Characterization of GhSERK2 and its expression associated with somatic embryogenesis and hormones level in Upland cotton
2018-03-07LlUZhengjieZHAOYanpengZENGLingheZHANGYuanWANGYumeiHUAJinping
LlU Zheng-jie, ZHAO Yan-peng, ZENG Ling-he, ZHANG Yuan, WANG Yu-mei, HUA Jin-ping
1 College of Agronomy and Biotechnology, China Agricultural University/Key Laboratory of Crop Heterosis and Utilization, Ministry of Education/Beijing Key Laboratory of Crop Genetic Improvement, Beijing 100193, P.R.China
2 United States Department of Agriculture- Agricultural Research Service (USDA-ARS), Crop Genetics Research Unit, Stoneville,MS 38776, USA
3 Research Institute of Cash Crop, Hubei Academy of Agricultural Sciences, Wuhan 430064, P.R.China
1.lntroduction
The effective regeneration system is a useful platform for genetic program, and the SE is the main factor influence the plant propagation.Hence, it’s vital for understanding the mechanism of SE during tissue culture of plants.The induction and growth of somatic embryos in vitro were first reported in carrot (Steward et al.1958).Somatic embryos have been widely used as a substitute model in studies of genetic transformation and zygotic embryos (Yang and Zhang 2010).However, the molecular mechanism of SE is not well understood.In the process of SE, many physiological, biochemical, and morphological changes occurred during development of the induced tissues(Sakhanokho et al.2001), and many factors such as hormones, developmental stages, and nutrition regime could influence SE.Previous research indicated that the process of SE was always accompanied by alteration of levels and patterns of genes expression (Wu et al.2009),especially for the differential expression genes involved in calli and SE induction.In Arabidopsis, the gene CLAVATA1 involved in the acquisition of embryogenic competence acted as a negative regulator (Stone et al.1998).In contrast, many genes acted as positive regulators with ectopic expression in the plant recipient for increasing embryogenesis competence of the cell without additional external hormones.The examples of these genes can be the genes of BABY BOOM (Boutilier et al.2002), WUSCHEL(Zuo et al.2002), LEAFY COTYLEDON 1 (LEC1) (Lotan et al.1998), LEC2 (Stone et al.2001), and SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 (SERK1) (Hecht et al.2001; Mahdavi-Darvari et al.2015).
A substantial amount of work has been conducted to analyze the SERK genes for their roles involved in the process of SE.SERKs are a subgroup of leucine-rich repeats and receptor-like kinases (LRR-RLKs) (Ma et al.2012a, b) that were first isolated as genes involving in signal transduction in embryonic cell development (Schmidt et al.1997).After that,the SERK homologs were isolated from both monocotyledons(Somleva et al.2000; Baudino et al.2001; Hu et al.2005;Bhumica et al.2008; Cueva et al.2012), and dicotyledons(Hecht et al.2001; Thomas et al.2004; Santos et al.2005;Schellenbaum et al.2008; Sharma et al.2008; Nolan et al.2009; Pérez-Núñez et al.2009; Hedayat et al.2010; Huang et al.2010; Yang et al.2011; Ma et al.2012b; Shi et al.2012,2014; Ahmadi et al.2016; Rocha et al.2016).SERKs located in membranes and these homologs were responsible for the cellular signaling and metabolic regulation, which have an important role in the acquisition of embryogenic competence in plant cells (Albrecht et al.2005).SERKs were shown to be characteristic markers of embryogenic cell cultures and somatic embryos in carrot, Dactylis glomerata, and Arabidopsis (Schmidt et al.1997; Somleva et al.2000; Hecht et al.2001).The expression of SERK was correlated with induction of SE and shoot organogenesis in M.truncatula and sunflower, and the status of SERK gene expression might influence the cell fate in organogenesis or SE (Thomas et al.2004; Nolan et al.2009).The transcripts of TaSERK genes could be induced in zygotic and somatic tissues, and SE was associated with high SERK expression, which could be auxin-enhanced and calcium-dependent in Triticum aestivum(Bhumica et al.2008).It was also reported that CiSERK was an essential gene for SE in cacao (Theobroma cacao)because its transcripts could only be detected in somatic embryos, but not in the simply proliferated callus (Santos et al.2005, 2009).In other studies, the expression of SERK suggested multi-roles for SERK homologs in plants and was detected not only in embryogenic cells, but also in different tissues such as the apical meristem, root, and leaves in different plant species (Thomas et al.2004; Albrecht et al.2005; Hu et al.2005; Santos et al.2005; Nolan et al.2009; Shi et al.2012, 2014; Pandey and Chaudhary 2014).AtSERK1 and AtSERK2 showed the redundancy by T-DNA mutational insertions and male sporogenesis in Arabidopsis because the double mutant of the two SERK homologs was sterile due to lack of mature pollen grains (Albrecht et al.2005).SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 (SERK1) and SERK2 were proved to interact and phosphorylate with EXCESS MICROSPOROCYTES1 (EMS1) in the anther development of Arabidopsis (Li et al.2016).GhSERK1,a SERK homolog in Gossypium hirsutum belonging to the Somatic Embryogenesis Receptor Kinase (SERK) family,showed variable levels in different tissues and was identified to play an important role during anther development (Shi et al.2012, 2014).However, GhSERK1 also supposed to modulate cellular redox levels to enhance the regeneration potential in cotton (Pandey and Chaudhary 2014).GhSERK3, another SERK homolog in G.hirsutum has been submitted to NCBI(accession no.AEG25668), but its functions have yet to determine.The SERK homolog also functions as a broader signal in response to biotic stress (Hu et al.2005; Santos et al.2009) or abiotic stress (Pandey and Chaudhary 2014; Ma et al.2016).Hormones level is vital for embryogenesis and somatic embryos development in plant regeneration (Jiménez et al.2005; Fraga et al.2016; Wu et al.2017), but there are no reports described the relationship clearly between SERK2 gene expression and the levels of hormones.
Upland cotton (G.hirsutum L.) is one of the species with most technical obstacles for in vitro regeneration.Hormone combination is one of the most important factors influencing embryogenesis and regeneration in cotton.It is vital for us to have a better understanding of the physiological and molecular mechanisms of SE in order to increase the efficiency of plant regeneration in cotton.GhSERK1 was first cloned in Upland cotton and was identified to influence anther formation and cotton regeneration (Shi et al.2012,2014; Pandey and Chaudhary 2014).The objectives of this study were to clone a novel G.hirsutum putative homolog of SERK (GhSERK2) and determine its expression pattern in different tissues as well as in vitro SE.The study was also designed to investigate the expression profiles of GhSERK2 gene in cultures treated with hormones during SE, and to detect hormone levels in calli of GhSERK2 transgenic plants in order to determine the relationships between GhSERK2 gene expression and the level of hormones.
2.Materials and methods
2.1.Plant materials and calli induction
Upland cotton cv.Coker 201 was planted at experimental field of China Agricultural University in Beijing under normal cultivation conditions.The hypocotyls were taken from 5 to 7 days old dark-grown sterile seedlings and placed in Murashige and Skoog (MS) medium (1962) containing 0.1 mg L-12,4-dichlorophenoxyacetic acid (2,4-D), 0.1 mg L-1indole-3-acetic acid (IAA), and 0.1 mg L-1kinetin (KT)for non-embryogenic calli (EC) initiation and proliferation.Subsequently, the loose and prolifically grown non-EC were picked and placed in medium supplemented with 0.3 mg L-1indole-3-butyric acid (IBA) and 0.1 mg L-1KT, as well as 0.5 g L-1glutamine and 0.5 g L-1asparagine for EC induction.The EC with high proliferation rate were selected and transferred to the suspension media for further treatments.
2.2.Material treatments and sampling
The different organs including ovaries, anthers, sepals,petals, ovules, roots, stems, and leaves from Coker 201 cultivated in field were collected and stored at -80°C for DNA and RNA extraction.The non-EC and EC were also sampled during callus formation.The samples of EC were treated with different hormones including 2,4-D, IBA, and KT at 0, 2, 4, 7, 14, and 21 days of suspension culture.The calli cultured without any hormone treatment were used as the negative control.The total RNA was extracted from all of the treated samples and their negative controls for determining the GhSERK2 expression level by semi-quantitative RTPCR or quantitative real-time PCR.
2.3.DNA, RNA isolation, and first-strand cDNA synthesis
Genomic DNA was extracted from young leaves of Coker 201 according to Paterson et al.(1993) and treated with RNase A to eliminate the presence of RNA.The total RNA was extracted using cetyltrimethyl ammonium bromidepolyvinyl pyrrolidone (CTAB-PVP) method (Zhang X et al.2011), and the first-strand cDNA was synthesized and finished using Qiagen Reverse Transcription Kit (Co: 70042,Germany) after gDNA digestion.
2.4.cDNA and gDNA isolation of GhSERK2
The putative full-length cDNA of GhSERK2 was identified by blasting NCBI expressed sequence tags (EST) database of G.hirsutum using AtSERK1 cDNA and amplified by RT-PCR from Coker 201 cDNA using primers P1F and P1R (Appendix A).The gDNA of GhSERK2 was also amplified using P1F and P1R primers, but could not be sequenced until the second intron.Thus, the eight couples of primers (P2-P9,Appendix A) designed for the conserved SERK exons were employed to amplify the remaining intron regions.The full length of genomic sequence of GhSERK2 was obtained after assembling all of the exon and intron regions.
2.5.Bioinformatics analysis of the sequences
Sequences were assembled, aligned, and analyzed using DNAMAN and Clustal software tools.The ORF Finder Program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to predict the open reading frame (ORF) of GhSERK2.Conserved Domains Database (CDD) database was used for searching for the conserved motifs of the predicted protein sequence (Zhang and Bryant 2009).The signal peptide and transmembrane region (TM) were predicted by using the ‘SignalP 3.0 Server’ tool (http://www.cbs.dtu.dk/servicesSignalP/) and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html), respectively.Multiple sequence alignment was performed using ClustalX.Phylogenetic trees were constructed using the MEGA 4 Program.
2.6.Semi-quantitive RT-PCR and quantitative realtime PCR analysis
To examine the GhSERK1 expression profiles in different tissues or organs, semi-quantitative RT-PCR and quantitative RT-PCR were performed.For semi-quantitative RT-PCR,cDNA samples were normalized using a pair of primers(P10F and P10R, Appendix A) designed for a G.hirsutum actin (GhActin) (Zhang X et al.2011).RT-PCR was performed using the normalized cDNA samples and the gene-specific primers P11F and P11R (Appendix A), under non-saturating PCR condition: initial denaturation at 94°C for 5 min; 22 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 1 min; followed by final extension at 72°C for 10 min.The obtained fragments were migrated together with a molecular marker in 0.8% agarose gel and visualized under UV light.The quantitative RT-PCR analyses of all cDNA samples were performed using SYBR Premix EX TaqTM(TaKaRa,Japan) with an ABI 7500 Real-Time PCR System (Bio-Rad Corporation, USA).The PCR for GhSERK2 gene was conducted using the following 25 μL reaction mixture: 12.5 μL Real-Time PCR Master Mix, 2 μL first-strand cDNA sample,1 μL gene-specific forward and reverse primers (P13F and P13R, Appendix A), and 9.5 μL nuclease-free water.The amplification parameters were as follows: 95°C for 10 min, 1 cycle; 95°C for 5 s, 60°C for 34 s, 40 cycles; 95°C for 15 s, 60°C for 60 s, 95°C for 5 s, 40 cycles.RT-qPCR expression levels of GhSERK2 were normalized to the ubiquitin housekeeping gene level for each tissue (P12F and P12R, Appendix A).Threshold (Ct) values in the analyses were based on three technical replicates for each biological sample.Standard deviation was calculated from the three biological replicates.
2.7.Expression vector construction and transformation in cotton
The ORF of GhSERK2 was amplified from Coker 201 cDNA using specific primers P14F and P14R (Appendix A).After purification, the fragment was digested with XbaI and SacI,and then linked with vector p2301M, which was digested with the same enzymes to form the expression vector p2301MSERK2 (Liu et al.2011).Then the vector p2301M-SERK2 was verified by double digestion and sequencing, and transformation of p2301M-SERK2 using cultivar Xu 244 as the recipient using pollen tube pathway method.The recipient plants were cultivated in an isolated field to avoid Bt cotton cross-pollination.The flowers pollinated 36 to 48 h were selected.A total of 10 μL plasmid DNA (0.01-0.02 μg μL-1) in water solution was injected into ovules, and the bolls injected with DNA were tagged.A total of 153 transformed bolls were obtained, and T1seeds were harvested.All T1seeds were planted in field, and the transformants were selected by kanamycin (2 g L-1) during cotyledon and three leaves stages.A total of 65 candidate T1plants with kanamycin resistance were obtained.
DNA was extracted from leaves of kanamycin resistant T1transformants for PCR and Southern blotting identification.Primers P14F and P14R (Appendix A) were designed based on gene sequence of neomycin phosphotransferase (NPTII)for PCR identification of transgenic plants.
2.8.DNA gel blot analysis
The genome DNA samples from leaves of transgenic and wild type (used as negative control) plants after PCR verification were purified and used for Southern blot with the PCR-amplified NPTII gene fragment as the probe.DIG-labeled DNA probe synthesis and Southern blot analysis were carried out using the DIG-High Prime DNA Labeling and Detection Starter Kit I according to the following protocol.Approximately 20 μg of total genomic DNA was digested with BamHI (NEB), separated by electrophoresis in 0.8% agarose gel including 25 ng of D15 000 plus DNA marker, and transferred to the nylon membrane (Hybond-N+;Amersham).PCR-amplified GhSERK2 fragment was used as a probe, and the hybridization of DIG-labeled DNA probes was performed at 42°C for 24 h.The blot nylon membrane was washed under high stringency with 2×SSC (sodium citrate buffer solution), 0.1% sodium dodecyl sulfate (SDS)twice for 5 min at room temperature, 0.5×SSC, and 0.1%SDS twice for 15 min at 68°C.After high stringency washing,the nylon membrane was dipped into 25 mL washing buffer for 2 min and treated with 10 mL blocking buffer for 30 min.After that, the blot nylon membrane was treated with antibody solution for 30 min and detection buffer for 3 min at room temperature.The nylon membrane was then soaked in color dye substrate solution for 15-30 min at room temperature and the results were recorded by photography.
2.9.Enzyme-linked immunosorbent assay (ELlSA)hormone assay
Seeds of T2transgenic lines S1-3-S3-7 as well as wild type Xu 244 (WT) were sterilized in 70% alcohol for 30 s and soaked in 0.1% HgCl2with shaking for 10 min, and then placed on half strength macronutrient MSB medium(MS medium containing B5vitamin) in dark at (28±2)°C for 5 to 7 days to germinate.Hypocotyls from the germinated T3transgenic lines and WT were cut into 5- to 8-mm segments to induce calli in MSB medium supplemented with 0.571 μmol L-1IAA, 0.465 μmol L-1KT, and 0.904 μmol L-12,4-D.Five independent plants were used to induce calli for each of the T2transgenic lines.After 10 days of calli induction, the levels of hormone combinations including IAA, ABA, gibberellin (GA), and BR (brassinolide for detected) were measured in the initial calli induced from GhSERK2 transgenic lines and WT plants using enzymelinked immunosorbent assay (ELISA) as described by Deng et al.(2008).The antibodies were received from Dr.Wang Baomin in College of Agronomy and Biotechnology, China Agricultural University.
2.10.Accession number
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession no.: JF430801.
3.Results
3.1.GhSERK2 isolation and identification
BlastN and TBlastX were employed to isolate the SERK protein coding genes in Upland cotton and NCBI was searched for available cotton cDNA clones or ESTs.Assembling the obtained sequences with the CAP3 software online (http://pbil.univ-lyon1.fr/cap3.php), all cloned fragments formed a single contig sequence of 2 131 bp,which displayed high similarities to those of SERK homologs or SERK-like genes in other species, and was used to design primers for RT-PCR in order to obtain cDNA clones from Coker 201.The total length of the sequence cloned by RT-PCR was 2 054 bp, which contained entire coding sequence and an additional 44 bp 3´-UTR and 147 bp 5´-UTR and verified by BLAST.The corresponding gene was designated as GhSERK2 and deposited in GenBank under accession no.: JF430801.The ORF encodes a protein of 620 amino acids (aa), with an estimated molecular mass of 68.596 kDa and a predicted isoelectric point of 5.43.
3.2.Sequence analysis of GhSERK2 gene
GhSERK2 amino acid sequence was aligned with 12 other SERK proteins reported from monocots and dicots with sequence identity ranging from 75 to 84% (Fig.1).Among them, the AtSERK3 was shared with the highest similarity of 84%.GhSERK2 contained a putative signal peptide followed by five leucine-rich repeats (LRRs), an SPP motif, a TM domain, and a serine/threonine kinase domain, which strongly indicated that GhSERK2 was one of the members of SERK gene family (Fig.2-A).As shown in Fig.2-B, the hydrophobic aa signal peptide located at the GhSERK2 N-terminal, and the peptide contained a possible cleavage site between 29 and 30 aa.Adjacent to signal peptide sequence, a leucine zipper domain of 43 aa was detected, although it was not existed in DcSERK (Schmidt et al.1997).Similar to SERK proteins in other species, there were five LRRs consensus sequences with typical 24 residues existing in GhSERK2 as defined by Hecht et al.(2001).Between the LRR and transmembrane regions there was a 37-aa Pro-rich domain containing a repeat SPP motif with the feature of SERK proteins (Hecht et al.2001).Using the TMHMM-2.0 software, it was found that there was a putative transmembrane domain located between 223 to 264 aa (Fig.2-C).Adjacent to the several basic amino acid residues, there was a conserved intracellular kinase domain containing 11 sub-domains located at the upstream of C-terminal region of the protein.
3.3.GhSERK2 genomic structure and phylogenetic analysis
Using the same primers (P1F&P1R, Appendix A) for amplifying cDNA, the full-length genomic sequence of GhSERK2 was isolated.Eight couples of primers were designed according to the conserved SERK exon regions for amplification of the sub-sequences of the genomic sequence.The sub-sequences were sequenced subsequently.The 6 558-bp full-length genomic sequences were obtained after assembling all sub-sequences (Fig.2-A).
A comparative alignment of the predicted GhSERK2 coding regions with the corresponding genomic sequence revealed that GhSERK2 consisted of 11 exons interspersed by 10 introns (Fig.2-A).Genomic sequences showed that GhSERK1 and GhSERK2 showed the conserved exon and intron structure (Appendix B).Unlike LRR2 and LRR3 shared a single exon, LRR1, LRR4, and LRR5 domains encoded by separate exons (Fig.2-A).The last three exons contained 11 kinase subdomains with the Xth subdomain which was separated by the last intron.This genomic sequence organization of GhSERK2 as described was quite similar with other previously reported SERKs(Hecht et al.2001; Sharma et al.2008).The phylogenetic relationships between GhSERK2 and other SERK proteins were assessed through an unrooted phylogenetic tree(Appendix C), obtained by the phylogenetic analysis of SERK full-length amino acid sequences.GhSERK2 was most related to AaSERK1 based on the phylogenetic relationships.As shown in Appendix B, the phylogenetic tree mainly represented four clades: clades 1 and 2 contained the dicot and monocot species, respectively; clade 3 formed by GhSERK2 and one Araucaria species; the last clade 4 contained AtSERK3/4/5.
3.4.Expression patterns of GhSERK2 gene in different tissues
Semi-quantitative RT-PCR was used to detect the expression of GhSERK2 in samples from different organs including ovary, anther, sepal, petal, ovule, root, stem, and leaves.The results showed that lower expression levels were exhibited in vegetative organs comparing with those in the reproductive organs (Fig.3-A).Similar results were also reported by Qi et al.(2010) in quantitative RT-PCR.The results were further verified by qRT-PCR method (Fig.3-B).The highest expression level of GhSERK2 was detected in ovary followed by the level of petal, whereas the lowest expression level was detected in stem.
3.5.Expression profile of GhSERK2 during embryo germination and development and response to hormone treatment
To identify relationship between GhSERK2 gene expression and SE, the expression levels of GhSERK2 during embryo germination and development were analyzed.As shown in Fig.3-C, the expression level of GhSERK2 increased more than 5-fold in globular embryos, the preliminary stage of embryo germination, than those in non-EC and EC.However, the expression level of GhSERK2 during the embryo development decreased sharply in tissues of torpedo and cotyledon embryos.

Fig.1 Alignment of multiple deduced amino acid sequences of Somatic Embryogenesis Receptor Kinase (SERK) family protein kinases.Identical amino acids were highlighted in black color, and similar ones in gray color.Accession numbers of SERK-homologs are listed as follows: AtSERK1 (Arabidopsis thaliana), NP_177328.1; AtSERK2, AF384969_1; AtSERK3, AF384970_1; AtSERK4,NP_178999; AtSERK5, NP_17900; MtSERK1 (Medicago truncatula), AAN64293.1; GmSERK (Glycine max), ACJ64717.1; CuSERK1(Citrus unshiu), BAD32780.1; ZmSERK1 (Zea mays), NP_001105132.1; ZmSERK2, CAC37641.1; OsSERK1 (Oryza sativa),AAU88198.1; ClSERK (Cyrtochilum loxense), CBV98085.1; AaSERK1 (Araucaria angustifolia), ACY91853.1.LRR, leucine-rich repeat; SPP, signal peptide peptidase; TM, transmembrane.

Fig.2 GhSERK2 sequence analysis.A, the genomic structure of GhSERK2.The boxes in different colors and lines represented the exon-intron organization feature of GhSERK2 gene.P2F&R-P9F&R, the primers position of introns amplification.Bar=1 kb.B, prediction of signal peptide sequence and its confirmatory analysis.C, the transmembrane region prediction.
The expression pattern of GhSERK2 gene in response to different hormones treatment for 3 weeks was analyzed using suspension culture.The EC cultured in suspension medium without hormone treatment was used as a control.The EC re-differentiation began after 14 days’ suspension culture, and embryos formed after 21 days of incubation in all treatments including the control (data not shown).GhSERK2 expressed in all EC treated with IBA after 0, 2,4, 7, 14, and 21 days, and its expression level increased following the time of IBA treatment, but remained unchanged when the EC were treated with KT from 0 to 21 days compared to that in EC treated without hormone (Fig.3-D).The expression level of GhSERK2 increased sharply in EC when treated with 2,4-D for 2 days, but decreased to the original level after 2 days as that in calli without hormone treatment (Fig.3-D).
3.6.Transformation of GhSERK2 and detection of hormones in transgenic cotton
After verification by double digestion and sequencing, the expression vector p2301M-SERK2 was used to transform Xu 244 with pollen tube pathway method.The positive resistant T1plants were tested by PCR amplification of NPT gene (Fig.4-A).RNA samples from immature embryos of T1transformed and WT plants were extracted and used for expression analysis by qRT-PCR, and we found the expression level of GhSERK2 in transgenic plants were higher than that in WT (Fig.5).DNA samples from T1transformed plants were extracted and digested by BamHI(NEB) and then hybridized with NPTII probe for copy number analysis.The copy numbers of the transgenic plants (S1-S5) ranged from 1 to 4, which indicated that T-DNA insertion was randomly integrated in cotton genome (Fig.4-B).
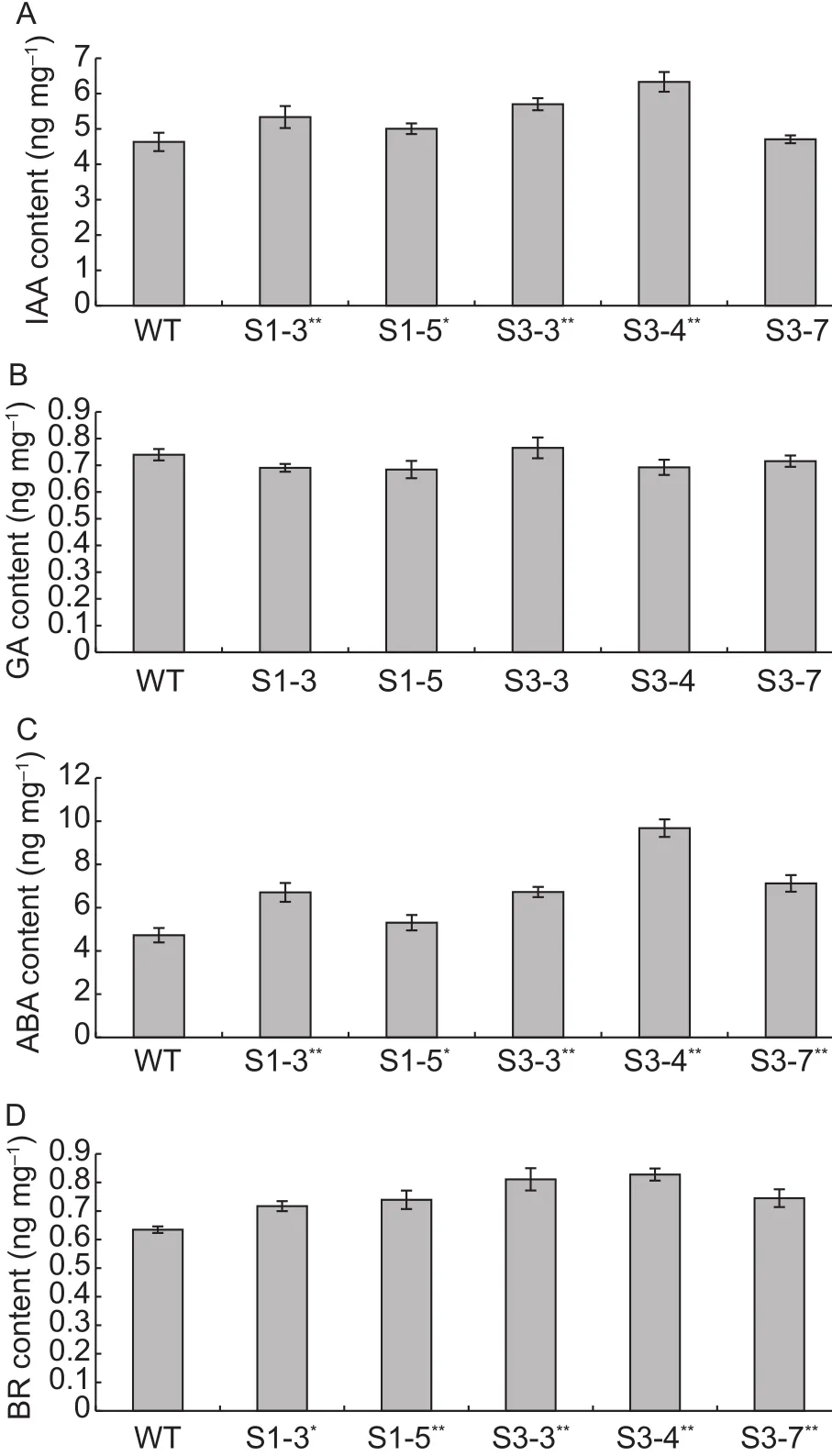
The phenotypic traits including plant height, branch number, the height of primary branch, boll number, and fiber length of transgenic plants were examined, but no change was detected between transgenic and wild type plants(data not shown).In order to investigate the influence of hormone level on over-expression of GhSERK2 in cotton,IAA, ABA, GA, and BR contents in the initial calli induced from hypocotyls of T3transgenic and WT plants were analyzed.As shown in Fig.6, the IAA contents in the initial calli induced from transgenic lines (6.33 ng mg-1in S3-4)were higher than that in the initial calli of the WT (4.63 ng mg-1).Similar results could be found in ABA contents,ranging from 4.72 ng mg-1in WT to 9.67 ng mg-1in S3-4,and in BR, ranging from 0.63 ng mg-1in WT to 0.83 ng mg-1in S3-4.However, there was no obvious difference of GA contents in the calli between the transgenic lines and WT.The IAA content in the transgenic line S3-7 has no obvious discrepancy comparing to WT (Fig.6-A), this result might be due to transgenic position or dosage effect.
The ratios of contents for different hormones, include IAA/GA, IAA/ABA, and IAA/BR, were further analyzed.Interestingly, we found that the ratios of IAA/GA in some transgenic plants were significant higher than that in WT, but in adversely, the ratios of IAA/ABA in transgenic plants were lower than that in WT.AtSERK1 enhanced embryogenic competence in the process of tissue culture (Hecht et al.2001), but needed more work to clarify the relation between expression level of GhSERK2 and hormone contents in transgenic lines, and if over-expression of GhSERK2 couldinfluence the SE in cotton or not.

4.Discussion
Since the first report of SE and regeneration in cotton(Davidonis and Hamilton 1983), a number of protocols for establishing regeneration and transformation systems in cotton via SE had been described (Sakhanokho et al.2001;Kumria et al.2003), and a tremendous amount of information on molecular mechanisms controlling embryogenesis in cotton was available (Wu et al.2009; Hu et al.2011).Considering the genotype limitation for regeneration in cotton tissue culture, a better understanding of the mechanisms of in vitro culture, cloning and characterization of genes related to SE and hormone levels could help us increase embryogenesis in calli to form embryos.In this study, the novel GhSERK2 gene encoding a leucine-repeat receptor protein kinase in Upland cotton was isolated and characterized.The putative amino acid sequence of GhSERK2 indicated that it may function as an LRR-RLK(Walker 1994).RLKs existing in plants had been reported to function as follows.It bound to an extracellular signal molecule and induced receptor dimerization, which brought the intracellular kinase domains of individual monomers into close proximity, caused trans-phosphorylation, and then activated kinase domains for the regulation of cellular response (Becraft 2002).As shown in Fig.1, GhSERK2 showed a high level of sequence and protein domain homology with other SERK proteins according to the multiple deduced amino acid sequence alignments.GhSERK2 was also equipped with the typical SERK protein domain distribution including a signal peptide, five LRRs, an SPPmotif, a TM domain, a serine/threonine kinase domain,and a C-terminal region.The GhSERK1 gene with 78% of sequence similarity to GhSERK2 also belonged to SERK family and played a critical role for fertility during anther formation (Shi et al.2012, 2014).In this study, GhSERK1 and GhSERK2 encoded the putative SERK-like protein of 624 and 620 aa, respectively.The sequence structures of GhSERK1 and GhSERK2 including all of the exons and introns were compared.The sequence lengths of all exons except the exons containing SPP and TM domains were identical (Appendix B).Interestingly, distinct differences existed among all the 10 introns.The sequence differences might result in the discrepancy of function betweenGhSERK1 and GhSERK2.It would be reasonable to expect multiple SERK genes in cotton to be identified since more than two SERK genes had been isolated in other species such as Arabidopsis thaliana (Hecht et al.2001), Zea mays(Zhang S Z et al.2011), Vitis vinifera (Schellenbaum et al.2008), and Medicago truncatula (Nolan et al.2011).Prior researches identified that SERK proteins in cotton genome had 2 to 3 copies (Shi et al.2012, 2014).In this study, we analyzed the copy number of SERK gene family in D subgenomes, the conversed domain of GhSERK2 sequence was blast to G.raimondii chromosome database, and hits could be found in five chromosomes (chr5, chr10, chr11,
chr12, and chr13) with an alignment length more than 200 bp(Appendix D).It was inferred that there were more than 5 copies of SERK protein-encoding genes in cotton genome.Therefore, the current study strongly supported the existence of a SERK family in cotton.


Fig.5 GhSERK2 gene expression level in immature embryos of T1 transformation plants (2-5) and wild type (1).Bars mean standard error.
SERK genes in plants have diverse functions and one associated with somatic and zygotic embryogenesis.DcSERK, first identified SERK gene in plants, was positively related to induction of somatic and zygotic embryogenesis, the transcription could be observed during the pro-embryogenic stage and the globular stage (Schmidt et al.1997).At the early stage of somatic and zygotic development, AtSERK1 gene could also be detected in Arabidopsis (Hecht et al.2001).The expression pattern of orthologous SERK genes in many other plants was similar to that of DcSERK or AtSERK1.Furthermore, it was observed in lettuce (Lactuca sativa L.) that the inhibition of endogenous SERK expression by antisense RNA induced a reduction in the formation of in vitro somatic embryonic structures (Santos et al.2009).In this paper, GhSERK2 expression level was highly induced in globular embryos at early phases of SE, and the expression pattern was consistent with those in other species (Che et al.2006;Talapatra et al.2014; Ahmadi et al.2016; Rajesh et al.2016).It was also found that Ser/Thr protein kinase-like protein might be involved in cellular communication or signal transduction in SE of cotton (Zeng et al.2006).Moreover, SERK expression was initiated in a small group of pro-embryogenic cells in Araucaria (Steiner et al.2012).In summary, SERK gene expression was correlated with SE and could be regarded as a potential molecular marker for embryogenic cell clusters (Santa-Catarina et al.2004).
The relationship between SERK gene expression and hormone level had not been well studied in previous researches.In present study, the expression pattern of GhSERK2 gene in response to hormone treatments during SE was examined in suspension culture.The fresh EC in basal liquid medium containing various hormones were cultured for 3 weeks.Although the growth regulator of 2,4-D was always used as an inducing agent for callus formation and proliferation in many species, it had disadvantages of inhibiting SE and embryo development in cotton tissue culture (Yang and Zhang 2010).In the current study,GhSERK2 gene expression level in cells with 2,4-D treatment was much higher than that of the control during the first 2-days incubation, but the expression level in treated cells decreased sharply relative to the control cells during the culture beyond 2 days.The possible explanation was that 2,4-D could induce the callus proliferation by increasing the expression of GhSERK2 at the early stage of treatment, but inhibit or influence SE and embryos initiation by decreasing GhSERK2 expression during extensive culture although this phenomenon was not observed in other species (Hu et al.2005; Nolan et al.2009; Zhang S Z et al.2011).Another growth regulator, IBA, had been widely used in cotton in vitro culture.In this study, IBA seemed to exert a great influence on the expression of the GhSERK2 gene.After culture in medium with IBA for 4 days, the expression level of GhSERK2 increased sharply and continued for up to 3 weeks.This result implied the existence of an IBA-dependent up-regulation for GhSERK2.Yang and Zhang(2010) also observed the active induction and proliferation of EC in the medium containing IBA during cotton somatic embryogenesis.Thus, it was concluded that IBA stimulated embryogenic cell formation and proliferation by increasing GhSERK2 expression.KT has been described as one of the most important cytokinins used in SE and regeneration of cotton (Yang and Zhang 2010).However, the expression level of GhSERK2 in embryogenic cells induced by kinetin (KT) was not significantly different from that of the control during treated 3 weeks with KT.Therefore, it was hypothesized that the function of KT for the induction of callus embryogenesis and proliferation was not regulated by the transcription of GhSERK2 and the high expression level of GhSERK2 triggered by 2,4-D maintained the calli proliferation, and induced the embryo initiation in the initial phase of SE, but the active induction and proliferation of embryogenic cells were maintained by IBA during in vitro culture.
In order to further investigate the connection between hormone level and GhSERK2 expression, the GhSERK2 gene was over-expressed in cotton, and we found that the over-expression of GhSERK2 increased hormone levels of IAA, ABA, and BR except GA compared with those in WT in the initial calli induced from the hypocotyls of transgenic plants (Fig.6).The results indicated that GhSERK2 could feedback regulate the level of endogenous hormones.The exogenous ABA had been widely used in plants tissue culture to improve the SE development (Li et al.2002;Srinivas et al.2006).Thus, the raised level of ABA detected in transgenic lines might increase the capacity in SE during tissue culture.GhSERK2 might be vital for the exogenous hormone regulation of the endogenous hormone level and the interaction between the exogenous and endogenous hormones during tissue culture of cotton plants.Further analysis of the ratios of different hormone contents reflected that in some transgenic plants, the ratios of IAA/GA were significant higher than that in WT, and the over-expression of GhSERK2 could significantly increase the ratios of IAA/GA in transgenic plants and further increased the ability of calli dedifferentiation.
In our study, a novel SERK-homology was cloned and characterized.The results supported the arguments that GhSERK2 gene expression profile at different developmental stages had a close relationship with embryo induction, and its expression could be induced by 2,4-D and IBA at certain phases of embryogenesis.GhSERK2 mediated acquisition of EC proliferation and embryo initiation needed to be further confirmed.Excepting the function involved in SE, SERK family also participated in the disease resistance responses and BR mediated signal transduction.For example, AtSERK3/BAK1 had been well studied for its role in BR signaling pathway in Arabidopsis (Li et al.2002;Hu et al.2005).Sequence analysis of GhSERK2 showed high sequence similarity and relatively close evolutionary relationship to AtSERK3 (Fig.4), and GhSERK2 may also take part in BR-mediated signal transduction.
5.Conclusion
In present study, a SERK-homolog, designated as GhSERK2, was cloned and characterized.Its expression was associated with the phases of SE and hormone treatments during SE.The over-expression of GhSERK2 influenced the levels of hormones in calli of the transgenic lines which had been reported in other plants.These results suggested that GhSERK2 gene expression was associated with the induction of SE and hormone level during tissue culture.
Acknowledgements
We thank Dr.Wang Baomin (China Agricultural University)for providing antibodies and help in ELISA technology.We thank Dr.Marina Naoumkina (Cotton Fiber Bioscience Research Unit, USDA-ARS-SRRC) for suggestive comments on manuscript.USDA is an equal opportunity provider and employer.This research was supported in part by the National Natural Science Foundation of China(31371666), and a grant from the National Key Specific Program to Hua Jinping (2016ZX08005-003).
Appendicesassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
Ahmadi B, Masoomi-Aladizgeh F, Shariatpanahi M E.2016.Molecular characterization and expression analysis of SERK1 and SERK2 in Brassica napus L.: implication for microspore embryogenesis and plant regeneration.Plant Cell Reports, 35, 185-193.
Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S.2005.The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis.The Plant Cell, 17, 3337-3349.
Baudino S, Hansen S, Brettschneider R, Hecht V R G,Dresselhaus T, Lorz H, Dumas C, Rogowsky P M.2001.Molecular characterization of two novel maize LRR receptor-like kinases, which belong to the SERK gene family.Planta, 213, 1-10.
Becraft P W.2002.Receptor kinase signaling in plant development.Annual Review of Cell and Developmental Biology, 18, 163-192.
Bhumica S, Jitendra P K, Paramjit K.2008.Characterization of three somatic embryogenesis receptor kinase genes from wheat, Triticum aestivum.Plant Cell Reports, 27, 833-843.
Boutilier K, Offringa R, Sharma V K, Kieft H, Ouellet T, Zhang L M, Hattori J, Liu C M, van Lammeren A A M, Miki B L A,Custers J B M, Campagne M M V.2002.Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth.The Plant Cell, 14, 1737-1749.
Che P, Love T M, Frame B R, Wang K, Carriquiry A L, Howell S H.2006.Gene expression patterns during somatic embryo development and germination in maize Hi II callus cultures.Plant Molecular Biology, 62, 1-14.
Cueva A, Concia L, Cella R.2012.Molecular characterization of a Cyrtochilum loxense Somatic Embryogenesis Receptorlike Kinase (SERK) gene expressed during somatic embryogenesis.Plant Cell Reports, 31, 1129-1139.
Davidonis G H, Hamilton R H.1983.Plant regeneration from callus tissue of Gossypium hirsutum L.Plant Science, 32,89-93.
Deng A, Tan W, He S, Liu W, Nan T, Li Z, Wang B, Li Q X.2008.Monoclonal antibody-based enzyme linked immunosorbent assay for the analysis of jasmonates in plants.Journal of Integrative Plant Biology, 50, 1046-1052.
Fraga H P, Vieira L D, Puttkammer C C, Dos Santos H P, Garighan J A, Guerra M P.2016.Glutathione and abscisic acid supplementation influences somatic embryo maturation and hormone endogenous levels during somatic embryogenesis in Podocarpus lambertii Klotzsch ex Endl.Plant Science, 253, 98-106.
Hecht V, Vielle-Calzada J P, Hartog M V, Schmidt E D L,Boutilier K, Grossniklaus U, de Vries S C.2001.The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture.Plant Physiology, 127, 803-816.
Hedayat Z, Bjarne M S, Henrik L, Renate M.2010.Isolation and characterization of four somatic embryogenesis receptorlike kinase (RhSERK) genes from miniature potted rose(Rosa hybrida cv.Linda).Plant Cell Tissue and Organ Culture, 101, 331-338.
Hu H, Xiong L, Yang Y.2005.Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection.Planta, 222,107-117.
Hu L S, Yang X Y, Yuan D J, Zeng F C, Zhang X L.2011.GhHmgB3 deficiency deregulates proliferation and differentiation of cells during somatic embryogenesis in cotton.Plant Biotechnology Journal, 9, 1038-1048.
Huang X, Lu X Y, Zhao J T, Chen J K, Dai X M, Xiao W, Chen Y P, Chen Y F, Huang X L.2010.MaSERK1 gene expression associated with somatic embryogenic competence and disease resistance response in banana (Musa spp.).Plant Molecular Biology Reporter, 28, 309-316.
Jiménez V M, Guevara E, Herrera J, Bangerth F.2005.Evolution of endogenous hormone concentration in embryogenic cultures of carrot during early expression of somatic embryogenesis.Plant Cell Reports, 23, 567-572.
Kumria R, Sunnichan V G, Das D K, Gupta S K, Reddy V S,Bhatnagar R K, Leelavathi S.2003.High frequency somatic embryo production and maturation into normal plants in cotton (Gossypium hirsutum) though metabolic stress.Plant Cell Reports, 21, 635-639.
Li J, Wen J, Lease K A, Doke J T, Tax F E, Walker J C.2002.BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling.Cell, 110, 213-222.
Li Z Y, Wang Y, Huang J, Ahsan N, Biener G, Paprocki J,Thelen J J, Raicu V, Zhao D Z.2016.Two SERK receptorlike kinases interact with EMS1 to control anther cell fate determination.Plant Physiology, 173, 326-337.
Liu Z J, Zhang Y, Wang Y X, Li P B, Su Y, Zhang X, Wang Y M, Hua J P.2011.Construction of seed specific expression vectors and genetic transformation for genes of heteromeric ACCase in Upland cotton.Molecular Plant Breeding, 9,270-277.(in Chinese)
Lotan T, Ohto M, Yee K M, West M A L, Lo R, Kwong R W,Yamagishi K, Fischer R L, Goldberg R B, Harada J J.1998.Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells.Cell, 93,1195-1205.
Ma J, He Y H, Hu Z Y, Kanakala S, Xu W T, Xia J X, Guo C H,Lin S Q, Chen C J, Wu C H, Zhang J L.2016.Histological analysis of somatic embryogenesis in pineapple: AcSERK1 and its expression validation under stress conditions.Journal of Plant Biochemistry and Biotechnology, 25, 49-55.
Ma J, He Y H, Hu Z Y, Xu W T, Xia J X, Guo C H, Lin S Q, Cao L,Chen C J, Wu C H, Zhang J L.2012a.Characterization and expression analysis of AcSERK2, a somatic embryogenesis and stress resistance related gene in pineapple.Gene,500, 115-123.
Ma J, He Y H, Wu C G, Liu H P, Hu Z Y, Sun G M.2012b.Cloning and molecular characterization of a SERK gene transcriptionally induced during somatic embryogenesis in Ananas comosus cv.Shenwan.Plant Molecular Biology Reporter, 30, 195-203.
Mahdavi-Darvari F, Noor N M, Ismanizan I.2015.Epigenetic regulation and gene markers as signals of early somatic embryogenesis.Plant Cell Tissue and Organ Culture, 120,407-422.
Nolan K E, Kurdyukov S, Rose R J.2009.Expression of the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1(SERK1) gene is associated with developmental change in the life cycle of the model legume Medicago truncatula.Journal of Experimental Botany, 60, 1759-1771.
Nolan K E, Kurdyukov S, Rose R J.2011.Characterisation of the legume SERK-NIK gene superfamily including splice variants: implications for development and defence.BMC Plant Biology, 11, 44-60.
Pandey D K, Chaudhary B.2014.Oxidative stress responsive SERK1 gene directs the progression of somatic embryogenesis in cotton (Gossypium hirsutum L.cv.Coker 310).American Journal of Plant Sciences, 5, 80-102.
Paterson A H, Brubaker C L, Wendel J F.1993.A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis.Plant Molecular Biology Reporter, 11, 122-127.
Pérez-Núñez M T, Souza R, Sáenz L.2009.Detection of a SERK-like gene in coconut and analysis of its expression during the formation of embryogenic callus and somatic embryos.Plant Cell Reports, 28, 11-19.
Qi J N, Yu S C, Zhang F L, Shen X Q, Zhao X Y, Yu Y J, Zhang D S.2010.Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in Chinese cabbage (Brassica rapa L.ssp.pekinensis).Plant Molecular Biology Reporter, 28, 597-604.
Rajesh M K, Fayas T P, Naganeeswaran S, Rachana K E,Bhavyashree U, Sajini K K, Karun A.2016.De novo assembly and characterization of global transcriptome of coconut palm (Cocos nucifera L.) embryogenic calli using Illumina paired-end sequencing.Protoplasma, 253,913-928.
Rocha D I, Monte-Bello C C, Aizza L C B, Dornelas M C.2016.A passion fruit putative ortholog of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 gene is expressed throughout the in vitro de novo shoot organogenesis developmental program.Plant Cell Tissue and Organ Culture, 125, 107-117.
Sakhanokho H F, Zipf A, Rajasekaran K, Sukumar S, Govind C S.2001.Induction of highly embryogenic calluses and plant regeneration in Upland (Gossypium hirsutum L.) and pima (Gossypium barbadense L.) cottons.Crop Science,41, 1235-1240.
Santa-Catarina C, Hanai L R, Dornelas M C, Viana A M, Floh E I S.2004.SERK gene homolog expression, polyamines and amino acids associated with somatic embryogenic competence of Ocotea catharinensis Mez.(Lauraceae).Plant Cell Tissue and Organ Culture, 79, 53-61.
Santos M O, Romano E, Vieira L S, Baldoni A B, Aragao F G L.2009.Suppression of SERK gene expression affects fungus tolerance and somatic embryogenesis in transgenic lettuce.Plant Biology, 11, 83-89.
Santos M O, Romano E, Yotoko K S C, Tinoco M L P, Dias B B A, Aragao F J L.2005.Characterisation of the cacao somatic embryogenesis receptor-like kinase (SERK) gene expressed during somatic embryogenesis.Plant Science,168, 723-729.
Schellenbaum P, Jacques A, Maillot P, Bertsch C, Mazet F,Farine S, Walter B.2008.Characterization of VvSERK1,VvSERK2, VvSERK3, and VvL1L genes and their expression during somatic embryogenesis of grapevine(Vitis vinifera L.).Plant Cell Reports, 27, 1799-1809.
Schmidt E D L, Guzzo F, Toonen M A J, deVries S C.1997.A leucine rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos.Development, 124, 2049-2062.
Sharma S K, Millam S, Hein I, Bryan G J.2008.Cloning and molecular characterization of a potato SERK gene transcriptionally induced during initiation of somatic embryogenesis.Planta, 228, 319-330.
Shi Y L, Guo S D, Zhang R, Meng Z G, Ren M Z.2014.The role of Somatic Embryogenesis Receptor-like Kinase 1 in controlling pollen production of the Gossypium anther.Molecular Biology Reports, 41, 411-422.
Shi Y L, Zhang R, Wu X P, Meng Z G, Guo S D.2012.Cloning and characterization of a Somatic Embryogenesis Receptorlike Kinase gene in cotton (Gossypium hirsutum).Journal of Integrative Agriculture, 11, 898-909.
Somleva M N, Schmidt E D L, de Vries S C.2000.Embryogenic cells in Dactylis glomerata L.(Poaceae) explants identified by cell tracking and by SERK expression.Plant Cell Reports,19, 718-726.
Srinivas L, Ganapathi T R, Suprasanna P, Bapat V A.2006.Desiccation and ABA treatment improves conversion of somatic embryos to plantlets in banana (Musa spp.)cv.Rasthali (AAB).Indian Journal of Biotechnology, 5,521-526.
Steiner N, Santa-Catarina C, Guerra M P, Cutri L, Dornelas M C, Floh E I S.2012.A gymnosperm homolog of SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE-1 (SERK1)is expressed during somatic embryogenesis.Plant Cell Tissue and Organ Culture, 109, 41-50.
Steward F C, Mapes M O, Mears K.1958.Growth and organized development of cultured cells: II.Organization in cultures grown from freely suspended cells.American Journal of Botany, 45, 705-708.
Stone J M, Trotochaud A E, Walker J C, Clark S E.1998.Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interaction.Plant Physiology, 117, 1217-1225.
Stone S L, Kwong L W, Yee K M, Pelletier J, Lepiniec L,Fischer R L, Goldberg R B, Harada J J.2001.LEAFY COTYLEDON2 encodes B3 domain transcription factor that induces embryo development.Proceedings of the National Academy of Sciences of the United States of America, 98,11806-11811.
Talapatra S, Ghoshal N, Raychaudhuri S S.2014.Molecular characterization, modeling and expression analysis of a somatic embryogenesis receptor kinase (SERK) gene in Momordica charantia L.during somatic embryogenesis.Plant Cell Tissue and Organ Culture, 116, 271-283.
Thomas C, Meyer D, Himber C, Steinmetz A.2004.Spatial expression of a sunflower SERK gene during induction of somatic embryogenesis and shoot organogenesis.Plant Physiology and Biochemistry, 42, 35-42.
Walker J C.1994.Structure and function of the receptor-like protein kinases of higher plants.Plant Molecular Biology,26, 1599-1609.
Wu X M, Li F G, Zhang C J, Liu C L, Zhang X Y.2009.Differential gene expression of cotton cultivar CCRI24 during somatic embryogenesis.Journal of Plant Physiology,166, 1275-1283.
Wu Y, Dor E, Hershenhorn J.2017.Strigolactones affect tomato hormone profile and somatic embryogenesis.Planta, 245,583-594.
Yang C, Zhao T J, Yu D Y, Gai J Y.2011.Isolation and functional characterization of a SERK gene from soybean(Glycine max (L.) Merr.).Plant Molecular Biology Reporter,29, 334-344.
Yang X Y, Zhang X L.2010.Regulation of somatic embryogenesis in higher plants.Critical Reviews in Plant Science, 29, 36-57.
Zeng F C, Zhang X L, Zhu L F, Tu L L, Guo X P, Nie Y C.2006.Isolation and characterization of genes associated to cotton somatic embryogenesis by suppression subtractive hybridization and macroarray.Plant Molecular Biology,60, 167-183.
Zhang N, Bryant S.2009.CDD: Specific functional annotation with the Conserved Domain Database.Nucleic Acids Research, 37, 205-210.
Zhang S Z, Liu X, Lin Y, Xie G, Fu F, Liu H, Wang J, Gao S,Lan H, Rong T.2011.Characterization of a ZmSERK gene and its relationship to somatic embryogenesis in a maize culture.Plant Cell Tissue and Organ Culture, 105, 29-37.Zhang X, Zhen J B, Li Z H, Kang D M, Yang Y M, Kong J, Hua J P.2011.Expression profile of early responsive genes under salt stress in upland cotton (Gossypium hirsutum L.).Plant Molecular Biology Reporter, 29, 626-637.
Zuo J, Niu Q W, Frugis G, Chua N H.2002.The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis.The Plant Journal, 30, 349-359.
杂志排行
Journal of Integrative Agriculture的其它文章
- Characteristic analysis of tetra-resistant genetically modified rice
- A wheat gene TaSAP17-D encoding an AN1/AN1 zinc finger protein improves salt stress tolerance in transgenic Arabidopsis
- GmNAC15 overexpression in hairy roots enhances salt tolerance in soybean
- Molecular cloning and functional characterization of a soybean GmGMP1 gene reveals its involvement in ascorbic acid biosynthesis and multiple abiotic stress tolerance in transgenic plants
- Responses of the antioxidant system to fluroxypyr in foxtail millet(Setaria italica L.) at the seedling stage
- Light interception and radiation use efficiency response to tridimensional uniform sowing in winter wheat
