A wheat gene TaSAP17-D encoding an AN1/AN1 zinc finger protein improves salt stress tolerance in transgenic Arabidopsis
2018-03-07XUQiaofangMAOXinguoWANGYixueWANGJingyiXIYajunJlNGRuilian
XU Qiao-fang, MAO Xin-guo, WANG Yi-xue, WANG Jing-yi, XI Ya-jun JlNG Rui-lian
1 College of Agronomy, Northwest A&F University, Yangling 712100, P.R.China
2 National Key Facility for Crop Gene Resources and Genetic Improvement/Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, P.R.China
1.lntroduction
As sessile organisms, plants are vulnerable to various abiotic stresses, such as drought, salinity, extreme temperature,high light intensity, and heavy metals.Crop production and productivity deteriorates under these adverse environmental conditions (Knight and Knight 2001).One important way to improve tolerances to multiple stresses is to overexpress transcription factor gene(s) that are able to control multiple genes from various pathways (Kasuga et al.1999; Shi et al.2015) or by overexpressing genes responsible for abiotic signal perception and transduction (Zhang et al.2012;Chakraborty et al.2015).The zinc finger protein (ZFP) is a multifunction protein that acts as a transcription factor,RNA-binding protein, as well as a protein modification enzyme protecting against environmental stresses (Jan et al.2013; Hichri et al.2014; Zhang et al.2014; Baek et al.2015).Stress-associated protein (SAP) is a group of ZFPs composed of two zinc-finger domains, an N-terminal A20 domain and/or a C-terminal AN1 domain.
The SAP gene family has been identified in eukaryotes and is well-characterized in animals (Gilles et al.2011).In plants, the first SAP gene OsiSAP1 isolated from rice and encodes an A20/AN1 ZFP induced by multiple abiotic stresses (Mukhopadhyay et al.2004).Since then, SAPs have been identified in many plant species, including tomato, maize, Arabidopsis, Aeluropus littpralis, Populus trichocarpa, Solanum lycopersicum, and Medicago truncatula (Vij and Tyagi 2008; Ben Saad et al.2010; Gilles et al.2011).The majority of SAPs are involved in abiotic stress response and several of them improve tolerances to biotic and abiotic stresses (Giri et al.2013).The AtSAP5 and OsSAP7 proteins have been shown to possess E3 ligase activity and increased drought tolerance (Kang et al.2013;Sharma et al.2015).In addition, AtSAP12 can function as a redox sensor by changing its oligomeric conformation depending upon the cellular redox potential (Ströher et al.2009).Plants transformed with the MusaSAP1 gene have demonstrated strengthened defense mechanisms against biotic stresses by upregulating polyphenol oxidase (PPO)at the transcriptional level (Sreedharan et al.2012).The TaSAP1 and TaSAP2 genes are found to be involved in multiple abiotic stress response, including drought, high salinity, cold, and exogenous abscisic acid (Wang 2011).Six haplotypes (Hap) of TaSAP1-A1 have been identified in 300 wheat accessions, among them TaSAP1-A1-HapIII is significantly associated with thousand-grain weight in multiple growing environments (Chang et al.2013).
Accumulating evidence shows that SAPs involved in various environmental stresses responses, while their underlying molecular and physiological mechanisms are poorly understood.Wheat is one of the most important staple crops yet only two SAPs (A20/AN1 domain) have been characterized (Wang 2011; Chang et al.2013).In this research, we identified a typical AN1/AN1 type SAP gene,TaSAP17, which is highly conserved in wheat nucleotide and amino acid sequences.The transcript of TaSAP17-D was induced by various environmental stresses.Overexpression of TaSAP17-D altered the expression patterns of stress responsive genes and led to strengthened salt stress tolerance in Arabidopsis.
2.Materials and methods
2.1.Plant materials, growth and treatment conditions
Wheat variety Hanxuan 10 that is known for its remarkable drought and salt tolerance was used in this study.For abiotic stress treatments, 11-d-old seedlings (at the two-leaf stage)grown in 1/2 Murashige and Skoog (MS) liquid culture at 23/18°C in a 16/8 h (light/dark) photoperiod were subjected to the following treatments: polyethylene glycol-6000 (16.1%PEG, -0.5 MPa), 250 mmol L-1NaCl, low temperature(4°C), and 50 μmol L-1abscisic acid (ABA).Leaf samples were harvested at 0.5, 1, 1.5, 2, 3, 6, 12, 24, 48, and 72 h.To investigate the expression of target genes at different developmental stages in wheat (germination, seedling,and heading stages), different tissues including plumule,root base, root, leaf, internode, flag leaf, stem, node, and depth of root at 0-30, 30-50, 50-70, 70-90, and 90-100 cm were collected.
Two wheat genotypes, the salt-tolerant variety Taishan 23 and the salt sensitive variety Nongda 20074, were selected to detect the expression patterns of TaSAP17-D under salt stress conditions (Peng et al.2017).
Arabidopsis thaliana ecotype Col-0 was used in the transgenic experiment.Arabidopsis plants were grown in the chamber at 23/18°C with a 16/8 h (light/dark) photoperiod and 70% humidity.
2.2.Real-time quantitative PCR
Real-time quantitative PCR (qRT-PCR) was performed to determine gene expression.The TaActin (forward primer,5´-CTCCCTCACAACAACAACCGC-3´; reverse primer,5´-TACCAGGAACTTCCATACCAAC-3´) and AtACTIN2(forward primer, 5´-AGCACTTGCACCAAGCAGCATG-3´;reverse primer, 5´-ACGATTCCTGGACCTGCCTCATC-3´)were used as an internal control to quantify the relative transcript level of wheat and Arabidopsis target genes,respectively.The qRT-PCR was performed in triplicate with a Roche LightCycler 96 Real-Time PCR System(Roche, Switzerland) using the SYBR Green PCR Master Mix Kit (TaKaRa, Japan).Thermal cycling conditions were pre-incubated at 95°C for 2 min, followed by 95°C for 10 s,60°C for 30 s, and 72°C for 30 s for 45 cycles.The relative transcription level for each gene was calculated using the 2-ΔΔCTmethod (Schmittgen and Livak 2008).
2.3.lsolation of the full-length cDNA of TaSAP17-D
The full-length cDNA of TaSAP17-D was amplified using primers TaSAP17 (forward primer, 5´-ATGGGCACG CCGGAGTTCC-3´; reverse primer, 5´-TTATGCTTTT GAAGTTCCTC-3´), and the PCR products were ligated with pEASY-Blunt vectors (TransGen, Beijing, China).The positive clones were selected after transformation, and then sequenced with an ABI 3730XL DNA Analyzer (Life Tech, USA).
2.4.Phylogenetic analyses
To understand the relationship of TaSAP17 and other SAP members, the amino acid sequence of TaSAP17 was used as a query in a BLASTX search to gather sequences with high similarity.The putative sequences with higher similarity were downloaded from NCBI for further analysis.Sequence alignments and comparisons were implemented by the MegAlign Program in DNAStar and DNAMAN.The neighbor-joining tree was built with 1 000 bootstrap replicates by MEGA 5.05 (Tamura et al.2011).
2.5.Generation of transgenic Arabidopsis
Full-length cDNA sequence of TaSAP17-D was introduced into XbaI and HindIII sites (forward primer,5´-TGCTCTAGAATGGGCACGCCGGAGTTCC-3´; reverse primer, 5´-CCCAAGCTTTTATGCTTTTGAAGTTCCTC-3´)of the pCAMBIA1300-35S vector.The construct was introduced into Agrobacterium tumefaciens strain (GV3101) and then transformed and random inserted into Arabidopsis by floral infiltration (Clough and Bent 1998).
2.6.Subcellular localization of TaSAP17-D protein
The fusion construct (pCAMBIA1300-TaSAP17-D-GFP) and control (pCAMBIA1300-GFP) were transformed into tobacco leaves by Agrobacterium tumefaciens (GV3101)-mediated transformation (Li et al.2016).After an incubation at 25°C for 2 d (16/8 h light/dark photoperiod), fluorescence signals were examined using a laser scanning confocal microscope(Leica TCSNT, Germany).GFP auto-fluorescence was collected in the range of 500-570 nm wavelengths.The chloroplast auto-fluorescence was measured in the wavelength ranged from 630 to 700 nm.
2.7.Salt treatment of TaSAP17-D transgenic plants
To probe the potential effects of TaSAP17-D overexpression in Arabidopsis, three T3homozygous lines were randomly selected for phenotyping.
To investigate the salt tolerance of transgenic plants,7-d-old seedlings were moved to MS medium with 0 or 200 mmol L-1NaCl for 5 d and then observed the phenotypes.
Germination assays were performed on MS medium supplemented with 0 and 150 mmol L-1NaCl.Eighty seeds were sown on medium, and the plates were placed at 4°C for 2 d in the dark, and then transferred to growth chambers at 23/18°C with a 16/8 h (light/dark) photoperiod and 70%humidity.The germination rate was calculated at the 4th and 6th d, respectively.
To investigate the physiological characterization of TaSAP17-D transgenic Arabidopsis, plants were cultured in soil for 5 wk, and then treated with 250 mmol L-1NaCl solution to measure abiotic stress-related physiological indices at the 4th d after treatment.These measurements included cell membrane stability (CMS), free proline content,osmotic potential (OP), chlorophyll content, and water loss rate.CMS, OP, chlorophyll content, and water loss rates of Arabidopsis of TaSAP17-D transgenic and WT plants were measured by methods as described by Mao et al.(2010).Free proline was extracted and quantified from fresh tissues of seedlings (0.5 g) as reported by Hu et al.(1992).
2.8.Expression analysis of the salt-related genes
Salt tolerance assays were conducted at the seedling stage.Arabidopsis seedlings were cultured as aforementioned.Five-week-old seedlings were irrigated with NaCl solution(250 mmol L-1) from the bottom of pots.When the soil was completely saturated with salt solution, NaCl solution was removed.The plants were cultured under normal condition for 2-d and the expression of salt responsive genes were analyzed by qRT-PCR.Specific primers were designed based on the conserved regions of genes (Appendix A).
2.9.Statistical analysis
All of the experiments were conducted with three replications.The data were presented as the mean±SD, and analyzed by Student’s t-test in a two-tailed analysis to compare the parameters obtained under normal and salt stress conditions.A P-value of <0.05 or <0.01 was considered to be statistically significant and highly significant, respectively.
3.Results
3.1.lsolation and sequence analysis of TaSAP17s
To obtain SAP candidate gene family members in wheat, the conserved sequences of the A20/AN1 domain were used to comprehensively screen the draft genomes of wheat putative diploid progenitors Triticum uratrtu (A genome) (Ling et al.2013) and Aegilops tauschii (D genome) (Jia et al.2013) via the HMMER of Hidden Markov model (HMM).The repetitive sequences were removed by comparing with previously obtained sequences (Han B T 2015, personal communication).The putative TaSAP17s have two AN1 domains and two conserved C2H2 motifs, structures specific to the SAP gene superfamily.Three copies of TaSAP17,originated from the A, B and D genomes, were cloned in common wheat.The three copies all have the same gene structure, each containing one intron and two exons.Sequence analysis showed that both TaSAP17-A and TaSAP17-D encode a protein containing 287 amino acids(AAs), five AAs more than TaSAP17-B.The sequence similarities of AA and cDNA in the three copies were as high as 99.03 and 98.35%, respectively.Since the three TaSAP17s showed extremely high similarities in nucleotide and amino acid sequence, the D genomic member, TaSAP17-D, was selected for further functional analysis.
To analyze the phylogenetic relationship, the putative amino acid sequences of TaSAP17s and members of SAPs in other plant species were used to construct the phylogenetic tree.Two types of SAP, class A20/AN1 and class AN1/AN1 were identified from plant species.Each type contained members from both monocots and eudicots,suggesting that the origination of SAP genes occurred before the divergence of monocots and eudicots.The deduced amino acid sequence of TaSAP17s showed high homology with the counterparts in Oryza sativa, Cynara cardunculus var.scolymus, Solanum lycopersicum, Glycine soja, and Medicago truncatula (Fig.1-A).Contrary to most SAP proteins, TaSAP17s have two AN1 zinc-finger domains.Among the annotated and characterized SAP proteins, TaSAP17s showed the highest homology with OsSAP16 (accession no.XP_015644892) (Fig.1-B).
3.2.Transcription of TaSAP17-D in various tissues and at different development stages in wheat
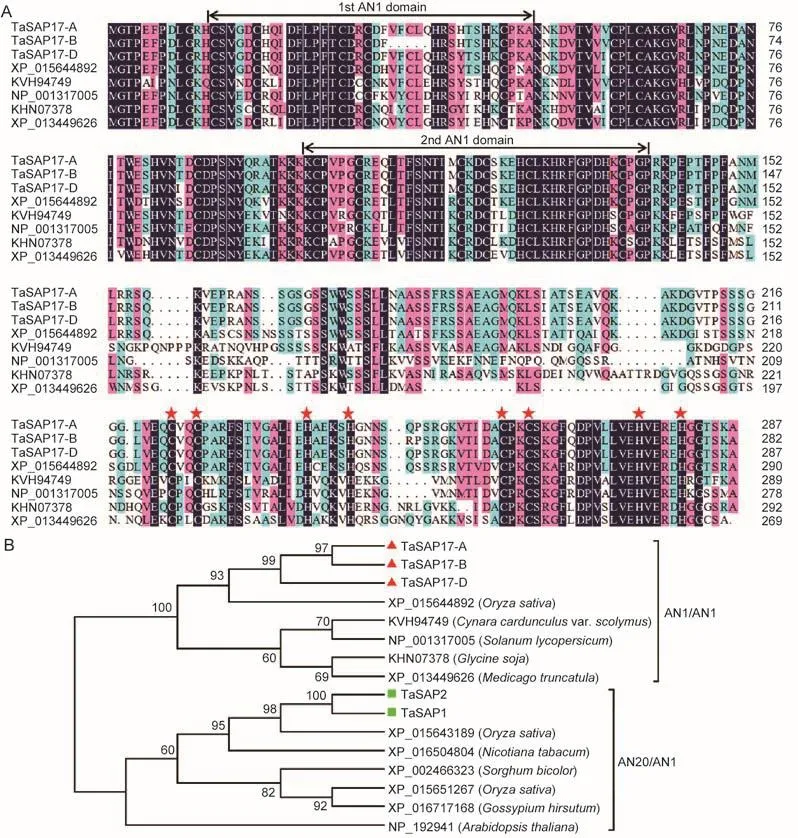
Fig.1 Sequence alignment of TaSAP17s and stress-associated proteins (SAPs) in other plant species.A, amino acid alignment of TaSAP17s and other SAPs.The numbers on the right indicate the amino acid position.Common identical amino acid residues are shown with black background.Dashed lines represent gaps introduced to the maximize alignment.The conserved AN1 domains are marked above the alignment with lines.Stars mark conserved C2H2-type zinc finger.B, phylogenetic tree of SAPs.The neighbor-joining tree was built with 1 000 bootstrap replicates by MEGA 5.05.Branch support bootstrap values are listed at each node.TaSAP17s and TaSAP1/2 in wheat are marked with triangle and filled rectangles, respectively.
The expression patterns of TaSAP17-D in wheat developmental stages (germination stage, seedling stage and heading stage) were examined by qRT-PCR.As shown in Fig.2-A, the transcript of TaSAP17-D was identified in all selected tissues.Higher expression levels were evident in the germination stage in plumule tissues, the seedling stages in leaf tissues, and the heading stage in stem tissues, respectively.The lowest expression was observed in the internode at heading stage.The expression level of TaSAP17-D in leaves at seedling stage was much higher than that in other tissues at different stages.Therefore, the present study mainly focused on the seedling stage for the subsequent analyses.
3.3.Expression patterns of TaSAP17-D under abiotic stresses
Expression patterns of TaSAP17-D were characterized under different stresses (high salinity, ABA, cold, and PEG).The transcript levels of TaSAP17-D were acutely activated by salt, while weakly by ABA, cold, and PEG (Fig.2-B-E).Compared with the control (0 h), the expression levels of TaSAP17-D peaked at 0.5 h and maintained to 2 h, and then quickly decreased under salt-stressed conditions (Fig.2-B).

Fig.2 Expression patterns (qRT-PCR) and subcellular localization of TaSAP17-D.A, expression patterns of TaSAP17-D in wheat tissues at different developmental stages.P, plumule; RB, root base; R, root; L, leaf; I, internode; F, flag leaf; S, stem; N, node;R1-R5, depths of root at 0-30, 30-50, 50-70, 70-90, and 90-100 cm, respectively.B-E, expression patterns of TaSAP17-D under NaCl (B), abscisic acid (ABA, C), cold (D), and polyethlene glycol-6000 (PEG, E) treatments in leaves at seedling stage,respectively.F-G, expression patterns of TaSAP17-D in leaves (F) and roots (G) of a salt-tolerant (Taishan 23) and a salt-sensitive(Nongda 20074) genotypes under NaCl treatments, respectively.Data represent mean±SD from three independent experiments.H, subcellular localization of TaSAP17-D in tobacco leaf cells.The vector control (35S::GFP) and 35S::TaSAP17-D::GFP fusion proteins were transiently expressed in tobacco leaves.35S::GFP, scale bar=50 μm; 35S::TaSAP17-D::GFP, scale bar=20 μm.
According to the expression patterns of TaSAP17-D under different abiotic stresses, we found that TaSAP17-D was remarkably induced by high salinity.In order to further confirm the expression pattern of TaSAP17-D under NaCl stress, we analyzed the expression of TaSAP17-D in two wheat varieties with different salt-tolerance.As we speculated, the expression levels of TaSAP17-D in salttolerant genotype (Taishan 23) were higher than that in saltsensitive genotype (Nongda 20074) in both salt-treated leaves (Fig.2-F) and roots (Fig.2-G).Expression patterns of TaSAP17-D in leaf and root were similar as both peaked at 1 h in the Taishan 23 wheat variety.
3.4.Subcellular localization of TaSAP17-D
Tobacco leaves were used to check subcellular localization of TaSAP17-D.The construct TaSAP17-D-GFP fusion protein driven by the CaMV 35S promoter was transiently expressed in living tobacco leaves.The green fluorescence was detected in the nucleus, cytoplasm, and cell membrane.Therefore, we preliminarily identified that TaSAP17-D-GFP was present in the nucleus, cytoplasm, and cell membrane(Fig.2-H).
3.5.Overexpression of TaSAP17-D in Arabidopsis results in enhanced salt tolerance
To examine the roles of TaSAP17-D in plant stress responses, WT and three homozygous lines of T3TaSAP17-D Arabidopsis transgenic plants were exposed to different abiotic stress conditions.Expression levels of TaSAP17-D in three transgenic lines varied observably, but all were significantly higher than that in the WT plants (Fig.3-B).We only found a significant phenotypic difference between TaSAP17-D lines and WT control under salt conditions and none of the other stress treatments.After a 5-d NaCl(200 mmol L-1) treatment, plants were severely stressed(Fig.3-A).The rosette leaves of WT plants were bleached and some plants were near death.By comparison, most TaSAP17-D plants were slightly wilted and only showed some chlorosis.The higher expression levels of TaSAP17-D in transgenic plants resulted in greater salt tolerance(Fig.3-A).Meanwhile, seedling survival rates of transgenic lines (>90%) were significantly higher than that of WT (66%)(Fig.3-C).
Salt stress often causes physiological changes in plants.Typical salt stress related physiological indices, including CMS, free proline content, OP, chlorophyll content, and water loss rate, were selected to monitor salinity tolerance in the present study.Compared to WT plants, the CMS,free proline contents, and chlorophyll contents of transgenic lines were significantly higher than that of WT (Fig.3-D,E and G).The TaSAP17-D expressing transgenic plants had significantly lower OP than WT plants under salt conditions (Fig.3-F).To evaluate water retention ability of TaSAP17-D-overexpressing Arabidopsis, 4-wk-old transgenic plants and WT were selected to perform a 9-h detached-rosette water loss rate assay.The three transgenic lines showed lower water loss rate than that in WT plants (Appendix B, not significantly).
3.6.Overexpression of TaSAP17-D transgenic plants possess lower germination rate under salt stress
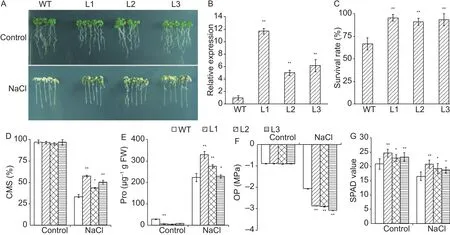
Fig.3 Overexpression of TaSAP17-D improves salt tolerance in transgenic Arabidopsis.A, phenotypes of three TaSAP17-D overexpressing Arabidopsis lines (L1-3) and wild type (WT).B, the expression level of TaSAP17-D in three transgenic lines (L1-3)overexpressing TaSAP17-D.C, comparison of seedling survival rate between WT and transgenic lines treated with 200 mmol L-1 NaCl.Comparison of cell membrane stability (CMS, D), free proline (Pro) contents (E), osmotic potential (OP, F), and chlorophyll content (SPAD value, G) of transgenic plants, and WT plants under 200 mmol L-1 salt stress.Data represent mean±SD (n=10)from three independent experiments.* and ** indicate significant difference levels at P<0.05 and 0.01, respectively.
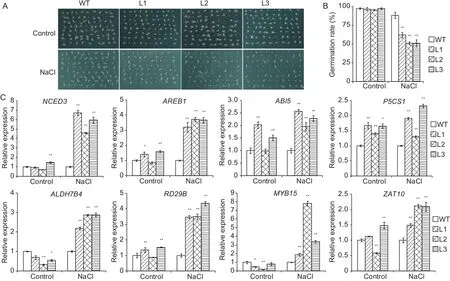
Fig.4 Germination rate and expression level of abiotic stress-responsive genes in TaSAP17-D transgenic Arabidopsis lines(L1-3) and wild type (WT) under salt-stressed conditions.A, the germination test of WT and TaSAP17-D transgenic plants.B,the germination rate of WT and TaSAP17-D transgenic plants.C, relative expression levels of stress-responsive genes were determined by qRT-PCR.Data represent mean±SD from three independent experiments.* and ** indicate significant difference levels at P<0.05 and 0.01, respectively.
To characterize the function of TaSAP17-D at the germination stage, we analyzed the germination rate of three transgenic lines and the WT on MS medium containing 150 mmol L-1NaCl.As shown in Fig.4-A and B, the germination rates of three transgenic lines were similar to the WT under normal conditions, about 100%, while the transgenic lines showed a significantly lower germination rate than that of WT plants on the medium containing 150 mmol L-1NaCl.
3.7.TaSAP17-D regulates the expression of a broad range of stress response genes
expression of membrane transporter genes (SOS1, HKT1 and NHX1) and antioxidant enzyme genes (SOD1, GPX2 and APX1) had no significant changes under salt treatment or normal conditions (Appendix C).It is likely that the TaSAP17-D enhanced tolerance to salt stress might not be attributed to Na+transportation and ROS scavenging in transgenic Arabidopsis.
To cope with high salt stress, plants have developed complex cellular signaling mechanism.In this study, we further confirmed some of the salt-inducible genes regulated by TaSAP17-D in the transgenic plants.The genes analyzed include ABA signaling pathway genes (NCED3, AREB1 and ABI5), high salt-inducible downstream genes (P5CS1,ALDH7B4 and RD29B), transcription factors (MYB15,ZAT10), membrane transporter genes (SOS1, HKT1 and NHX1), and antioxidant enzyme genes (SOD1, GPX2 and APX1) (Fig.4-C, Appendix C).The expression level of the NCED3, AREB1, ABI5, P5CS1, ALDH7B4, RD29B, MYB15,and ZAT10 genes were highly upregulated under saline conditions by 1.5- to 8-fold in the WT (Fig.4-C).However,in our study, compared to the WT transcription levels, the
4.Discussion
To cope with salt stress, plants have developed three major surviving strategies.One is maintaining ionic equilibrium by strengthening the ability to transport Na+across the cell membrane.The second one is regulating the activity of enzymes involved in scavenging reactive oxygen species(ROS) to relief oxidative damages.The third is accumulating more osmolytes, i.e., proline and glycine betaine to reduce osmotic stress (Yu et al.2012).Under high salt stress,plants suffer from hyperosmotic stress, ion poisoning, and oxidative damage.Membrane transporters and antioxidant enzymes are important for salt tolerance.The SOS1,HKT1, and NHX1 proteins are important Na+carriers that work cooperatively to reduce salt toxicity at the cellular and whole plant levels.Plants have developed various protective mechanisms to eliminate or reduce the deleterious effects of ROS through antioxidant enzymes such as SOD1 (superoxide dismutase 1), GPX2 (glutathione peroxidase 2),APX1 (ascorbate peroxidase 1), and non-enzymatic antioxidants (Hasanuzzaman et al.2014).However, in our study,the transcription levels of these genes had no significant changes compared to WT under salt-stressed and normal conditions.However, the transcriptional level of P5CS1 in TaSAP17-D transgenic plants was upregulated by salt.The increased expression of P5CS1 may lead to proline use augmentation, functioning as an osmolyte to balance the water potential within plant cells.It was suggested that TaSAP17-D enhanced salt stress tolerance in transgenic Arabidopsis via osmotic adjustment, rather than Na+transportation and ROS scavenging.
Previous studies indicate that ABA signal production is controlled by sustained activation of NCED3 and the synergistic regulation of ABA biosynthesis and catabolism(Ren et al.2007).The ABI5 and AREB1 genes control the transcription of downstream ABA-dependent and salt-responsive genes.The ABRE (ABA responsive elements)were found in the promoter region of many stress-induce genes, including RD29B, ALDH7B4, MYB15, and ZAT10(Chien et al.2015).We found the expression levels of the NCED3, AREB1, ABI5, RD29B, ALDH7B4, MYB15, and ZAT10 genes were highly upregulated in transgenic plants under salt condition.Additionally, ABA can promote seed dormancy and delay germination (Ali-Rachedi et al.2004;Finch-Savage et al.2006).We speculate that NCED3 promotes the synthesis of ABA resulting in lower germination rates of TaSAP17-D expressing transgenic plants under salt stress.Taken together, our results suggest that TaSAP17-D might act as a positive regulator in the ABA-dependent signaling pathways to synthesize ABA and upregulate salt-tolerance genes involved in response to salt stress.However, it remains to be determined whether TaSAP17-D binds to DNA and regulates transcription directly or whether the observed transcriptome changes are the result of the transcription factor activation by TaSAP17-D.
Overexpression of OsSAP16 can regulate the expression of a set of stress-associated genes, including pathogenesis-related (PR) proteins, signaling proteins, secondary metabolism proteins, MYB transcription factor, and peroxidases(Wang et al.2016).We found that the amino acid sequence of OsSAP16 showed the highest homology with TaSAP17 and interestingly, the overexpression of TaSAP17-D upregulated a broad range of stress responsive genes.We suggest that this upregulation enhances salt tolerance by participating in the ABA-dependent signaling pathway and accumulating osmolytes in transgenic Arabidopsis plants.Ultimately, TaSAP17-D expressing transgenic plants maintained higher physiological levels of cell membrane stability,free proline content, chlorophyll content, lower osmotic potential, and a lower water loss rate than WT plants under the same salt stress conditions.These physiological adaptations may reflect a better abiotic stress tolerance in transgenic plants at seedling stages.
5.Conclusion
According to our results, we proposed that the enhanced salt tolerance by TaSAP17-D overexpressing plants is attributed to osmolyte accumulation, as well as the upregulation of salt-response related genes expression.The TaSAP17-D gene could be instrumental in improving salt tolerance in wheat and other plants.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2016YFD0100605),and the Agricultural Science and Technology Innovation Program, China (ASTIP).
Appendicesassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
Ali-Rachedi S, Bouinot D, Wagner M H, Bonnet M, Sotta B, Grappin P, Jullien M.2004.Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: Studies with the cape verde islands ecotype, the dormant model of Arabidopsis thaliana.Planta, 219, 479-488.
Baek D, Cha J Y, Kang S, Park B, Lee H J, Hong H, Chun H J,Kim D H, Kim M C, Lee S Y, Yun D J.2015.The Arabidopsis a zinc finger domain protein ARS1 is essential for seed germination and ROS homeostasis in response to ABA and oxidative stress.Frontiers in Plant Science, 6, doi: 10.3389/fpls.2015.00963
Chakraborty N, Singh N, Kaur K, Raghuram N.2015.G-protein signaling components GCR1 and GPA1 mediate responses to multiple abiotic stresses in Arabidopsis.Frontiers in Plant Science, 6, doi: 10.3389/fpls.2015.01000
Chang J Z, Zhang J N, Mao X G, Li A, Jia J Z, Jing R L.2013.Polymorphism of TaSAP1-A1 and its association with agronomic traits in wheat.Planta, 237, 1495-1508.
Chien P S, Nam H G, Chen Y R.2015.A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis.Journal of Experimental Botany, 66, 5301-5313.
Clough S J, Bent A F.1998.Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana.The Plant Journal, 16, 735-743.
Finch-Savage W E, Leubner-Metzger G.2006.Seed dormancy and the control of germination.The New Phytologist, 171,501-523.
Gilles G C, Gervais M L, Planchet E, Satour P, Limami A M,Lelievre E.2011.A stress-associated protein containing A20/AN1 zing-finger domains expressed in Medicago truncatula seeds.Plant Physiology and Biochemistry, 49,303-310.
Giri J, Dansana P K, Kothari K S, Sharma G, Vij S, Tyagi A K.2013.SAPs as novel regulators of abiotic stress response in plants.Bioessays, 35, 639-648.
Hasanuzzaman M, Alam M M, Rahman A, Hasanuzzaman M,Nahar K, Fujita M.2014.Exogenous proline and glycine betaine mediated upregulation of antioxidant defense and glyoxalase systems provides better protection against salt-induced oxidative stress in two rice (Oryza sativa L.)varieties.BioMed Research International, 2014, 757219.
Hichri I, Muhovski Y, Zizková E, Dobrev P I, Franco-Zorrilla J M, Solano R, Lopez-Vidriero I, Motyka V, Lutts S.2014.The Solanum lycopersicum zinc finger2 cysteine-2/histidine-2 repressor-like transcription factor regulates development and tolerance to salinity in tomato and Arabidopsis.Plant Physiology, 164, 1967-1990.
Hu C A A, Delauney A J, Verma D P.1992.A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase)catalyzes the first two steps in proline biosynthesis in plants.Proceedings of the National Academy of Sciences of the United States of America, 89, 9354-9358.
Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, Shinozaki K, Nakashima K, Yamaguchi-Shinozaki K.2013.OsTZF1, a CCCH-tandem zinc finger protein,confers delayed senescence and stress tolerance in rice by regulating stress-related genes.Plant Physiology, 161,1202-1216.
Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, Jing R, Zhang C, Ma Y, Gao L, Gao C, Spannagl M, Mayer K F, Li D, Pan S, Zheng F, et al.2013.Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation.Nature, 496, 91-95.
Kang M, Abdelmageed H, Lee S, Reichert A, Mysore K S, Allen R D.2013.AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5.The Plant Journal, 76,481-493.
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K.1999.Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor.Nature Biotechnology, 17, 287-291.
Knight H, Knight M R.2001.Abiotic stress signalling pathways:Specificity and cross-talk.Trends in Plant Science, 6,262-267.
Li B, Liu D, Li Q R, Mao X G, Li A, Wang J Y, Chang X P, Jing R L.2016.Overexpression of wheat gene TaMOR improves root system architecture and grain yield in Oryza sativa.Journal of Experimental Botany, 67, 4155-4167.
Ling H Q, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D,Dong L, Tao Y, Gao C, Wu H, Li Y, Cui Y, Guo X, Zheng S,Wang B, Yu K, Liang Q, Yang W, et al.2013.Draft genome of the wheat A-genome progenitor Triticum urartu.Nature,496, 87-90.
Mao X G, Zhang H Y, Tian S J, Chang X P, Jing R L.2010.TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis.Journal of Experimental Botany, 61, 683-696.
Mukhopadhyay A, Vij S, Tyagi A K.2004.Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco.Proceedings of the National Academy of Sciences of the United States of America, 101, 6309-6314.
Peng Z, Li L, Liu Y P, Liu H M, Jing R L.2017.Evaluation of salinity tolerance in wheat genotypes at germination and seedling stages.Journal of Plant Genetic Resources, 18,638-645.
Ren H B, Fan Y J, Gao Z H, Wei K F, Li G F, Liu J, Chen L, Li B B, Hu J F, Jia W S.2007.Roles of a sustained activation of NCED3 and the synergistic regulation of ABA biosynthesis and catabolism in ABA signal production in Arabidopsis.Chinese Science Bulletin, 52, 484-491.
Ben Saad R, Zouari N, Ben Ramdhan W, Azaza J, Meynard D, Guiderdoni E, Hassairi A.2010.Improved drought and salt stress tolerance in transgenic tobacco overexpressing a novel A20/AN1 zinc-finger “AlSAP” gene isolated from the halophyte grass Aeluropus littoralis.Plant Molecular Biology, 72, 171-190.
Schmittgen T D, Livak K J.2008.Analyzing real-time PCR data by the comparative CTmethod.Nature Protocols, 3,1101-1108.
Sharma G, Giri J, Tyagi A K.2015.Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water-deficit stress in Arabidopsis.Plant Science, 237,80-92.
Shi H, Jiang C, Ye T T, Tan D X, Reiter R J, Zhang H, Liu R Y,Chan Z L.2015.Comparative physiological, metabolomic,and transcriptomic analyses reveal mechanisms of improved abiotic stress resistance in bermudagrass[Cynodon dactylon (L).Pers.] by exogenous melatonin.Journal of Experimental Botany, 66, 681-694.
Sreedharan S, Shekhawat U K S, Ganapathi T R.2012.MusaSAP1, a A20/AN1 zinc finger gene from banana functions as a positive regulator in different stress responses.Plant Molecular Biology, 80, 503-517.
Ströher E, Wang X J, Roloff N, Klein P, Husemann A, Dietz K J.2009.Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana.Molecular Plant, 2, 357-367.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.2011.MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Molecular Biology and Evolution, 28, 2731-2739.
Vij S, Tyagi A K.2008.A20/AN1 zinc-finger domain-containing proteins in plants and animals represent common elements in stress response.Functional & Integrative Genomics, 8,301-307.
Wang C X.2011.Isolation and functional analysis of stressresponse gene TaABC1 and TaSAP1/2 from wheat(Triticum aestivum L.).Ph D thesis, Chinese Academy of Agricultural Sciences, Beijing.(in Chinese)
Wang F, Coe R A, Karki S, Wanchana S, Thakur V, Henry A, Lin H C, Huang J, Peng S, Quick W P.2016.Overexpression of OsSAP16 regulates photosynthesis and the expression of a broad range of stress response genes in rice (Oryza sativa L.).PLoS ONE, 11, e0157244.
Yu S, Wang W, Wang B.2012.Recent progress of salinity tolerance research in plants.Russian Journal of Genetics,48, 497-505.
Zhang H, Liu Y P, Wen F, Yao D M, Wang L, Guo J, Ni L,Zhang A Y, Tan M P, Jiang M Y.2014.A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice.Journal of Experimental Botany,65, 5795-5809.
Zhang L, Li Y Z, Lu W J, Meng F, Wu C A, Guo X Q.2012.Cotton GhMKK5 affects disease resistance, induces HR-like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana.Journal of Experimental Botany, 63, 3935-3951.
杂志排行
Journal of Integrative Agriculture的其它文章
- Characteristic analysis of tetra-resistant genetically modified rice
- Characterization of GhSERK2 and its expression associated with somatic embryogenesis and hormones level in Upland cotton
- GmNAC15 overexpression in hairy roots enhances salt tolerance in soybean
- Molecular cloning and functional characterization of a soybean GmGMP1 gene reveals its involvement in ascorbic acid biosynthesis and multiple abiotic stress tolerance in transgenic plants
- Responses of the antioxidant system to fluroxypyr in foxtail millet(Setaria italica L.) at the seedling stage
- Light interception and radiation use efficiency response to tridimensional uniform sowing in winter wheat
