dilution; Abcam plc ab93926, Cambridge Science Park,Cambridge, CB4 0FL, United Kingdom) and anti-synaptophysin (SYN) (rabbit monoclonal, 38 kDa; 1:1000 dilution; Abcam plc 32127). After three rinses in TBST buffer, the secondary antibody (HPR-conjugated anti-mouse; 1/1000 dilution; Dako,Glostrup, Denmark) was added, and the membranes were incubated for 1 h at room temperature. After the membranes were rinsed, protein expression levels were detected by enhanced chemiluminescence and visualized by autoradiography (ECL).Beta-tubulin (mouse monoclonal, 54 KDa; 1:4000 dilution; Sigma) was utilized as the loading control.
Statistical analysis
Statistical procedures were performed using SPSS 20.0 software (SPSS, Chicago, IL,United States). The results were reported as the mean ± SEM. Statistical analyses of the data were performed by the Student’s t-test. Differences were set as significant at P < 0.05.
RESULTS
Gastric mucosal damage induced by RWIS
A macroscopic analysis showed no gastric mucosal damage in the control group,whereas scattered spotty and linear haemorrhage was observed in the gastric mucosa of rats in the RWIS group (Figure 1). The damage index was significantly different between the control (0.0 ± 0.0) and RWIS groups (47.5 ± 11.1) (P < 0.01).
Identification of differentially expressed proteins
Relative protein expression values were compared between the control and RWIS groups to identify the differentially expressed proteins. This analysis indentified 2853 proteins (Table S1), including 65 unique (31 significantly upregulated and 34 downregulated) proteins that were dysregulated (fold change ratio ≥ 1.2, with P <0.05) (Figure 2A). These upregulated proteins (e.g., cell division cycle-16, CDC16)might be involved in cell division, and the downregulated proteins (e.g., GSK3B)might be related to motor neuron axon guidance, nutrient delivery to the nervous system, and neurotransmitter regulation (Table S2). Based on the premise that each dysregulated protein was assigned at least one term, GO annotation was applied to reveal the CC, MF, and BP of the identified proteins. Of all the dysregulated proteins,18% (eight proteins) were annotated as cytoplasm-associated proteins, and the other main location categories of the proteins are the extracellular exosomes, cytoplasm,and cytosol (13%) and the nucleus, membrane, and plasma membrane (11%) (Figure 2B). The molecular functions of the dysregulated proteins are associated with poly(A)RNA binding, ATP binding, metal binding, protein homodimerization activity, and other processes (Figure 3A). GO annotation analysis showed that the 65 dysregulated proteins are mainly involved in biological processes, such as response to oxidative stress, inflammatory response, response to peptide hormones, translation, protein phosphorylation, protein oligomerization, response to glucose, and pigmentation(Figure 3B).
IPA network analysis of differentially expressed proteins
Because the molecular function of proteins in organisms is not accomplished by proteins alone but rather in coordination with many biological molecules, the roles of differentially expressed proteins were further analysed using the IPA tool according to -log (P-value). The IPA analysis identified 123 signalling pathways, and the highest-ranked canonical IPA pathways are shown in Table 2. Specifically, protein kinase A signalling was found to be significantly negative. The signalling pathways of the dysregulated proteins are mainly associated with cardiovascular system development and function and organ morphology, which comprise the first tier. The second tier consists of infectious diseases, digestive system development and function, gastrointestinal disease, organismal development, and organismal injury and abnormalities, and the third tier comprises cellular growth and proliferation,neurological disease, and haematological system development and function. Notably,GSK3B participates in five of the top ten most enriched pathways (Table 2) and is related to all the above-mentioned functions. Moreover, the dysregulated proteins were co-enriched into five networks, three of which are related to nerve signalling pathways (Figure 4). The highest-scoring IPA network contained 34 focus molecules and six upregulated and 10 downregulated proteins that met the significance criterion, including GSK3B; these proteins are mostly involved in cardiovascular system development and function, organ morphology, and neurological disease signalling pathways (Figure 4A).

Figure 1 Gastric mucosal damage induced by restraint water-immersion stress. A: Control group. The gastric mucosal surface was intact and smooth; B:Stressed group. The gastric mucosal surface showed spotty and linear haemorrhages after 4 h of restraint water-immersion stress.
Western blot validation
According to both the GO and IPA network analyses, GSK3B was downregulated(0.71-fold change) and might affect synaptic plasticity[45]. SYN, which is involved in the process of synaptic neurotransmission, was not among the 2853 proteins showing significant expression changes between the groups. The GSK3B and SYN expression levels were validated at the protein level by Western blot, and beta-tubulin was included as a control for protein loading quantification. As shown in Figure 5, the protein level of GSK3B was significantly downregulated (n = 9, P = 0.037) in rats after RWIS for 4 h, whereas the protein level of SYN did not differ between the two groups(n = 9, P = 0.069). These results are closely consistent with the iTRAQ analysis results.Because the proteomic and WB data are quite comparable and consistent with each other, GSK3B might be related to the central mechanism of gastrointestinal dysfunction induced by RWIS, but the underlying molecular mechanisms need to be further dissected.
DISCUSSION
Many previous studies focused on the peripheral and primary central regulatory mechanisms of SGU[9-15], but little is known about the higher central mechanism[17-20,46].This study constitutes the first analysis of the molecular differences in the MD at the proteome level between control and RWIS-exposed rats and thus, provides further insights into the molecular mechanisms and pathogenesis of SGU. Screening potential curative drugs for SGU has created an urgent need to identify new targets and iTRAQ becomes a powerful tool to explore the response in proteins to stress. Among the 2853 identified proteins, 65 were dysregulated, and these included 31 upregulated and 34 downregulated proteins. Functional analysis indentified 29 unique proteins, such as Rab31, GSK3B, and Lynx1, and these 29 proteins were primarily related to the nervous system, due to their involvement in synaptic vesicle exocytosis and endocytosis, neuron morphogenesis, and particularly neuritis, which indicates that some differences in synaptic plasticity might exist between the control and stressed groups[47-49].
To further verify the data acquired by iTRAQ proteomics, the expression of the downregulated protein GSK3B and the unchanged protein SYN, which are used as specific markers to reflect changes in the synaptic plasticity of the presynaptic terminal, was assessed by Western blot using commercially available antibodies. The Western blot results are consistent with the iTRAQ findings. The 0-3 fold change in expression obtained by Western blot analysis, which was notably greater than the changes obtained in the the proteomic analysis, might be due to the cascade amplification of optical signals during the Western blot analysis for GSK3B using the primary and secondary antibodies.
GO annotation analysis showed that the molecular functions of the 65 identified proteins are mostly associated with protein binding, including poly(A) RNA binding and ATP binding. RNA-binding proteins are dynamic posttranscriptional regulators of gene expression, whereas ATP-binding proteins are known to play a fundamental role in biological processes, which indicates that synthetic and metabolic processes undergo changes in response to RWIS. The biological processes involving the dysregulated proteins were found to be associated with oxidative stress, inflammatory response, and protein phosphorylation, indicating that inflammatory disorders and abnormal metabolic functions occur in rats after RWIS for 4 h.Furthermore, some of the dysregulated proteins, such as glutathione S-transferases(GSTs), have dehydrogenase activity, which also indicates that the organism has evolved a mechanism that protects cells against reactive oxygen species during exposure to RWIS, similarly to that found in response to other stressors[50-52].

Figure 2 Features of the proteomic data from rats subjected to restraint water-immersion stress for 0 h and 4 h. The data were obtained by isobaric tags for relative and absolute quantitation shotgun analysis. A: Volcano plot indicating the changes in protein abundance in the mediodorsal thalamic nucleus between the control and stressed rats. A total of 65 dysregulated proteins with at least a ±1.2-fold change and P < 0.05 were identified. Orange dots indicate the upregulatedproteins, and blue dots indicate the downregulated proteins; B: Subcellular localizations of the identified proteins.
According to IPA pathway analysis, the dysregulated proteins are mainly involved in pathways related to oxidative stress, lipid and amino acid metabolism, hepatotoxicity, and kidney damage. The protein kinase A signalling pathway is the only one of the top-ranked IPA pathways that was significantly downregulated in rats after RWIS. This kinase signalling pathway plays a critical role in multiple cellular processes through the post-translational phosphorylation of various proteins,including signalling and metabolic enzymes, transcription factors, and ion channels.The altered proteins involved in this signalling pathway include two downregulated proteins, calcium/calmodulin-dependent kinase II (CAMK2D) and GSK3B, and two upregulated proteins, protein tyrosine phosphatase non-receptor type 9 (PTPN9) and cell division cycle-16 (CDC16). CAMK2, one of the most abundant protein kinases in the brain, plays a key role in neurotransmission, synaptic plasticity, and brain synapse development and showed neuroprotective activity both in vitro and in vivo[53,54].Previous studies have found that CAMK2 can be rapidly activated and then inactivated after focal ischaemia and that the degree of inactivation is correlated with the extent of neuronal damage[55,56]. In the present study, CAMK2D was found to be downregulated in the stressed group, which indicates that the neurons in the MD were damaged during the 4-h exposure to RWIS. GSK3B is the major kinase phosphorylating tau protein and plays an important role in various physiological and pathological processes, such as glycometabolism, cell proliferation, immune response,inflammatory response, tumourigenesis, and neural development, degeneration, and regeneration[57-59]. The inhibition of GSK3 activity can promote neurite regeneration[60,61]. In this study, GSK3B was found to be downregulated, which indicates that RWIS injures neurons in the MD. However, because GSK3B can participate in many enriched pathways involved in energy metabolism, inflammation, endoplasmic reticulum stress, mitochondrial dysfunction and apoptosis pathways and so on, the molecular regulatory mechanism of GSK3 in gastric stress should be further studied.PTPN9, a cytoplasmic protein tyrosine phosphatase, is considered a marker of a tumour suppressor[62]. CDC16 was found in eukaryotic organisms and has been demonstrated to be involved in cell division[63,64]. The upregulation of PTPN9 and CDC16 in rats after RWIS, as observed in the present study, will be an interesting topic that should be investigated in future studies using other methods[65].

Figure 3 Features of the proteomic data from the mediodorsal thalamic nucleus of rats exposed to restraintwater-immersion-stress. The data were obtained by isobaric tags for relative and absolute quantitation shotgun analysis. A: Gene ontology (GO) analysis of the involved molecular functions; B: GO analysis of the involved biological processes. Only the top ten terms are shown.
In conclusion, an iTRAQ-based quantitative proteomic approach has been applied to detect and quantitatively analyse the alteration in the protein expression profile of the MD of rats subjected to RWIS. RWIS-induced proteomic alterations indicate that the neurons in the MD are disordered and show an abnormal function after 4 h of RWIS. Because heavy metabolic activity indicates great potential for plasticity and regeneration, the altered proteins might play important roles in cellular energy conservation and translation regulation to prevent the synthesis of unwanted proteins that could interfere with the cellular response to stress. Therefore, this study provides resources for identifying the biological functions of dysregulated proteins and dissecting pathways that could aid the identification of the molecular regulatory mechanism of SGU. The functions of key proteins linked to gastric ulcer, e.g., GSK3B,during RWIS should later be verified by QRT-PCR or immunoblotting, and further functional studies using RNAi.
ACKNOWLEDGEMENTS
The mass technical service was supported by Jiyun Biotech.
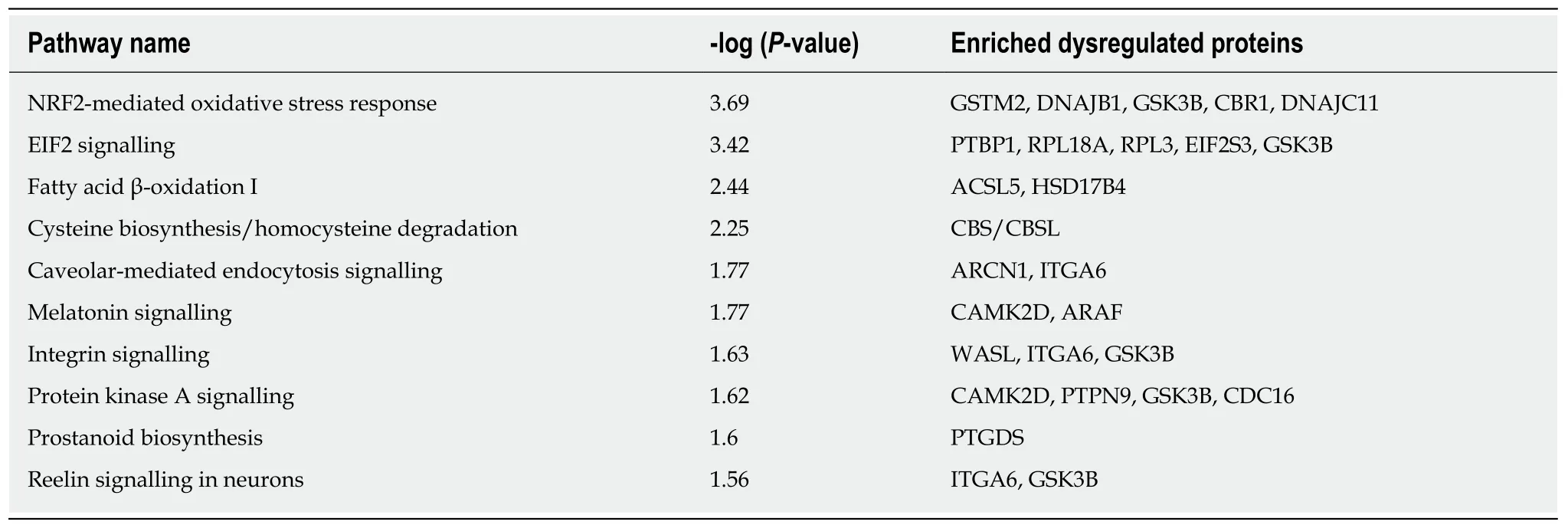
Table 2 Top ten most enriched pathways indentified by an ingenuity pathway analysis of the dysregulated proteins according to -log (Pvalue)

Figure 4 Key signalling pathways involving the proteins that were differentially expressed in the mediodorsal thalamic nucleu of rats exposed to restraint water-immersion stress. A: Cardiovascular system development and function, organ morphology, and neurological disease; B: Cellular assembly and organization,nervous system development and function, and tissue morphology; C: Cellular development, cellular growth and proliferation, and nervous system development and function. Orange labels indicate the upregulated proteins, and blue labels indicate the downregulated proteins.

Figure 5 Verification of glycogen synthase kinase-3 beta and synaptophysin expression by Western blot. After brain dissection, the proteins in the mediodorsal thalamic nucleu were separated by SDS-PAGE, and the expression levels of glycogen synthase kinase-3 beta (GSK3B) and synaptophysin (SYN) were detected by Western blot using antibodies against GSK3B and SYN. The bars represent the changes in the total GSK3B and SYN levels. Each value is the mean ±SD from at least three independent experiments; aP < 0.05. The expression of GSK3B was increased in the stressed group, whereas SYN showed no significant difference, which is in agreement with the isobaric tags for relative and absolute quantitation results.
ARTICLE HIGHLIGHTS
Research background
Stress-induced gastric ulcer (SGU) is one of the most common visceral complications after trauma. Restraint water-immersion stress (RWIS) can cause serious gastrointestinal dysfunction and has been widely used to study the pathogenesis of SGU to identify medications that can cure the disease. We have focused on providing a resource for determining the molecular regulatory mechanisms of stress-induced gastric mucosal lesion since 1990s. The mediodorsal thalamic nucleus (MD) is the centre integrating visceral and physical activity. There is remarkable Fos expression in the MD of rats subjected to RWIS.
Research motivation
iTRAQ becomes a powerful tool to explore the response in proteins to stress. A comparative proteomic analysis of RWIS-exposed and control rats might not only shed light on the role of the MD in gastrointestinal dysfunction induced by RWIS but also contribute to the detection of proteomic differences and the identification of targets for more specific therapies.
Recearch objectives
To screen for differentially expressed proteins in the MD of the RWIS rats to further elucidate the molecular mechanisms of SGU.
Research methods
Male Wistar rats were selected randomly and divided into two groups, namely, a control group and an RWIS group. Gastric mucosal lesions of the sacrificed rats were measured using the erosion index (EI) and the proteomic profiles of the MD were generated through isobaric tags for relative and absolute quantitation (iTRAQ) coupled with two-dimensional liquid chromatography and tandem mass spectrometry (LC-MS/MS). Additionally, iTRAQ results were verified by Western blot analysis.
Research results
A total of 2853 proteins were identified, and these included 65 dysregulated (31 upregulated and 34 downregulated) proteins (fold change ratio ≥ 1.2). Gene Ontology (GO) analysis showed that most of the upregulated proteins are primarily related to cell division, whereas most of the downregulated proteins are related to neuron morphogenesis and neurotransmitter regulation.Ingenuity Pathway Analysis (IPA) analysis revealed that the dysregulated proteins are mainly involved in the neurological disease signalling pathways. Furthermore, our results indicated that glycogen synthase kinase-3 beta (GSK3B) might be related to the central mechanism through which RWIS gives rise to SGU.
Research conclusions
Quantitative proteomic analysis elucidates the molecular targets associated with the production of SGU and provides insights into the effects of the MD. The underlying molecular mechanisms need to be further dissected.
Research perspectives
This study provides resources for identifying the biological functions of dysregulated proteins and dissecting pathways that could aid the identification of the molecular regulatory mechanism of SGU. The functions of key proteins linked to gastric ulcer, e.g., GSK3B, during RWIS should later be verified by qrt-pCR or immunoblotting, and further functional studies using RNAi.
杂志排行
World Journal of Gastroenterology的其它文章
- Modified FOLFlRlNOX for resected pancreatic cancer: Opportunities and challenges
- Role of cytochrome P450 polymorphisms and functions in development of ulcerative colitis
- Role of epigenetics in transformation of inflammation into colorectal cancer
- Postoperative complications in gastrointestinal surgery: A “hidden”basic quality indicator
- The role of endoscopy in the management of hereditary diffuse gastric cancer syndrome
- Predicting systemic spread in early colorectal cancer: Can we do better?
