Comparison of the Tellgenplex HPV DNA test with the PCR-reverse dot blot assay for human papillomavirus genotyping
2018-03-08YaChaoYaoNanLiLiangShanHuYaHongLiZhiZhang
Ya-Chao Yao, Nan Li, Liang-Shan Hu, Ya-Hong Li, Zhi Zhang
Department of Laboratory Medicine and Central Laboratories, Guangdong Second Provincial General Hospital, Guangzhou 510317, China
1. Introduction
Cervical cancer is the 4thmost common female cancer worldwide.More than 120 different human papillomavirus (HPV) types have been identified, of which have been divided into low-risk (LR)and high-risk (HR) type. LR HPVs are associated with genital warts, whereas HR HPVs are related to invasive cervical cancer[1].Recently increasing evidences show that persistent infection with the same genotype increases the risk of developing cervical cancer[2]. Twelve HR HPVs (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52,56, 58 and 59) are classified as carcinogenic HPV genotypes by the International Agency for Research on Cancer (IARC) and illuminate virtually all of cervical cancers[3]. It is widely known that HPV 16 and 18 are the main HR HPV types discovered in cervical cancer around the world; however, the prevalence of the other HR HPVs display geographical and regional differences[4-6].
HPV testing has already been recommended into cervical cancer screening guidelines in many countries[7], as well as an accessory examination to cytological screening such as cervical Papanicolau(Pap) smear. HPV testing alone can help to triage curative effect in women with minor cytological abnormalities and cervical intraepithelial neoplasia[8].
Therefore, several molecular technologies are now available for detecting HPV genotyping such as hybridization with RNA probes, DNA chip, PCR, and sequencing[9-11]. The tests approved by the U.S. Food and Drug Administration (FDA) for detecting the HPV DNA are the hybrid capture 2 system (Digene Corporation,Gaithersburg, Md) and the Roche Cobas HPV test (Roche Molecular Diagnostics, Pleasanton, CA)[12,13]. The hybrid capture 2 assay can detect 13 HR HPVs (type 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58,59, and 68); however, it doesn’t recognize individual HPV types.The Roche Cobas HPV test, based on the real-time PCR method,can detect 14 types of HR HPVs (type 16, 18, 31, 33, 35, 39, 45,51, 52, 56, 58, 59, 66, and 68) with only providing HPV16/HPV18 genotypes. Whatever their merits, the two assays provide limited genotyping information.
Nowadays a lot of commercial kits are available to identify HR and LR HPVs in cervical cytology specimens in China[14-17]. Among them, the Tellgenplex HPV DNA test and the polymerase chain reaction-reverse dot blot (PCR-RDB) assay were the representative HPV genotyping assays which were widely used[14,18,19]. The main drawbacks of the PCR-RDB assay are time consuming. Meanwhile,the Tellgenplex HPV DNA test, based on flow cytometry fluorescence hybridization method, only takes less than four hours during the detection. Moreover, the Tellgenplex HPV DNA test has the advantage of being automated, and it can detect more HPV genotypes. In our study, we first evaluated the performance of the Tellgenplex HPV DNA test compared to the PCR-RDB assay for the molecular genotyping of HPV.
2. Materials and methods
2.1. Study population
Cervical samples were collected with cervical swabs from women undergoing routine cervical cancer screening by the gynecological practitioners between June 2016 and November 2016 in the Guangdong Second Provincial General Hospital. The study was approved by the Hospital Ethics Committee of Guangdong Second Provincial General Hospital. The population eligible for this study was randomly selected including 60 women (age range: 25-63 years old) with positive results by the SNIPER HR HPV assay (Genetel Pharmaceuticals Ltd. Shenzhen, China)[16]. All patients provided informed consent.
2.2. DNA isolation
The cervical swabs were placed in a standard transport medium and were oscillated with a swirl. Then, 1 mL of this suspension was used as described using the SNIPER assay according to the recommendation of the manufacturer. Then, 2 μL of HPV DNA used for the SNIPER™ HR HPV assay as described in the text. The rest 1 mL of suspension was used to extract HPV DNA using the Tellgenplex 26 HPV genotyping panel nuclear acid detection kit(TELLGEN Life Science Co. Ltd., Shanghai, China). The extraction HPV DNA was utilized for the Tellgenplex HPV DNA test and the PCR-RDB assay.
2.3. Tellgenplex HPV DNA test
The 26 HPV Genotyping Panel kit (TELLGEN Life Science Co.Ltd., Shanghai, China), based on the multiplex technique, was performed according to the manufacturer’s instructions for HPV genotyping. The HPV DNA was amplified by using biotin-labeled PCR primers. Then the PCR products are hybridized to series of beads with coated HPV type-specific probes. At the end, beads are read on a Luminex 200 system (Luminex Corporation, Texas). This assay detects 17 HR HPVs (HPV 16, 18, 26, 31, 33, 35, 39, 45, 51,52, 53, 56, 58, 59, 66, 68, and 82), and 10 LR HPVs (HPV 6, 11, 40,42, 43, 44, 55, 61, 81, and 83).
2.4. PCR-RDB assay
The PCR-RDB assay (Yaneng Biotech, Shenzhen, China) can distinguish 18 HR HPVs (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 53,56, 58, 59, 66, 68, 73, 82, and 83) and 5 LR HPVs (HPV 6, 11, 42,43, and 81). The L1 HPV PGMY09/PGMY11 primer pair was used to amplify the extracted HPV DNA or positive or negative control.The HPV DNA was amplified in a thermal cycler with the following conditions: The pre-denaturation and denaturation was performed at 50 ℃ for 15 min, and 95 ℃ for 10 min, followed by 40 cycles each at 94 ℃ for 10 s, 45 ℃ for 90 s, 72 ℃ for 30 s. After amplification,hybridization and RDB on the strips fixed with 23 different typespecific probes were processed for HPV genotyping. The positive result was judged by the blue spots on the membrane by the naked eye.
2.5. Sequencing
Each Sample showing discrepancy was genotyped using sequencing. HPV L1 region (LG life Science, Seoul, Korea) is internal standards represented by nonhomologous DNA fragments with primer templates that are recognized by MY/GP primers. PCR amplification was performed using Takara PCR Thermal Cycler Dice(Takara Bio Inc., Shiga, Japan). Then, 5 μL of extracted template DNA were used as template. Then QIAquick PCR purification kit(Qiagen, Hilden, Germany) was utilized for the purification of PCR products. Subsequent the Applied Biosystems (ABI) 3730XL DNA analyzer (Life Technologies Co., Carlsbad, CA, USA) was used to read the HPV DNA regions. The specific HPV genotype was aligned with the Basic Local Alignment Search Tool (BLAST) database.
2.6. Statistical analysis
Only the genotypes detected by both assays were considered for comparison. SPSS for Windows, version 19.0 (SPSS Inc., Chicago,IL) were used for statistical analyses.P<0.05 was considered statistically significant. The concordance between the results of two different tests was estimated by the agreement rate, kappa coefficient,proportion of positive agreement (Ppos), proportion of negative agreement (Pneg), and McNemar’sPvalue (in cases of more than ten positive results)P<0.05 was considered statistically significant.Ppos, was calculated as twice the number of agreed positives/(total number of specimens + number of agreed positives-number of agreed negatives); and Pnegwas calculated as twice the number of agreed negatives / (total number of specimens-number of agreed positives + number of agreed negatives). And we also calculated the relative sensitivity and specificity of the two assays. The sensitivity of the Tellgenplex HPV DNA test relative to that of the PCR-RDB assay is the proportion of the Tellgenplex HPV DNA test-positive samples among those that are the PCR-RDB assay-positive, and the specificity of the Tellgenplex HPV DNA test relative to that of the PCR-RDB assay is the proportion of the Tellgenplex HPV DNA test-negative samples to the number of the PCR-RDB assay-negative samples, and vice versa.
3. Results
3.1. Identification of HPV genotypes by the Tellgenplex HPV DNA test and the PCR-RDB assay
Of the 62 selected samples, two were excluded due to negative results by both assays. In the remaining 60 samples, four samples were positive by only the PCR-RDB assay and three samples were positive by only the Tellgenplex HPV DNA test. The results of each assay are summarized in Table 1. The Tellgenplex HPV DNA test and the PCR-RDB assay detected multiple HPV genotypes in 25 (41.67%) and 32 (53.33%) samples, respectively. The numbers of genotypes per sample were 1.79 and 2.09, respectively. Among them, the PCR-RDB assay detected more HPV genotypes compared to the Tellgenplex HPV DNA test.
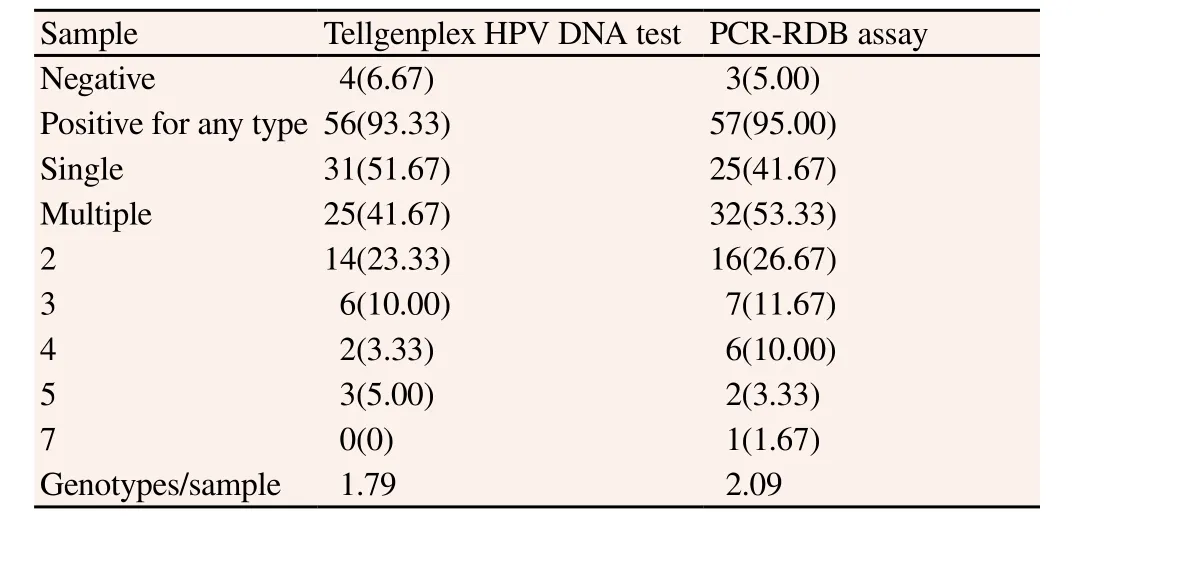
Table 1Identification of genotypes by the Tellgenplex HPV DNA test andPCR-RDB assay [n(%)].
3.2. Individual genotype agreement between the Tellgenplex HPV DNA test and the PCR-RDB assay
Only the HPV genotypes detected by both assays including HPV genotypes 6, 11, 16, 18, 31, 33, 35, 39, 42, 43, 45, 51, 52, 53,56, 58, 59, 66, 68, 81, and 82 were considered for comparison. A comparison of the genotyping results of the two assays is shown in Table 2. The agreement rates of the two assays ranged from 83.3%to 100%. Analyzing kappa values showed that the two assays had almost perfect agreement (>0.8) for HR HPV genotypes 35, 39, 45,53, 56, 59, 66, 68, and 82, and substantial agreement (0.61-0.80) forHPV genotypes 16, 18, 33, 43, 52, and 58. The detection rates of the two assays for frequent HPV genotypes (16, 35, 39, 45, 52, 53,58, 59, 66, and 82) were not statistically different, but the PCR-RDB assay showed higher detection rates than the Tellgenplex HPV DNA test for HPV genotypes 81 (P<0.05).

Table 2Agreement between the Tellgenplex HPV DNA test and the PCR-RDB assay.
3.3. Relative sensitivity and specificity of individual genotypes
The sensitivities and specificities for the four HPV genotypes (16,52, 58, and 81) of the Tellgenplex HPV DNA test relative to the PCR-RDB assay ranged from 50.0% to 88.2% and 90.7% to 100%,respectively. The sensitivities and specificities for the four HPV genotypes (16, 52, 58, and 81) of the Tellgenplex HPV DNA test relative to the PCR-RDB assay ranged from 78.9% to 100.0% and 82.0% to 92.2%, respectively (Table 3).

Table 3Agreement between Tellgenplex HPV DNA test and PCR-RDB assay.
3.4. HPV genotypes detected by only the Tellgenplex HPV DNA test
The Tellgenplex HPV DNA test could detect additional HPV genotypes compared to the PCR-RDB assay, including HPV genotypes 44 and 61. All HPV genotypes that could be detected by only the Tellgenplex HPV DNA test were confirmed by sequencing;HPV genotypes 44 (n=1), and 61 (n=1).
3.5. Analysis of discrepancy between the Tellgenplex HPV DNA test and the PCR-RDB assay
The discordant results between the Tellgenplex HPV DNA test and the PCR-RDB assay are summarized in Table 4. Most of the 84 types in 25 specimens showing discrepancy were observed in cases of infection by multiple HPV genotypes. According to the HPV genotypes determined by sequencing analysis, the Tellgenplex HPV DNA test yielded more false-negative results for various HPV genotypes, including HPV genotypes 16 (n=4), 31 (n=4), 52 (n=5),and 81 (n=9). The PCR-RDB assay yielded more false-negative results for HPV genotypes 58 (n=4).

Table 4Discordant results of Tellgenplex HPV DNA test and PCR-RDB assay.
4. Discussion
In some hospitals in China, HPV DNA was routinely tested by the Tellgenplex HPV DNA test or the PCR-RDB assay[14,18,19].The Tellgenplex HPV DNA test, which based on flow cytometry fluorescence hybridization method, has been utilized for simultaneous genotyping and can detect 17 HR HPV genotypes and 10 LR HPV genotypes. Meanwhile, the PCR-RDB assay allows for genotyping of 17 HR HPV genotypes and 6 LR HPV genotypes.However, the main drawbacks of the PCR-RDB assay are time consuming. In the contrast, the Tellgenplex HPV DNA test has the advantage of being automated, compared with the PCR-RDB assay,and can be completed in less than 4 h. We decided to identify a better assay which will be widely used to monitor disease progression and to examine the impact of widespread vaccination on prevalent HPV types in the future.
Overall, there was a good agreement between the Tellgenplex HPV DNA test and the PCR-RDB assay in this study. The agreement rates of the two assays were high, ranging from 83.3% to 100.0%.Analyzing k values showed that the two assays had almost perfect agreement (>0.8) for HR HPV genotypes 35, 39, 45, 53, 56, 59,66, 68, and 82, and substantial agreement (0.61-0.80) for HPV genotypes 16, 18, 33, 43, 52, and 58. In a previous study, 10 442 women were evaluated using the liquid-based cytology (Thinprep cytologic test, TCT) and the PCR-RDB assay. There was 99.2%concordance between HPV PCR-RDB testing and sequencing[14].Previous study has reported that a total coincidence of 90.5%between the Tellgenplex HPV DNA test and the HC II was determined with a kappa value of 0.88[19].
The detection rates of the two assays for frequent HPV genotypes(16, 35, 39, 45, 52, 53, 58, 59, 66, and 82) were not statistically different. For HPV genotypes 81, significant differences were observed (P<0.05), likely due to the higher limit of detection of the Tellgenplex HPV DNA test compared to the PCR-RDB assay.
Most discordant results were likely due to co-infection of multiple HPV genotypes. The Tellgenplex HPV DNA test and the RCRRDB assay detected multiple HPV genotypes in 25 (41.67%) and 32 (53.33%) samples, respectively, and the numbers of genotypes per sample were 1.79 and 2.09, respectively. Previous study have reported that there is limited concordance for infection of multiple HPV genotypes[10,12,13]. The selective amplification of one HPV DNA over another occur when multiple HPV genotypes are present.In this study, sensitivity and specificity were calculated relative to one another. For HR HPV genotypes 16, 18, 31, and 33, sensitivities of the Tellgenplex HPV DNA test relative to the RCR-RDB assay were 66.7%, 100.0%, 42.9%, and 57.1%, respectively, and specificities of the Tellgenplex HPV DNA test relative to the RCRRDB assay ranged from 97.9% to 100.0%. Our results indicate that the PCR-RDB assay showed better relative sensitivity and specificity than the Tellgenplex HPV DNA test. It is known that DNA sequencing is the “gold standard” method for HPV genotyping. The Tellgenplex HPV DNA test yielded more false-negative results in this study, likely due to co-infection of multiple HPV genotypes or low viral load. The limit of detection of the Tellgenplex HPV DNA test was higher than those of the PCR-RDB assay for most HPV genotypes. On the other hand, in HPV genotypes 40, the PCR-RDB assay yielded more false-negative results. Each limit of detection of the PCR-RDB assay was greater than 1 000 copies, which was higher than those of the Tellgenplex HPV DNA test (20 copies,according to the instructions provided by the manufacturer). The Tellgenplex HPV DNA test yielded false-negative results in four cases each for HPV genotypes 16 and 31, and the PCR-RDB assay yielded a false-negative result in one case for HPV genotype 18.Overall, for the HR HPV genotypes 16, 18, and 33, the Tellgenplex HPV DNA test and PCR-RDB assays showed substantial agreement;for the HR HPV genotype 31, they showed moderate agreement.
The potential benefits of using the HPV genotyping assay for primary cervical cancer screening are increasingly apparent[20].There currently had no study to evaluate the Tellgenplex HPV DNA test and the PCR-RDB assay for detecting the HPV genotyping. We selected samples with HPV-positive results by the SNIPER HR-HPV assay and then directly compared the results of the Tellgenplex HPV DNA test and the PCR-RDB assay.
In conclusion, the Tellgenplex HPV DNA test shows substantial agreement with the PCR-RDB assay. However, the Tellgenplex HPV DNA test had lower detection rates than the PCR-RDB assay for HPV genotypes 42, 73, and 81. The study showed that the PCRRDB assay which could detect more multiple HPV genotypes in one sample showed more sensitivity and specificity than the Tellgenplex HPV DNA test, which makes it a useful and valid alternative for HPV genotyping in clinical laboratories.
Conflict of interest statement
The authors have declared that they have no conflict of interest.
This work was supported by the National Nature Science Foundation of China, Grant Number: 81400639; and the Science Foundation for Youth Scientists of the Second People’s Hospital of Guangdong Province of China, Grant Number: YQ2015-002.
[1] Cricca M, Marasco E, Alessandrini F, Fazio C, Prossomariti A,Savini C, et al. High-throughput genotyping of high-risk human papillomavirus by MALDI-TOF mass spectrometry-based method.New Microbiol2015; 38(2): 211-223.
[2] Brown B. High HPV prevalence and need for ancillary care.Braz J Infect Dis2015; 19(1): 110.
[3] Mendes D, Bains I, Vanni T, Jit M. Systematic review of modelbased cervical screening evaluations.BMC Cancer2015; 15:334.
[4] Lin H, Moh JS, Ou YC, Shen SY, Tsai YM, ChangChien CC, et al. A simple method for the detection and genotyping of high-risk human papillomavirus using seminested polymerase chain reaction and reverse hybridization.Gynecol Oncol2005; 96(1):84-91.
[5] Carozzi F, Bisanzi S, Sani C, Zappa M, Cecchini S, Ciatto S, et al. Agreement between the AMPLICOR human papillomavirus test and the hybrid capture 2 assay in detection of high-risk human papillomavirus and diagnosis of biopsy-confirmed highgrade cervical disease.J Clin Microbiol2007; 45(2): 364-369.
[6] Ejegod DM, Rebolj M, Bonde J. Comparison of analytical and clinical performance of CLART HPV2 genotyping assay to linear array and hybrid capture 2: A split-sample study.BMC Cancer2015; 15: 216.
[7] Saslow D, Runowicz CD, Solomon D, Moscicki AB, Smith RA,Eyre HJ, et al. American cancer society guideline for the early detection of cervical neoplasia and cancer.J Low Genit Tract Dis2003; 7(2): 67-86.
[8] Yu S, Kwon MJ, Lee EH, Park H, Woo HY. Comparison of clinical performances among Roche Cobas HPV, RFMP HPV PapilloTyper and hybrid capture 2 assays for detection of highrisk types of human papillomavirus.J Med Virol2015; 87(9):1587-1593.
[9] Kim KH, Yoon MS, Na YJ, Park CS, Oh MR, Moon WC.Development and evaluation of a highly sensitive human papillomavirus genotyping DNA chip.Gynecol Oncol2006;100(1): 38-43.
[10] Lin CY. Evaluation of using composite HPV genotyping assay results to monitor human papillomavirus infection burden through simulation.BMC Infect Dis2015; 15: 123.
[11] Chung HS, Lee M. Comparison of the AdvanSure HPV GenoBlot assay with the INNO-LiPA HPV genotyping assay for human papillomavirus genotyping.J Clin Virol2014; 60(1): 34-38.
[12] Khunamornpong S, Settakorn J, Sukpan K, Srisomboon J,Suprasert P, Siriaunkgul S. Performance of HPV DNA testing with hybrid capture 2 in triaging women with minor cervical cytologic abnormalities (ASC-US/LSIL) in Northern Thailand.Asian Pac J Cancer Prev2014; 15(24): 10961-10966.
[13] Arbyn M, Roelens J, Cuschieri K, Cuzick J, Szarewski A,Ratnam S, et al. The APTIMA HPV assay versus the hybrid capture 2 test in triage of women with ASC-US or LSIL cervical cytology: A meta-analysis of the diagnostic accuracy.Int J Cancer2013; 132(1): 101-108.
[14] Sun P, Song Y, Ruan G, Mao X, Kang Y, Dong B, et al. Clinical validation of the PCR-reverse dot blot human papillomavirus genotyping test in cervical lesions from Chinese women in the Fujian province: A hospital-based population study.J Gynecol Oncol2017; 28(5): e50.
[15] Zhang L, Dai Y, Chen J, Hong L, Liu Y, Ke Q, et al. Comparison of the performance in detection of HPV infections between the high-risk HPV genotyping real time PCR and the PCR-reverse dot blot assays.J Med Virol2017; 90(1): 177-183.
[16] Belinson SE, Wulan N, Li R, Zhang W, Rong X, Zhu Y, et al.SNIPER: A novel assay for human papillomavirus testing among women in Guizhou, China.Int J Gynecol Cancer2010; 20(6):1006-1010.
[17] Zeng Z, Yang H, Li Z, He X, Griffith CC, Chen X, et al.Prevalence and genotype distribution of HPV infection in China:Analysis of 51,345 HPV genotyping results from China’s Largest CAP Certified Laboratory.J Cancer2016; 7(9): 1037-1043.
[18] Wang R, Guo XL, Wisman GB, Schuuring E, Wang WF, Zeng ZY, et al. Nationwide prevalence of human papillomavirus infection and viral genotype distribution in 37 cities in China.BMC Infect Dis2015; 15: 257.
[19] Zheng B, Li Z, Griffith CC, Yan S, Chen C, Ding X, et al. Prior high-risk HPV testing and Pap test results for 427 invasive cervical cancers in China’s largest CAP-certified laboratory.Cancer Cytopathol2015; 123(7): 428-434.
[20] Cornall AM, Poljak M, Garland SM, Phillips S, Tan JH,Machalek DA, et al. Anyplex II HPV28 detection and Anyplex II HPV HR detection assays are highly concordant with other commercial assays for detection of high-risk HPV genotypes in women with high grade cervical abnormalities.Eur J Clin Microbiol Infect Dis2017; 36(3): 545-551.
杂志排行
Asian Pacific Journal of Tropical Medicine的其它文章
- A hiddenly high hepatitis C virus related liver disease burden among Chinese patients with non-liver disease complaints: A hospital based study from 2013 to 2017
- Visceral leishmaniasis: An immunological viewpoint on asymptomatic infections and post kala azar dermal leishmaniasis
- In vitro fungistatic activity of 36 traditional oriental medicines and their synergistic effect against Trichophyton rubrume
- Antidiabetic effects of galactomannans from Adenanthera pavonina L. in streptozotocin-induced diabetic mice
- Syzygium aromaticum ethanol extract reduces AlCl3-induced neurotoxicity in mice brain through regulation of amyloid precursor protein and oxidative stress gene expression
- Phlebotomus (Adlerius) kabulensis (Diptera: Psychodidae) a new record sand fly species from Iran: Morphological and molecular aspects
