Isolation and Characterization of E. Coli O157 : H7 from Infected Newborn Calves in Northeast China
2019-01-09ZhiYongLiuNaZhangPeiFanYuyingJiaHaotianGeRuidongMuJingLeiLeiandLiuYun
Zhi Yong, Liu Na, Zhang Pei, Fan Yu-ying, Jia Hao-tian, Ge Rui-dong, Mu Jing, Lei Lei, and Liu Yun
Heilongjiang Provincial Laboratory of Laboratory Animal and Comparative Medicine (Surgery), College of Veterinary Medicine, Northeast Agricultural University, Harbin 150030, China
Abstract: Escherichia coli O157 : H7 is a foodborne pathogen that poses a major threat to public health. Epidemiologic investigations have identified dairy cows, especially calves, are the principal reservoir of E. coli O157 : H7. In this study, based on the results,E. coli O157 : H7 was the main cause of E. coli disease outbreak in late October, 2015, and more than 90% of newborn calves died of serious diarrhea. Through further experiments, the drug sensitivity and resistance of the strain, the expression of the virulence gene and virulence pathogenicity were studied. E. coli O157 : H7 isolates were resistant to 12 antibiotics including penicillin, tetracycline and ampicillin, and were sensitive to eight antibiotics including cefoperazone, ceftazidime and amikacin. Resistance genes included tetB, strB, aadB, aphA, floR, TEM and virulence genes included stx1, eaeA and hlyA. Using specific pathogen free mice, the result showed that the isolate was pathogenic with a median lethal dose of 7.9×107 CFU • mL-1. This study described the pathogenesis and clinical manifestations of E. coli O157 : H7 infection. These results guided the use of antibiotics in prevent and control of bacterial infections in the future.
Key words: E. coli O157 : H7, drug sensitivity test, pathogenicity, resistance gene, virulence gene
Introduction
Escherichia coli (E. coli) serotype O157 : H7 is a foodborne pathogen. It is a serotype of the enterohemorrhagic E. coli (EHEC) pathogenic type and is a major cause of sudden abdominal pain, diarrhea and gastrointestinal disease in humans (Wang et al., 1996; Söderlund et al., 2014). EHEC infection may require an unusually low infectious dose, and 50-100 colony forming units (CFUs) are sufficient to cause disease in healthy individuals (Sánchez et al.,2010).
Epidemiologic investigations have demonstrated that dairy cows, particularly calves, are the principal reservoir of E. coli O157 : H7 (Rasmussen and Casey,2001). Multiple factors, including diet, housing,season, stress and age, can contribute to E. coli O157 : H7 infection in cows (Rasmussen and Casey,2001; Hallewell et al., 2014). E. coli O157 : H7 can be detected by using serology (Khalil et al., 2015),immunoassay (Aydin et al., 2014) and other molecular biology techniques (de Boer and Beumer, 1999).Many virulence factors contribute to the pathogenicity of E. coli O157:H7, including the production of Shiga toxin (Stx) and Stxs (Kudva et al., 1997). Stx induces cell death in endothelial cells, especially in the urinary tract, causing hemolytic uremic syndrome(HUS). The capacity of bacteria to cause attachingand-effacing lesions in the host intestine is associated with the presence of eae. Additionally, the "newer β-lactamases", including plasmid-mediated AmpC β-lactamases (e.g. CMY), extended-spectrum β-lactamases (e.g. CTX-M) and carbapenemases (e.g. NDM)produced by E. coli O157 : H7, are considered to be particularly dangerous (Pitout, 2012).
Antibiotics have been widely used to treat E. coli O157 : H7 infection, so that it may lead to drug resistance. Prevalence of antibiotic resistance and increased frequency spectrum are global public health issues and also threaten sustainable development of animal husbandry in China. The increasing popularity of drugresistant bacteria requires effective diagnostic tools. Studying the molecular mechanism of antibio-tic resistance might provide a scientific basis for the development of new antibiotics, and could better control the occurrence and spread of drugresistant strains.
In this study, in late October, 2015, diarrhea occurred in a dairy farm in Heilongjiang Province in northeast China, resulted in the death of more than 90% of newborn calves. The infection was caused by E. coli O157 : H7. The study further evaluated the antibiotic sensitivity, identified the genes responsible for drug resistance, virulence in the strain and studied its pathogenicity.
Materials and Methods
Sample collection
Samples were collected from a farm in Heilongjiang Province in northeast China in late October 2015,where diarrhea broke out, and 90% (61/68, calves/total) newborn calves died of serious diarrhea. Necropsy suggested E. coli was the major reason of the diarrhea. The lungs, mesentery, pleura and peritoneum from 30 dead calves were collected in sterile bags,transferred in iceboxes and investigated immediately after arriving at the laboratory. All the experiments were approved by the Institutional Animal Care and Use Committee of the Northeast Agricultural University, Harbin, China.
lsolation and identification of E. coli O157 : H7
Collected samples were homogenized and inoculated onto Sorbitol-MacConkey agar (Hangzhou Base Bio-Tech Co., China) supplemented with cefixime(0.05 mg • L-1) and potassium tellurite (2.5 mg • L-1)(Harbin Pharmaceutical Group Holding Co., China).Plates were incubated at 37℃ for 18 h. The cleat and colorless colonies with the characteristic E.coli O157 : H7 morphology were selected from the plates, screened on eosin methylene blue agar(Hangzhou Base Bio-Tech Co, China) at 37℃ for 18 h. Colonies with a metallic luster were gram stained. All the E. coli isolates were confirmed by biochemical tests. The bacteria were cultured in solutions containing mannose, glucose, lactose, maltose,raffinose, sorbitol, fructose, mushroom, nitrate, Voges-Proskauer (VP), dulcitol, sucrose, H2S, indole or citrate (Hangzhou Tianhe Microbe Reagent Co., Ltd.)for 24 h at 37℃. Agarose was shaken in 5 mL brain heart infusion (BHI) broth (Hangzhou Base Bio-Tech Co., Zhejiang China) and enriched for E. coli isolates at 37℃ for 18 h. As previously mentioned, the isolates were further confirmed by PCR assay (Yuan et al.,2010).
Detection of resistance and virulence genes
The major resistance and virulence genes were detected by PCR as described by Boerlin et al (2005).PCR amplification was conducted using 16S rRNA universal primers (Table 1) from Shanghai Invitrogen Reagent Company and using E. coli standard strain ATCC 25922 (purchased from China industrial culture collection center) as a positive control. E. coli O157 : H7 isolates were serotyped for O157 (rfbEO157)and H7 (E. coli structural flagellum antigen H7)antigens as described previously (Wang et al., 1996).Enterohemorrhagic E. coli standard strain CICC 21530(purchased from China Industrial Culture Collection Center) was used as a positive control for EHEC.
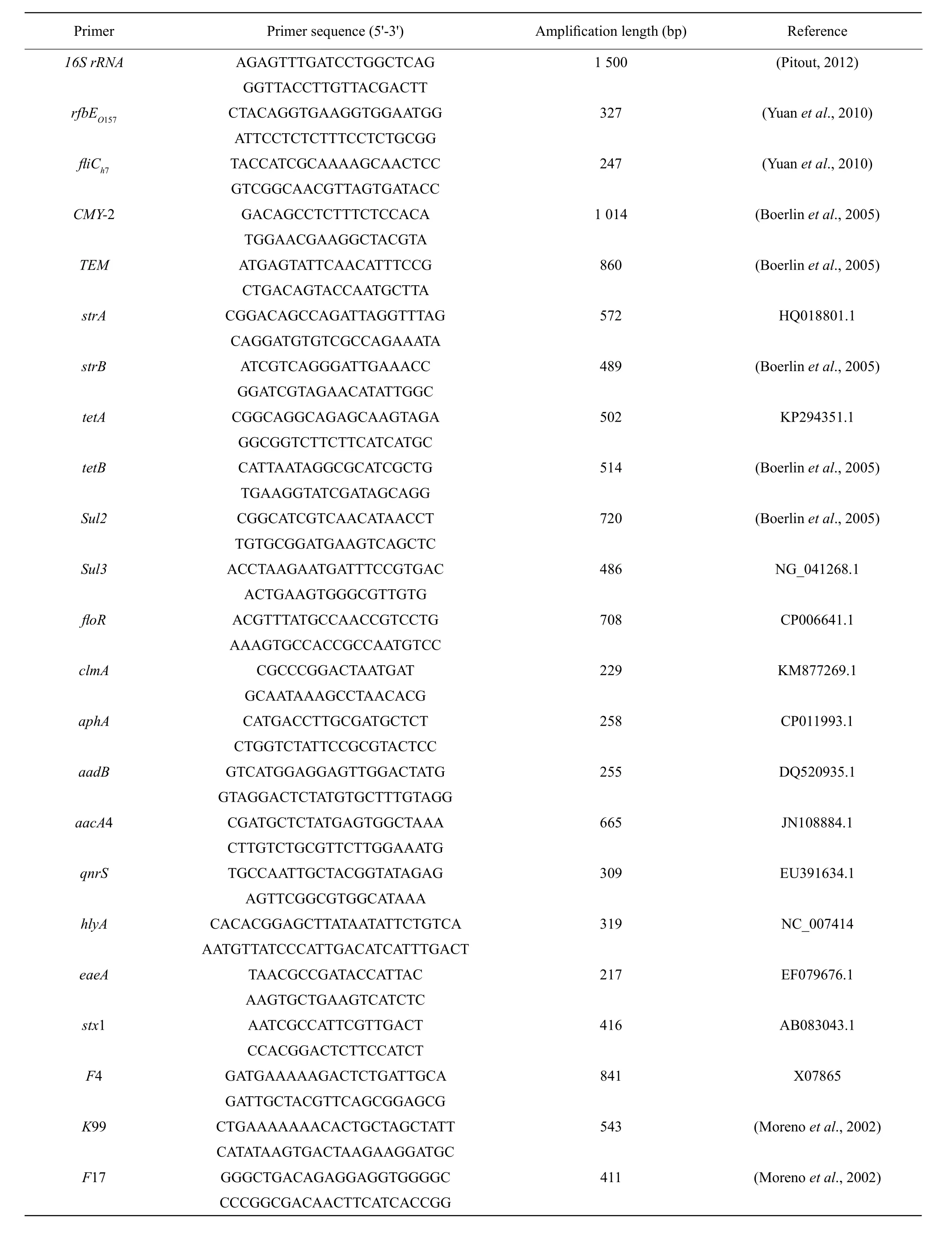
Table 1 Primers of multiplex PCR condition
Antibiotic sensitivity testing
Antimicrobial susceptibility testing (AST) was performed using the Mueller-Hinton agar diffusion method (Reller et al., 2009). Bacteria were cultured in BHI medium at 37℃ for 10 h under constant shaking (220 r • min-1). Bacterial suspend in sterile saline (pH 7.4) was compared to 0.5 McFarland standards solutions, and then was spread evenly on the surface of Mueller-Hinton agar using a sterile cotton swab. Then, agar plates were incubated at 37℃ for 18 h. Ampicillin, penicillin, cefoperazone,amikacin, erythromycin, cefalothin, carbenicillin,nitrofurantoin, tetracycline, trimethoprim, cefotaxime, gentamicin, streptomycin, kanamycin, chloramphenicol, ceftazi-dime, rifampicin, tobramycin,ciprofloxacin and ceftriaxone (all purchased from Harbin Pharmaceutical Group Co., Harbin, China)were evaluated.
Virulence test of E. coli O157 : H7 strain
Specific pathogen free (SPF) mice (Kunming mice,five weeks of age, female) were purchased from the Chinese Academy of Agricultural Sciences of Harbin Veterinary Research Institute. Mice were housed in sterile environment and fed with sterilized food and water. Thirty-six mice (weight: 20±5 g female mice)were randomly divided into six groups. Each group was injected intraperitoneally with 0.5 mL of 1×1010,1×109, 1×108, 1×107and 1×106CFU • mL-1bacteria.The control group was administered the same volume of saline. Clinical symptoms and mortality of the mice were recorded daily, and the half lethal dose (LD50)was calculated, according to the following formula:LD50=log-1[Xm–i(Pm–0.5)]. Xm, represented the maximum dose of logarithm; i, represented the group interval and the two groups were adjacent to the log difference; Pm, represented the sum of the mortality of each group.
Pathogenicity
SPF mice werefirst fed with water containing 50 μg • mL-1nalidixic acid for two days, then fasted for two days before intraperitoneal injection of 0.5 mL E. coli O157 : H7 (1×1010CFU • mL-1). The control group was injected with the same volume of saline and were sacrificed 24 h after injection, their major organs were collected for histological examination. Collected organs werefixed with 4% formaldehyde in PBS for 24 h, dehydrated and embedded in paraffin. Sections(4 μm) were cut using a microtome (Leitz 1512), then stained with hematoxylin and eosin (HE) and observed using a light microscope (Nikon, Tokyo, Japan).
Statistical analysis
The data provided represented at least two independent experiments. Statistical analysis was conducted by ANOVA (PROC MIXED) and the significance difference was determined using least squares (SAS Statistical Analysis Software, version 8.0, SAS Institute, Cary, N.C.). p<0.05 was considered statistically significant.
Results
lsolation and identification of E. coli O157 : H7
The newborn calves showed symptoms of hemorrhagic pleurisy or peritonitis and intestinal hemorrhage,calves with severe diarrhea appeared dehydration,nonambulatory and asthenia universalis, suggesting E. coli infection. Colonies produced by bacterial culture were mostly clear and colorless, further indicating that they were E. coli O157 : H7. When grown on eosin methylene blue plates, colonies exhibited a dark purple metallic luster. Gram staining was negative (Fig. 1), and biochemical tests (Table 2)also showed that strains were E. coli, according to Berger's Manual of Determinative Bacteriology (Boone et al., 2005).
To further confirm that the isolate was indeed E. coli, the experiment used 16S rRNA primers(Table 1) to amplify rRNA gene. Sequencing revealed up to 99% homology with E. coli 16S rRNA sequence included in GenBank. Next, using E. coli CICC 21530 as a positive control, rfb EO157gene and fli CH7gene were detected by PCR. Result indicated that the isolate contained the same bands as CICC 21530(Fig. 2).
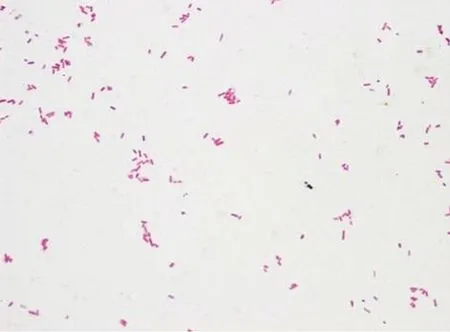
Fig. 1 Bacterial gram staining results (×1 000)
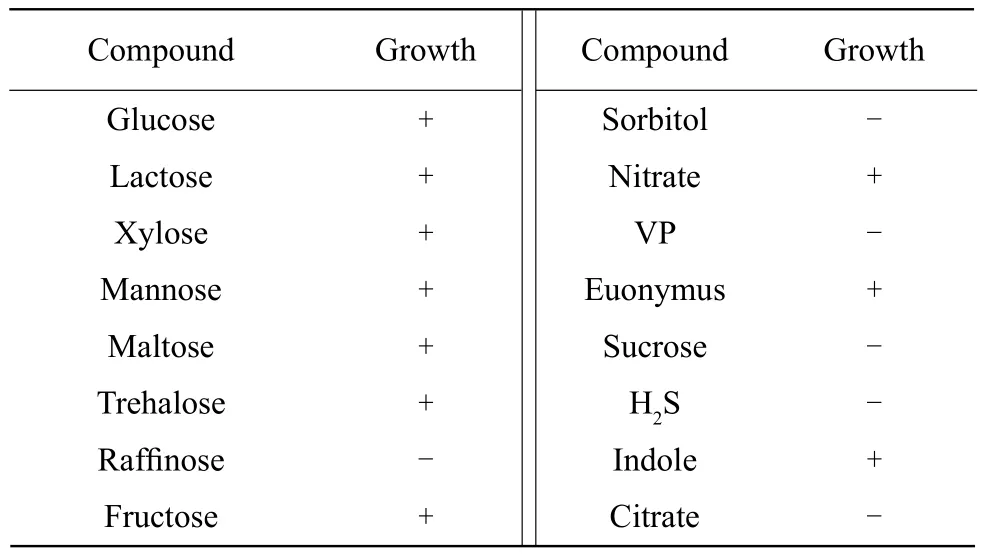
Table 2 Biochemical test of isolate

Fig. 2 PCR test results of isolated plant specific gene rfb E. coli O157 and fli CH7
Antibiotic sensitivity
Table 3 showed the separation sensitivity to various antibiotics. Among the 20 antibiotics tested, isolates were sensitive to eight antibiotics, but not to 12 antibiotics.
Resistance and virulence genes
PCR was used to detect resistance and virulence genes. The isolate contained six of the fourteen tested resistance genes, including tetB, strB, aadB, aphA,floR and TEM, while did not contain qnrs, sul3,CMY-2, clmA, strA, aacA4, sul2 and tetA (Fig. 3).In addition, the isolate contained all the three of the virulence genes for stx1, eaeA and hlyA (Fig. 4).

Table 3 Results of E. coli O157 : H7 for antibiotics sensitivity

Fig. 3 PCR amplification of E. coli O157 : H7 resistance genes

Fig. 4 Identification of stx1, eaeA and hlyA genes by PCR
E. coli O157 : H7 isolate virulence
The experiment used SPF mice further investigated the virulence of E. coli O157 : H7 isolate. SPF mice were intraperitoneally injected with 0.5 mL of difference concentrations of the isolate. Mice administered 1×1010, 1×109and 1×108CFU • mL-1E. coli O157 : H7 began to die after 18 h. Mice injected with 1×107CFU • mL-1started to die at 48 h. No saline-injected mice died in this period (Table 4). The calculated median lethal dose of E. coli O157 : H7 was 7.9×107CFU • mL-1.
Pathogenicity
After 7 h of intraperitoneal injection of 0.5 mL(1×1010CFU • mL-1), the systemic symptoms induced by E. coli O157 : H7 were observed, including lethargy,anorexia, dehydration, hair disorder and shortness of breath, and all the six mice were administered 1×1010CFU • mL-1E. coli O157 : H7 died within 18 h.No abnormal symptoms were observed in salineadministered mice. In the necropsy of dead mice,swelling, or bleeding of the lungs, heart and kidneys were observed; yellow mucous in the small intestine mucosa and intestinal mucosal congestion in the intestine were expanded.
Histological evaluation
Histopathological examination of the heart revealed fractured myocardialfibers, ruptured muscle cells,interstitial hyperemia and venous congestion (Fig. 5A).Liver sections were fragile with granulation (Fig. 5B).Nuclear fragmentation, a sign of cell necrosis, and red cell infiltration of the spleen could also be found(Fig. 5C). No apparent changes were observed in mice administered saline (Fig. 5D-F).

Table 4 Mortality of mice administered E. coli O157 : H7 by intraperitoneal injection

Fig. 5 HE staining showing histopathological changes in cardiac (A, D), liver (B, E) and spleen (C, F)
Degeneration of lung bronchiolar epithelial cells was clearly observed (Fig. 6A). Glomerular epithelial cells were swollen, ruptured and degenerated, but no necrosis (Fig. 6B). Shedding duodenal epithelial cells and goblet cells were destroyed and cytoplasmic degeneration was reduced (Fig. 6C). No pathological changes were observed in saline-administered mice(Fig. 6D-F).

Fig. 6 HE staining of histopathological changes in lung (A, D), kidney (B, E) and intestine (C, F)
Discussion
E. coli O157 : H7 was identified as the main cause of severe intestinal disease and hemolytic uremic syndrome (Rabinovitz et al., 2014). In this study,traditional culture methods and PCR were used to isolate and characterize E. coli O157 : H7 strain from infected calves in northeast China. PCR had become a valuable tool for investigation of virulence genes. In this study, PCR confirmd the presence of the virulence genes stx1, stx2, eaeA and hlyA and it was found that the isolated strain carried stx1+hlyA+eaeA virulence genes. It had previously been shown that stx1 producing strains were closely associated with HUS.
Several virulence genes had been linked with pathogenicity of EPEC. These genes were frequently encoded by pathogenic islands and other mobile DNA islands (Mathers et al., 2015). Pathogenicity depended not only on the presence of virulence-associated genes,but also on their expression levels, which could vary between pathogenic and non-pathogenic isolates.Consistent with other reports (Bosilevac et al., 2014),it could be found the expression of several virulence genes, including stx1, eaeA and hlyA in this isolate.
Excessive use of antibiotics had contributed to emergence and spread of antibiotic-resistant strains. In China, for example, several antibiotics,such as penicillin, ampicillin and tetracycline, were commonly used in the production of veterinary and animal husbandry to promote disease prevention or promote growth (Zhang et al., 2015). E. coli O157 : H7 isolates were found to be resistant to these three antibiotics through experiments, and were different from the strain isolated in India. It was reported that this strain was mainly resistant to amikacin, amoxicillin, cefixime and streptomycin, but sensitive to chloramphenicol (Rehman et al., 2014).
E. coli was a large and diverse bacterial population.Most were harmless to mammals, and were naturally found in human intestinal tract, but others could be deadly. In this work, the results showed that E. coli O157 : H7 caused pleurisy in calves and had strong resistance to cephalosporins, tetracycline, rifampicin, cotrimoxazole, ciprofloxacin and erythromycin, but sensitive to cefoperazone, ceftazidime and amino-glycoside antibiotics.
Previous reports also suggested that E. coli could cause severe and potentially lethal diseases characterized by hemorrhagic colitis and hemolytic uremic syndrome (Ferreira et al., 2014). In this experiment, only a low dose of E. coli O157 : H7 was required to cause infection, in accordance with the previous report (Viazis et al., 2011). The high incidence of E. coli in dairy cows posed a threat to public health,since these microorganisms might human contaminate products intended for human consumption, including water, raw and pasteurized milk, meat products, dairy products and plant products (Ferreira et al., 2014).Future researches should focus on the microbiological and ecological factors that affect the growth of E. coli O157 : H7, and better understand the dynamics of transmission and drug resistance.
Conclusions
This study isolated and identified E. coli O157 : H7 from infected calves from a farm in northeast China and characterized the pathogenesis and clinical manifestations of E. coli O157 : H7 infection, which might provide information for future diagnosis and treatment of dairy cows. It also provided a theoretical basis for the prevention and control of drug-resistant E. coli O157 : H7.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Improvement of Temperature Uniformity of Instant Rice Inside Plastic Rectangular Container Under Microwave Reheating
- GIS Based Aroma Characteristic for Regional Distribution of Tobacco Using Kriging Interpolation Method Set in Henan Province of China
- Hepatoprotective Effects of Folium syringae Extracts Against Ethanolinduced Acute Liver Injury
- Porcine Parvovirus Inducing Autophagy to Benefit Its Replication
- Function of RanGAP1 in Mouse Oocyte Fertilization
- FSH Promoting Proliferation of Calf Sertoli Cells Through Wnt/β-catenin Signaling Pathway with CDC25B Being Involved
