细菌对有机污染物的趋化性及其对降解的影响
2018-01-12王慧胡金星秦智慧徐新华沈超峰
王慧,胡金星,秦智慧,徐新华,沈超峰*
工农业生产的迅速发展给人类生产和生活带来了许多方便,同时,也使各种污染物被释放到环境中。尤其是大量使用的环境异生物质,因其潜在毒性强,具有生物富集、生物放大等特点,给全球生态环境和人类健康带来严重威胁[1]。对异生物质污染治理刻不容缓,其治理手段主要有物理、化学和生物修复等。微生物修复方法是最具有前景的修复手段之一,具有安全、经济、二次污染少等特点。研究表明,环境中的大部分有机污染物可被微生物降解[2]。污染物的生物降解效率除了与降解菌本身的降解能力有关之外,还依赖于污染物的生物可利用性[3]。土壤污染物的生物可利用性除受土壤介质、污染物性质影响外,也受土壤微生物移动性的影响[4]。大多数具有移动性的细菌能通过趋化过程感知和寻找污染物[5-6],具有运动和降解能力的细菌主动移向吸附态污染物,可以提高污染物的生物可利用性和生物降解效率[4,7-9]。因此,研究者开始关注细菌对环境污染物的趋化性。本文对细菌趋化性及趋化信号传导机制、细菌对典型有机污染物的趋化、细菌趋化对降解的影响以及趋化与降解的分子关联进行了归纳、总结,以期为进一步的研究提供思路。
1 细菌趋化性
趋化性是指具有运动能力的细菌对物质化学浓度梯度做出的响应,顺浓度梯度迁移叫正趋化,逆浓度梯度迁移叫负趋化[10]。最早报道细菌趋化现象的是ENGELMANN和PFEFFER,随后,ADLER[11]和HAZELBAUER[12]深入研究了大肠杆菌对氨基酸及糖类物质的趋化性机制。近年来,越来越多的研究者开始从事细菌趋化性研究。
细菌能够对随时改变的化学浓度梯度做出反应,是因为细菌细胞膜表面有专一的化学受体蛋白,细菌能够利用这些受体蛋白感知外界刺激物信号[13]。以大肠杆菌为例(图1),趋化反应通过处于细胞两极的受体复合体和随机分布于细胞四周、埋于细胞膜中的鞭毛-马达复合体调节。甲基趋化受体蛋白(methylaccepting chemotaxis proteins,MCPs)感知刺激物信号,信号通过趋化性组氨酸激酶A(chemotaxis histidine kinase A,CheA)和CheY传递给鞭毛马达。CheA可发生自身磷酸化,从而使调节蛋白CheY磷酸化,磷酸化的CheY易与鞭毛马达蛋白结合,调节鞭毛的旋转方向。磷酸化的CheA同时调节甲基酯酶CheB的磷酸化,磷酸化的CheB和甲基转移酶CheR分别调节MCPs的去甲基化和甲基化,使细菌适应化学物质浓度梯度,做出运动改变[13-15]。
2 细菌对典型有机污染物的趋化性
虽然细菌的趋化性在大肠杆菌中研究的比较深入,但是大肠杆菌主要对简单的氨基酸、糖类等物质具有趋化性。近几十年的研究发现,细菌对很多化合物都具有趋化性(表1),趋化物主要包括萘[16]、甲苯[17-18]、联苯[19-20]、多氯联苯[19]、苯甲酸[19]、氯代苯甲酸[21]、硝基芳香化合物[22]、甲基对硫磷[23]、阿特拉津[24]、2,4-D[5]、呋喃类化合物[25]等。涉及的细菌种属主要有假单胞菌(Pseudomonas sp.)、罗尔斯通菌(Ralstonia sp.)、固氮螺菌(Azospirillum sp.)、根瘤菌(Rhizobium sp.)、伯克氏菌(Burkholderia sp.)和节杆菌(Arthrobacter sp.)等。细菌能以大部分趋化物为碳源和能源,在碳源和能源相对缺乏的环境中,细菌对环境异生物质的趋化性无疑是有益于其生存的行为。

图1 大肠杆菌趋化信号传导机制Fig.1 Signal transduction of chemotaxis in Escherichia coli
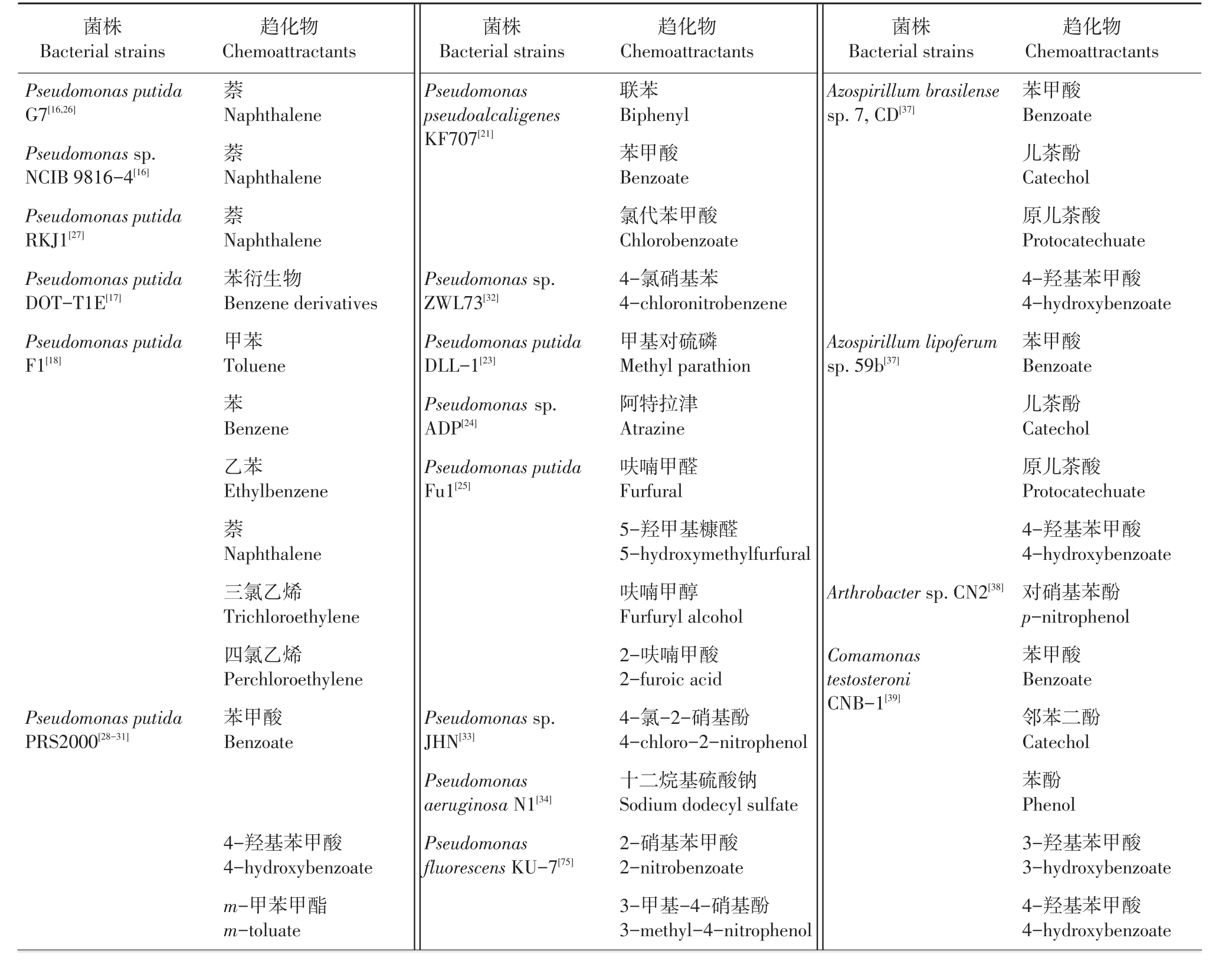
表1 对环境污染物有趋化性的细菌Table 1 Chemotaxis to pollutants by bacteria
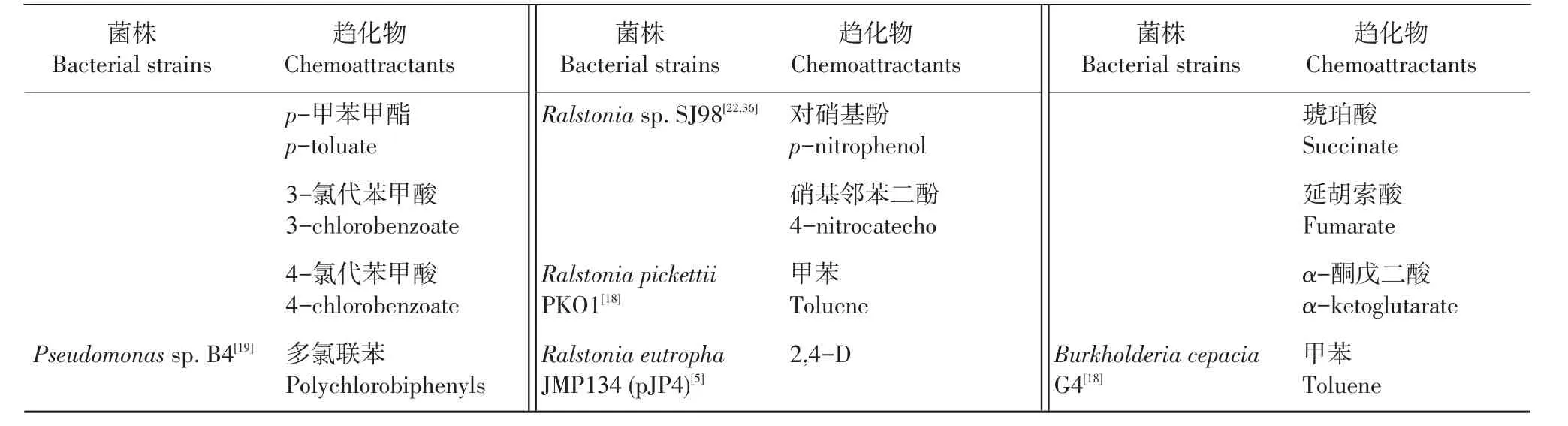
表1 (续) Continuation of Table 1
3 细菌趋化性对污染物降解的影响
有机污染物的生物可利用性被认为是影响生物降解效率的主要限制因子之一。环境中的一些有机污染物水溶性较低,常常被吸附到固体颗粒表面。尤其在土壤环境里,高疏水性的有机污染物通常被吸附到土壤颗粒或者非水相液体(nonaqueousphase liquid,NAPL)里,细菌难以接触到污染物,生物可利用性低。趋化作用使降解菌有效感应并靠近污染物,增强污染物的生物可利用性,提高生物降解效率,在污染物的微生物降解过程中发挥着重要作用[7]。
大量研究表明,细菌趋化性能通过各种机制提高污染物的生物可利用性,其中,恶臭假单胞菌(Pseudomonas putida)G7对萘的趋化性及其对降解的影响研究比较深入。早在1999年,GRIMM等[26]研究发现萘的受体蛋白NahY由代谢质粒编码时,就猜测细菌的趋化可能提高生物降解效率。MARX等[40]的研究证明了这一猜想,他们通过比较野生菌株Pseudomonas putida G7、趋化突变株和移动突变株对萘的降解,发现野生菌株的降解速率最快,如果要达到相同的降解效果,突变株需要更长的降解时间。LAW等[41]在非水相液体2,2,4,4,6,8,8-七甲基壬烷中也证明了野生型菌株Pseudomonas putida G7的趋化性能明显提高萘的降解效率。
除了液相体系,在土壤等高异质性介质里,细菌的趋化性在生物降解中也发挥着重要作用。Pseudomonas putida DLL-1是甲基对硫磷的高效降解菌株[42],该菌株对甲基对硫磷也具有趋化性[23]。通过基因打靶使Pseudomonas putida DLL-1染色体上单拷贝的cheA基因失活,成功获得与野生菌株生长没有显著差异的趋化突变株Pseudomonas putida DAK。cheA基因的失活中断了整个趋化信号传导,使菌株DAK丧失了对甲基对硫磷的趋化性。在摇瓶实验中,野生和突变株对甲基对硫磷的降解效率几乎没有差异,说明在液态环境中,菌体与污染物充分接触对降解影响不大。但是,在土培实验中,趋化突变株DAK对甲基对硫磷的降解效率比野生菌株DLL-1的降解效率低约20%~30%[23],揭示出在原位条件下,趋化性在生物降解过程中具有重要作用。PAUL等[43]以土壤为介质,采用能降解对硝基苯酚但无趋化性的洋葱伯克霍尔德菌(Burkholderia cepacia)RKJ200作为阴性对照,发现Ralstonia sp.SJ98对对硝基酚具有趋化性,并且有明显的降解现象,而Burkholderia cepacia RKJ200则无明显的降解现象。他们的研究均证明了在土壤介质里细菌对污染物的趋化,说明细菌趋化性确实能提高土壤和沉积物里污染物的生物降解效率。
生物膜是细菌细胞附着在非生物或者生物机体表面、并由自身产生的胞外多聚物(extracellular polymeric substance,EPS)包裹所形成的多细胞聚合体[44],是细菌在自然界中常见的存在形式,使细菌在不利环境中生存,抵抗外界不利条件。生物膜在污水处理、原位污染治理与生态修复等过程中发挥重要作用[45-46]。研究表明,趋化性在生物膜的形成过程中发挥重要作用[47-49]。细菌通过趋化作用感知环境中的营养物质,通过鞭毛、菌毛黏附在附着物表面,趋化运动使细菌沿着附着物表面生长,膜不断扩散开并加厚,最后成熟脱落,再形成新的生物膜[45,50-51]。因此,趋化性可能通过影响生物膜的形成,进而影响污染物的生物降解效率。
大量研究证明,趋化性可以提高污染物的生物可利用性,在污染物的降解,尤其是在原位修复过程中发挥重要作用。细菌趋化性对降解的影响表明,趋化性可能是降解的重要属性之一。
4 细菌趋化与降解的内在关联
从以上分析可以看出,趋化可促进降解,反过来,降解对趋化也有影响。研究降解对趋化的影响有助于进一步揭示趋化与降解的内在联系。从细菌对污染物趋化性的表观现象可以发现,很多细菌只对能降解的底物具有趋化性,而且有些趋化需要底物诱导。Ralstonia sp.SJ98能降解某些硝基芳香族化合物(nitroaromatic compounds,NACs);且该菌株只对能降解的硝基芳香族化合物有趋化性,而对不能降解的硝基芳香族化合物则无趋化性[22]。Pseudomonas sp.JHN也只对能代谢的4-氯-2-硝基酚具有趋化性,对不能代谢的底物没有趋化性[33]。氯代苯甲酸是多氯联苯在好氧降解过程中易积累的中间代谢产物,该类化合物能通过抑制降解菌的生长而影响多氯联苯的降解效率[52-53]。GORDILLO等[19]的研究表明,经联苯或苯甲酸诱导的联苯降解菌Pseudomonas sp.B4对苯甲酸具有趋化作用,经苯甲酸诱导的B4对4-氯代苯甲酸具有趋化作用,说明Pseudomonas sp.B4对这些底物的趋化属于诱导趋化。TREMAROLI等[21]的研究也表明,类产碱假单胞菌(Pseudomonas pseudoalcaligenes)KF707对苯甲酸及氯代苯甲酸的趋化也属于诱导趋化。
某些降解菌的趋化基因位于代谢质粒上,趋化和代谢基因共转录。如:Pseudomonas putida G7对萘的趋化受体蛋白NahY由代谢质粒NAH7编码,由nahY和降解基因共转录,且同受转录调控因子NahR的调控[26]。Pseudomonas sp.ZWL73是4-氯硝苯基的完全降解菌,其降解由质粒控制。Pseudomonas putia PaW 340不能利用也不能转化4-氯硝基苯,而菌株ZP8是将ZWL73的质粒转入无质粒的Pseudomonas putia PaW 340得到的质粒转移接合子,该菌获得了以4-氯硝基苯为唯一碳、氮源及能源生长的能力。ZP8和ZWL73对受试化合物具有相同趋化谱,说明ZWL73对4-氯硝基苯的趋化基因也由其质粒编码[32]。
研究发现,某些细菌趋化、转运和代谢之间有内在联系。LUU等[54]证明了Pseudomonas putida F1对4-羟基苯甲酸的趋化受体基因pcaY的表达受转录激活因子PcaR的调控,同时需要4-羟基苯甲酸代谢中间产物β-酮己二酸的诱导,并且和代谢基因共转录。该研究还发现,超家族转运蛋白PcaK能通过促进4-羟基苯甲酸的吸收,累积大量的β-酮己二酸,诱导PcaR对pcaY的调控,从而间接影响pcaY的表达(图2A)。罗尔斯通菌(Ralstonia eutropha)JMP134含有自主转移质粒pJP4,该质粒编码2,4-D的降解基因族tfd。研究发现,对质粒pJP4的改造导致该菌株丧失了对2,4-D的趋化性,说明菌株JMP134对2,4-D的趋化基因也由该质粒编码。tfd基因簇同时编码2,4-D的转运蛋白TfdK,且tfdK缺失突变株对2,4-D没有趋化现象[5],因此推测TfdK可能间接影响JMP134对2,4-D的趋化。可见,细菌对物质的转运、趋化和降解基因之间的协调表达,可能是细菌选择最佳生存环境的一般机制,也体现了细菌趋化和降解的分子关联。
虽然鞭毛介导的细菌趋化信号传导的核心机制相对保守,但是负责感受趋化信号的甲基趋化受体蛋白在不同种属间差异较大。在某些情况下,受体蛋白不能直接感知趋化物,而是通过感知代谢中间产物实现趋化过程。NI等[39,55]研究发现,菌株睾丸酮丛毛单胞菌(Comamonas testosteronei)CNB-1的甲基趋化受体蛋白MCP2201和MCP2983不能直接与芳环类化合物结合,而是与三羧酸循环中间产物结合(图2B)。他们的研究揭示了一类新的趋化途径,即细菌可以通过感知三羧酸循环中间化合物实现对芳香类化合物的趋化。代谢突变株对芳环类化合物没有趋化性,说明CNB-1对芳环类化合物的趋化属于代谢依赖趋化,这更深入地揭示了降解和趋化之间的内在关联。
5 结语
关于细菌趋化性与降解性之间关系的研究备受关注,目前的研究主要集中在趋化现象的表征上,很多细菌对污染物的探测机制尚不清楚,包括化学受体的鉴定、反应调控以及趋化、降解和转运之间的关系等研究相对较少。深入研究细菌对污染物的趋化机制,阐明趋化、转运和降解之间的分子关联,对于提高污染物的生物降解效率有重要意义。本文从以下几个方面提出展望:
1)趋化受体蛋白作为细菌感应外界刺激信号的关键分子,到目前为止,只有少部分趋化受体蛋白被发现与细菌对污染物的趋化相关,多数受体蛋白对污染物的探测机制依然未知。因此,以后的研究工作仍需继续鉴别细菌对污染物的趋化受体蛋白,以阐明细菌对污染物的感应机制。
2)研究者虽然进行了大量的趋化研究,但大多是在半固体培养基里进行的,在土壤等实际固体环境中研究趋化性的报道鲜见。今后应在实际污染的土壤环境中进行更多的实验,利用生化与分子手段评估降解菌是否完全适应胁迫环境。明确细菌对污染物的探测机制,有目的地调控细菌对污染物的趋化行为,对污染土壤的生物修复具有重要意义。
3)目前研究较多的是鞭毛介导的运动与趋化行为,除此之外,也有细菌借助菌毛的伸缩在固体表面进行蹭行运动[56-59]及黏性物质介导的滑行运动[60-70]等。但在实际污染物环境里,无鞭毛降解菌占有一定的生境[71-73],它们如何克服有机污染物的高疏水性和低生物可利用性,如何与污染物分子相互接触,及其在土壤环境中的分散机制等是否与趋化性相关,目前尚不清楚。因此,有必要进行更多降解菌运动模式与趋化机制的研究。

图2 Pseudomonas putida F1(A)和Comamonas testosteronei CNB-1(B)对4-羟基苯甲酸趋化信号探测示意图Fig.2 Schematic representation of 4-hydroxybenzoate chemotaxis in Pseudomonas putida F1(A)and Comamonas testosteronei CNB-1(B)
[1] 王庆仁,刘秀梅,崔岩山,等.土壤与水体有机污染的生物修复及其应用研究进展.生态学报,2001,21(1):159-163.WANG Q R,LIU X M,CUI Y S,et al.Concept and advances of applied bioremediation for organic pollutants in soil and water.Acta Ecologica Sinica,2001,21(1):159-163.(in Chinese with English abstract)
[2] PIEPER D H,REINEKE W.Engineering bacteria for bioremediation.Current Opinion in Biotechnology,2000,11(3):262-270.
[3] YANG X Q,ERICKSON L E,FAN L T.A study of the dissolution rate-limited bioremediation of soil contaminated by residual hydrocarbons.Journal of Hazardous Materials,1995,41(2/3):299-313.
[4] 罗启仕,张锡辉,王慧,等.生物修复中有机污染物的生物可利用性.生态环境,2004,13(1):85-87.LUO Q S,ZHANG X H,WANG H,et al.Bioavailability of organic contaminants during bioremediation.Ecology and Environment,2004,13(1):85-87.(in Chinese with English abstract)
[5] HAWKINSA A C,HARWOOD C S.Chemotaxis of Ralstonia eutropha JMP134(pJP4)to the herbicide 2,4-dichlorophenoxyacetate.Applied and Environmental Microbiology,2002,68(2):968-972.
[6] LACAL J,REYES-DARIAS J A,GARCÍA-FONTANA C,et al.Tactic responses to pollutants and their potential to increase biodegradation efficiency.Journal of Applied Microbiology,2013,114(4):923-933.
[7] KRELL T,LACAL J,REYES-DARIAS J A,et al.Bioavailability of pollutants and chemotaxis.Current Opinion in Biotechnology,2013,24(3):451-456.
[8] PANDEY G,JAIN R K.Bacterial chemotaxis toward environmental pollutants:Role in bioremediation.Applied and Environmental Microbiology,2002,68(12):5789-5795.
[9] PARALES R E,LUU R A,HUGHES J G,et al.Bacterial chemotaxis to xenobiotic chemicals and naturally-occurring analogs.Current Opinion in Biotechnology,2015,33:318-326.
[10]蒋建东,张瑞福,何健,等.细菌对环境污染物的趋化性及其在生物修复中的作用.生态学报,2005,25(7):1764-1771.JIANG J D,ZHANG R F,HE J,et al.Bacterial chemotaxis to environmental pollutants and its significance in bioremediation.Acta Ecologica Sinica,2005,25(7):1764-1771.(in Chinese with English abstract)
[11]ADLER J.My life with nature.Annual Review of Biochemistry,2011,80:42-70.
[12]HAZELBAUER G L.Bacterial chemotaxis:The early years of molecular studies.Annual Review of Biochemistry,2012,66:285-303.
[13]李茹,陈鹏.细菌趋化性的信号传导及调节机制研究进展.生物技术通报,2011(11):54-57.LI R,CHEN P.Progress in mechanism of signal transduction and regulation in bacterial chemotaxis.Biotechnology Bulletin,2011(11):54-57.(in Chinese with English abstract)
[14]MICALI G,ENDRES R G.Bacterial chemotaxis:Information processing,thermodynamics,and behavior.Current Opinion in Microbiology,2016,30:8-15.
[15]WADHAMS G H,ARMITAGE J P.Making sense of it all:Bacterial chemotaxis.Nature Reviews:Molecular Cell Biology,2004,5(12):1024-1037.
[16]GRIMM A C,HARWOOD C S.Chemotaxis of Pseudomonas spp.to the polyaromatic hydrocarbon naphthalene.Applied and Environmental Microbiology,1997,63(10):4111-4115.
[17]LACAL J,MUÑOZ-MARTÍNEZ F,REYES-DARÍAS J A,et al.Bacterial chemotaxis towards aromatic hydrocarbons in Pseudomonas.Environmental Microbiology,2011,13(7):1733-1744.
[18]PARALES R E,DITTY J L,HARWOOD C S.Toluene-degrading bacteria are chemotactic towards the environmental pollutants benzene, toluene, and trichloroethylene. Applied and Environmental Microbiology,2000,66(9):4098-4104.
[19]GORDILLO F,CHÁVEZ F P,JEREZ C A.Motility and chemotaxis of Pseudomonas sp.B4 towards polychlorobiphenyls and chlorobenzoates.FEMS Microbiology Ecology,2007,60(2):322-328.
[20]TREMAROLI V,FEDI S,TAMBURINI S,et al.A histidinekinase cheA gene of Pseudomonas pseudoalcaligens KF707 not only has a key role in chemotaxis but also affects biofilm formation and cell metabolism.Biofouling,2011,27(1):33-46.
[21]TREMAROLI V,SUZZI C V,FEDI S,et al.Tolerance of Pseudomonas pseudoalcaligenes KF707 to metals,polychlorobiphenyls and chlorobenzoates:Effects on chemotaxis-,biofilm-and planktonic-grown cells.FEMS Microbiology Ecology,2010,74(2):291-301.
[22]SAMANTA S K,BHUSHAN B,CHAUHAN A,et al.Chemotaxis of a Ralstonia sp.SJ98 toward different nitroaromatic compounds and their degradation.Biochemical and Biophysical Research Communications,2000,269(1):117-123.
[23]文阳,蒋建东,邓海华,等.趋化信号转导基因cheA突变对Pseudomonas putida DLL-1甲基对硫磷的趋化性及原位降解的影响.微生物学报,2007,47(3):471-476.WEN Y,JIANG J D,DENG H H,et al.Effect of mutation of chemotaxis signal transduction gene cheA in Pseudomonas putida DLL-1 on its chemotaxis and methyl parathion biodegradation.Acta Microbiologica Sinica,2007,47(3):471-476.(in Chinese with English abstract)
[24]LIU X X,PARALES R E.Bacterial chemotaxis to atrazine and related S-triazines.Applied and Environmental Microbiology,2009,75(17):5481-5488.
[25]NICHOLS N N,LUNDE T A,GRADEN K C,et al.Chemotaxis to furan compounds by furan-degrading Pseudomonas strains.Applied and Environmental Microbiology,2012,78(17):6365-6368.
[26]GRIMM A C,HARWOOD C S.NahY,a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene.Journal of Bacteriology,1999,181(10):3310-3316.
[27]SAMANTA S K,JAIN R K.Evidence for plasmid-mediated chemotaxis of Pseudomonas putida towards naphthalene and salicylate.Canadian Journal of Microbiology,2000,46(1):1-6.
[28]HARWOOD C S,RIVELLI M,ORNSTON L N.Aromatic acids are chemoattractants for Pseudomonasputida.Journalof Bacteriology,1984,160(2):622-628.
[29]HARWOOD C S,NICHOLS N N,KIM M K,et al.Identification ofthe pcaRKF gene clusterfrom Pseudomonas putida:Involvement in chemotaxis,biodegradation,and transport of 4-hydroxybenzoate.Journal of Bacteriology,1994,176(21):6479-6488.
[30]HARWOOD C S.A methyl-accepting protein is involved in benzoate taxis in Pseudomonas putida.Journal of Bacteriology,1989,171(9):4603-4608.
[31]HARWOOD C S,PARALES R E,DISPENSAL M.Chemotaxis of Pseudomonas putida toward chlorinated benzoates.Applied and Environmental Microbiology,1990,56(5):1501-1503.
[32] 赵非,刘虹,王淑君,等.Pseudomonas sp.ZWL73对4-氯硝基苯及多种芳香烃化合物的趋化性.浙江大学学报(农业与生命科学版),2004,30(6):624-627.ZHAO F,LIU H,WANG S J,et al.Chemotaxis of Pseudomonas sp.strain ZWL73 to 4-chloronitrobenzene and various aromatic compounds.Journal of Zhejiang University(Agriculture and Life Sciences),2004,30(6):624-627.(in Chinese with English abstract)
[33]ARORA P K,BAE H.Biotransformation and chemotaxis of 4-chloro-2-nitrophenol by Pseudomonas sp.JHN.Microbial Cell Factories,2014,13:110.
[34]CHATURVEDI V,KUMAR A.Metabolism dependent chemotaxis of Pseudomonas aeruginosa N1 towards anionic detergent sodium dodecyl sulfate.Indian Journal of Microbiology,2014,54(2):134-138.
[35]IWAKI H,MURAKI T,ISHIHARA S,et al.Characterization of a pseudomonad 2-nitrobenzoate nitroreductase and its catabolic pathway-associated 2-hydroxylaminobenzoatemutaseand a chemoreceptor involved in 2-nitrobenzoate chemotaxis.Journal of Bacteriology,2007,189(9):3502-3514.
[36]BHUSHAN B,SAMANTA S K,CHAUHAN A,et al.Chemotaxis and biodegradation of 3-methyl-4-nitrophenol by Ralstonia sp.SJ98.Biochemical and Biophysical Research Communications,2000,275(1):129-133.
[37]LOPEZ-DE-VICTORIA G,LOVELL C R.Chemotaxis of Azospirillum species to aromatic compounds.Applied and Environmental Microbiology,1993,59(9):2951-2955.
[38] 乔铖,任磊,贾阳,等.节杆菌(Arthrobacter sp.)CN2对对硝基苯酚的趋化性研究.农业环境科学学报,2016,35(10):1945-1952.QIAO C,REN L,JIA Y,et al.Chemotaxis of Arthrobacter sp.CN2 towards p-nitrophenol.Journal of Agro-Environment Science,2016,35(10):1945-1952.(in Chinese with English abstract)
[39]NI B,HUANG Z,FAN Z,et al.Comamonas testosteroni uses a chemoreceptor for tricarboxylic acid cycle intermediates to trigger chemotactic responses towards aromatic compounds.Molecular Microbiology,2013,90(4):813-823.
[40]MARX R B,AITKEN M D.Bacterial chemotaxis enhances naphthalene degradation in a heterogeneous aqueous system.Environmental Science&Technology,2000,34(16):3379-3383.
[41]LAW A M J,AITKEN M D.Bacterial chemotaxis to naphthalene desorbing from a nonaqueous liquid.Applied and Environmental Microbiology,2003,69(10):5968-5973.
[42]刘智,孙建春,李顺鹏.甲基对硫磷降解菌DLL-1的分离、鉴定及降解性研究.应用与环境生物学报,1999,5(增刊1):147-150.LIU Z,SUN J C,LI S P.Isolation,identification and characters of methyl parathion degrading bacterium.Chinese Journal of Applied&Environmental Biology,1999,5(Suppl.1):147-150.(in Chinese with English abstract)
[43]PAUL D,SINGH R,JAIN R K.Chemotaxis of Ralstonia sp.SJ98 towards p-nitrophenol in soil.Environmental Microbiology,2006,8(10):1797-1804.
[44]COSTERTON J W,STEWART P S,GREENBERG E P.Bacterial biofilms:A common cause of persistent infections.Science,1999,284(5418):1318-1322.
[45]SINGH R,PAUL D,JAIN R K.Biofilms:Implications in bioremediation.Trends in Microbiology,2006,14(9):389-397.
[46]ABBASNEZHAD H,GRAY M,FOGHT J M.Influence of adhesion on aerobic biodegradation and bioremediation of liquid hydrocarbons.Applied Microbiology and Biotechnology,2011,92(4):653-675.
[47]PRATT L A,KOLTER R.Genetic analysis of Escherichia coli biofilm formation:Roles of flagella,motility,chemotaxis and type I pili.Molecular Microbiology,1998,30(2):285-293.
[48]ALEXANDRE G. Chemotaxis control of transient cell aggregation.Journal of Bacteriology,2015,197(20):3230-3237.
[49]HE K,BAUER C E.Chemosensory signaling systems that control bacterial survival.Trends in Microbiology,2014,22(7):389-398.
[50]DONLAN R M.Biofilms:Microbial life on surfaces.Emerging Infectious Diseases,2002,8(9):881-890.
[51]ALEXANDRE G,ZHULIN I B.Different evolutionary constraints on chemotaxis proteins CheW and CheY revealed by heterologous expression studies and protein sequence analysis.Journal of Bacteriology,2003,185(2):544-552.
[52]AHMED M,FOCHT D D.Degradation of polychlorinated biphenyls by two species of Achvornobacter.Canadian Journal of Microbiology,1973,19(1):47-52.
[53]FURUKAWA K,MATSUMURA F,TONOMURA K.Alcaligenes and Acinetobacter strains capable of degrading polychlorinated biphenyls.Agricultural and Biological Chemistry,1978,42(3):543-548.
[54]LUU R A,KOOTSTRA J D,NESTERYUK V,et al.Integration of chemotaxis,transport and catabolism in Pseudomonas putida and identification ofthe aromatic acid chemoreceptor PcaY.Molecular Microbiology,2015,96(1):134-147.
[55]NI B,HUANG Z,WU Y F,et al.A novel chemoreceptor MCP2983 from Comamonas testosteroni specifically binds to cisaconitate and triggerschemotaxistowardsdiverse organic compounds.Applied Microbiology and Biotechnology,2015,99(6):2773-2781.
[56]MILLER R M,TOMARAS A P,BARKER A P,et al.Pseudomonas aeruginosa twitching motility-mediated chemotaxis towards phospholipids and fatty acids:Specificity and metabolic requirements.Journal of Bacteriology,2008,190(11):4038-4049.
[57]KLAUSEN M,HEYDORN A,RAGAS P,et al.Biofilm formation by Pseudomonas aeruginosa wild type,flagella and typeⅣpili mutants.Molecular Microbiology,2003,48(6):1511-1524.
[58]MATTICK J S.TypeⅣpili and twitching motility.Annual Review of Microbiology,2002,56:289-314.
[59]KENNAN R M,LOVITT C J,HAN X Y,et al.A two-component regulatory system modulates twitching motility in Dichelobacter nodosus.Veterinary Microbiology,2015,179(1/2):34-41.
[60]KIRBY JR.Chemotaxisgenes,fibrilsandPE taxisin Myxococcus xanthus.Trends in Microbiology,2001,9(5):205.
[61]SPORMANN A M,KAISER A D.Gliding movements in Myxococcus xanthus.Journal of Bacteriology,1995,177(20):5846-5852.
[62]KEARNS D B,SHIMKETS L J.Chemotaxis in a gliding bacterium.Proceedings of the National Academy of Sciences of the USA,1998,95(20):11957-11962.
[63]DARZINS A.Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility:Sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus.Molecular Microbiology,1994,11(1):137-153.
[64]KRELL T,LACAL J,MUÑOZ-MARTÍNEZ F,et al.Diversity at its best:Bacterial taxis.Environmental Microbiology,2011,13(5):1115-1124.
[65]HARSHEY R M.Bacterial motility on a surface:Many ways to a common goal.Annual Review of Microbiology,2003,57:249-273.
[66]RECHT J,MARTÍNEZ A,TORELLO S,et al.Genetic analysis of sliding motility in Mycobacterium smegmatis.Journalof Bacteriology,2000,182(15):4348-4351.
[67]SUN M Z,WARTEL M,CASCALES E,et al.Motor-driven intracellular transport powers bacterial gliding motility.Proceedings of the National Academy of Sciences of the USA,2011,108(18):7559-7564.
[68]RISSER D D,CHEW W G,MEEKS J C.Genetic characterization of the hmp locus,a chemotaxis-like gene cluster that regulates hormogonium development and motility in Nostoc punctiforme.Molecular Microbiology,2014,92(2):222-233.
[69]COZY L M,CALLAHAN S M.The hmp chemotaxis cluster regulates gliding in the filamentous cyanobacterium Nostoc punctiforme.Molecular Microbiology,2014,92(2):213-216.
[70]POLLITT E J G,CRUSZ S A,DIGGLE S P.Staphylococcus aureus forms spreading dendrites that have characteristics of active motility.Scientific Reports,2015,5:17698.
[71]SUNGTHONG R,VAN WEST P,HEYMAN F,et al.Mobilization of pollutant-degrading bacteria by eukaryotic zoospores.Environmental Science&Technology,2016,50(14):7633-7640.
[72]WAGNER-DÖBLER I,BENNASAR A,VANCANNEYT M,et al.Microcosm enrichment of biphenyl-degrading microbial communities from soils and sediments.Applied and Environmental Microbiology,1998,64(8):3014-3022.
[73]UYTTEBROEK M,BREUGELMANS P,JANSSEN M,et al.Distribution of the Mycobacterium community and polycyclic aromatic hydrocarbons(PAHs)among different size fractions of a long-termPAH-contaminated soil.Environmental Microbiology,2006,8(5):836-847.
