Genetic Diversity of Wild Begonia fimbristipulataHance Revealed by ISSR Analysis
2017-11-16ZHAOBoMAOShizhongLIJingjianHUANGShixunZHONGShuhua
ZHAO Bo, MAO Shi-zhong, LI Jing-jian, HUANG Shi-xun, ZHONG Shu-hua*
(1. Guangxi Institute of Botany, The Chinese Academy of Sciences, Guangxi Guilin 541006, China; 2. Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China)
GeneticDiversityofWildBegoniafimbristipulataHanceRevealedbyISSRAnalysis
ZHAO Bo1, 2, MAO Shi-zhong1, LI Jing-jian1, HUANG Shi-xun1, ZHONG Shu-hua1*
(1. Guangxi Institute of Botany, The Chinese Academy of Sciences, Guangxi Guilin 541006, China; 2. Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China)
【Objective】 Genetic diversity of wildBegoniafimbristipulataHance would offer practical advices for its further breeding, utilization and conservation. 【Method】 To assess the genetic diversity ofB.fimbristipulataHance in 3 wild populations, the molecular marker technique ISSR was used to investigate the genetic diversity of 55 samples from 3 populations in Guangdong Zhaoqing dinghusha, Guangxi Guilin Dayeshenjing and Liutang Qingshitan. Data was analyzed by POPGEN32, and a cluster diagram was presented by UPGMA. 【Result】One hundred and twelve amplified fragments were obtained using 12 ISSR primers. 102 polymorphic loci were identified, with a percentage of 91.07 %. Nei's genetic diversity index (He) was 0.3383, Shannon diversity index (I) was 0.5011, and the coefficient of gene differentiation (Gst) was 0.1507. The genetic similarity coefficient among the populations ranged from 0.8949 to 0.9162 in an average of 0.9026. Based on the clustering analysis, the individuals of each population group together related to geographic distribution. 【Conclusion】B.fimbristipulataHance holds high genetic diversity and the majority of genetic variation occurs in the population with a low level of genetic differentiation among populations. In order to increase the genetic diversity, we suggested that seeds or seedlings collected from each wild populations and cross-breeding.
BegoniafimbristipulataHance; ISSR analysis; Genetic diversity
【Significance】Begoniais a genus of perennial flowering plants in the family Begoniaceae. With more than 1600 species, placing it in the top ten most speciose angiosperm genera in the world[1-2]. TheBegoniasare native to moist subtropical and tropical climates, in South and Central America, Africa, and Southern Asia[3]. These plants are monoecious, with unisexual male and female flowers occurring separately on the same plant, the male contains numerous stamens, and the female has a large inferior ovary and two to four branched or twisted stigmas.Begoniasare classified taxonomically into about 80 sections from their stem/root shapes and origin of phyletic line[4]. Even though this genus has a large species, most of its were commercial cultivated for their bright colourful leaves and flowers, only relatively few of the species have an important contribution to medicine.BegoniafimbristipulataHance B. cyclophylla Hook. F., a small size species belongs toBegoniaonly distributed in China (Jiangxi, Hainan, Guangdong, Hunan, Guangxi and Fujian provinces)[5].B.fimbristipulataHance has a long history in Chinese traditional medicine and used for treatment of bronchitis, inflammatory diseases and diabetic nephrophathy[6-8]. It also used as soft drink in Guangdong province[9-11]. Recently, it has also been reported that the effective ingredients (Flavonoids) inB.fimbristipulataHance have anticancer effects on human cancer cells. However,B.fimbristipulataHance is being destroyed at an alarming rate due to serious ecological environmental destruction, and has been listed as the national protection plant[12]. Investigation of germplasm resources and evaluation of genetic diversity ofB.fimbristipulataHance will help ascertain the present condition of this medicinal plant and thus offer practical advices for its further breeding, utilization and conservation.
【Research Progress】Genetic diversity analysis has proven to be a useful strategy for revealing genetic backgrounds and relationships of germplasm resources. In the past decade, research onB.fimbristipulataHance have been carried out. However, most of these studies are only focused on morphology, cytology and biochemistry, not involving genetics diversity[13-16]. Molecular markers such as inter-simple sequence repeats (ISSR) have been proposed an economical and reliable DNA marker system[17], it has a number of advantages, e.g. low amount of template DNA required, markers randomly distributed throughout the plant genome, acceptable stability and high reproducibility. And widely used for analyses on genetic diversity, gene mapping and phylogenetic relationship in multiple species[18-21].
【Breakthrough Point of this study】In this study, ISSR marks were applied to analyze the genetic diversity and relationships among individuals and populations ofB.fimbristipulataHance germplasm resources from three different populations in Guilin China. 【Key issues to be solved】 Genetic diversity of wildB.fimbristipulataHance would offer practical advices for its further breeding, utilization and conservation.
1 Materials and Methods
A total of 55 samples from 3 populations were originally collected throughout the distribution areas ofB.fimbristipulataHance in China (including Dinghushan Zhaoqing Guangdong province, Dayeshenjing Lingchuan county and Qingshitang Liutang Gulin China). Fresh leaves in squaring period were randomly collected in each population and desiccated in silica gel for molecular analyses. The detailed locations of the studied populations are summarized in Table 1.
Genomic DNA was extracted and purified from leaves desiccated in silica gel following the improving CTAB protocol described by Doyle and Doyle[22], with little modifications to improve the DNA quality. The DNA concentration was estimated by standard spectrophotometric methods at 260 and 280 nm UV lengths by Thermo Scientific Nanodrop 2000 and the integrity by gel electrophoresis in a 0.8 % agarose gel. Samples were then diluted to 30 ng/L work concentration.
A total of 100 ISSR primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (China), according to the public biotechnology website of University of British Columbia. These primers were used initially for amplification to optimize the PCR conditions, twelve (12) of them yielded clear, reproducible and relatively high polymorphism bands (Table 2). ISSR-PCR amplifications were performed in 25 ul reactions containing 30 ng genomic DNA templates, 1×PCR Buffer, 0.15 mM Mg2+, 0.2 mM of each dNTP, 10 pmol primers, and 2 U ofTaqDNA polymerase. Amplification reactions were performed on a PCR Thermal Cycler under the following conditions: initial denaturation at 94 ℃ for 3 min followed by 40 cycles of 94 ℃ for 30 s, annealing at optimal temperature for 45s and 72 ℃ for 2 min, and a final 7 min elongation step at 72 ℃. The PCR products were analyzed electrophoretically on 1.5 % (w/v) agarose gels in 1×TAE Buffer at 110 V for 50 min. A total of 1 μl 6×Loading buffer (Sangon Biotech, China) was added to each reaction before electrophoresis. Gels with amplification fragments were visualized and photographed under UV light and molecular weights were estimated based on the DNA molecular size markers (λDNA/HindIII +EcoRI).

Table 1 The description of the experimental material
The PCR products at each allele produced by polymorphic ISSR markers were manually scored as binary data: present (1) or absent (0) across 55 samples for each primer. POPGENE v1.32 was used to calculate various genetic diversity parameters, including the percentage of polymorphic bands (PPB), the observed total number of alleles (Na), effective allele number (Ne), Nei’s gene diversity (He) and Shannon’s information index (I)[23]. A cluster dendrogram was constructed based on the average genetic distances using the unweighted pair group method with arithmetic average (UPGMA) with the SAHN module of NTSYS-pc 2.20[24].
2 Results and Analysis
Total 100 ISSR primers were screened and 12 primers yielded clear, reproducible and relatively high polymorphism bands were selected for further study across 55 individual samples. ISSR primers produced different numbers of DNA fragments, depending upon their simple sequence repeat motifs. The 12 ISSR primers produced 112 bands across all samples, of which 102 were polymorphic, accounting for 91.07 % polymorphism. The number of bands ranged from 8 to 11, and the amplicon size varied from 200 to 2000 bp. Average number of bands and polymorphic bands per primer were 9.3 and 8.5, respectively.
The number of alleles is one of the most important genetic components for genetic diversification in populations. The percentage of polymorphic bands (PPB) at the population level ranged from 71.43 %-78.57 % with an average of 74.40 %. In the 55 plant germplasm resources, we found the number of alleles (Na) ranged from 1.7143 to 1.7857 with a mean of 1.7440 in these 3 populations. The mean effective number of alleles (Ne) was ranged from 1.4926 to 1.5200 with an average of 1.5071. Interestingly,Nawas highest (1.7857) for Qingtang populations from Liutang and lowest for plants from Dayeshenjing (1.7143). But highest and lowest Ne values were found in Dinghushan (1.5200) and Dayeshenjing (1.4926) populations, respectively. As an index of the genetic diversity of populations, Nei's gene diversity (H) was measured, and ranged from 0.2809 to 0.29290, with an average of 0.2885. Shannon's information index (I) ranged from 0.4120 to 0.4329, with an average of 0.4232. Similar to PPB, Nei’s gene diversity (H) and Shannon’s Information Index (I) found for Qingshitang populations were the highest of the values determined in the three populations (H=0.2929;I=0.4329).
The total gene diversity (Ht) and the gene diversity within populations (Hs) were 0.3396 and 0.2885 respectively. According toGst=Ht-Hs/Ht, the populations from three different locations had a coefficient of genetic diversification (Gst) of 0.1507, which indicates that 15.07 % genetic variation occurred between any two populations and 84.93 % of the genetic variation occurred intra-population. Thus, the genetic diversification within a population is far greater than that between populations, indicated that most of the genetic differentiation were occurred within population. The estimate of gene flow (Nm=1.4089) exists among these populations stating clearly that gene migration was limited among widely distributed populations.
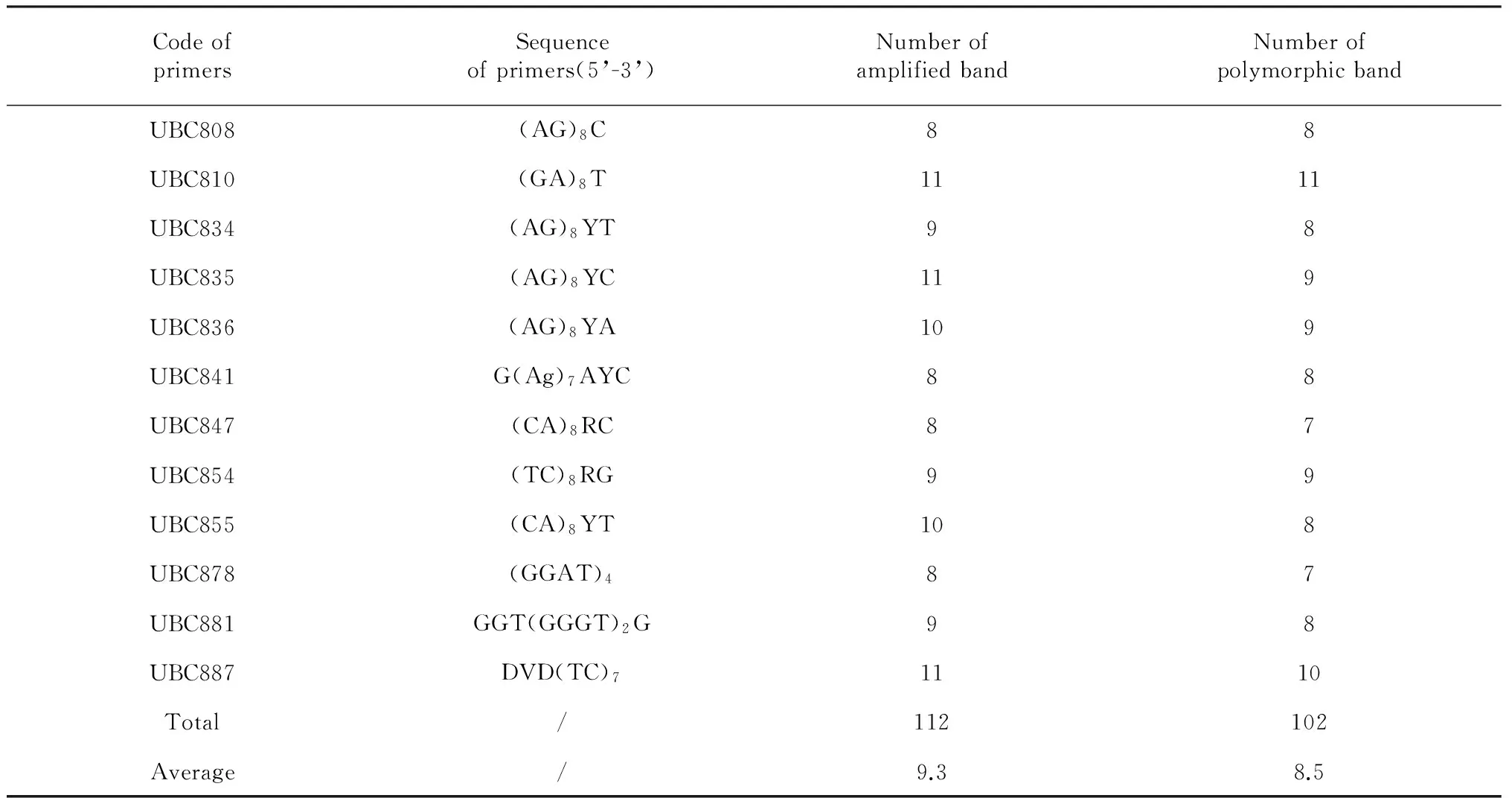
Table 2 12 ISSR primer sequences and amplification results
Table 4 shows the Nei's genetic distance (D) and the genetic identity (I) among populations ofB.fimbristipulataHance. The Nei's genetic distance (D) ranged from 0.0875 to 0.1111, with the nearest (0.0875) being between Qingshitang and Dayeshenjing population, and the farthest (0.1111) being between Qingshitang and Dinghushan population. An inverse relationship was found for genetic identity (I).
Based on similarity coefficient among 55 samples, cluster analysis was carried out using UPGMA method and resulted in a phylogenetic dendrogram shown in Fig. 2. All the samples could be classified into three groups with a similarity coefficient value of 0.68. Most of these samples were clustered into a big group and then further divided into two sub-groups, all of which were from Dayeshenjing and Qingshitang populations. In addition, the rest samples from Dinghushan were classified into the same group, and it appeared to be distinct from the other two population genotypes. The clustering result ofB.fimbristipulataHance samples corresponded generally with the geographical distribution of their collections (Fig.2).
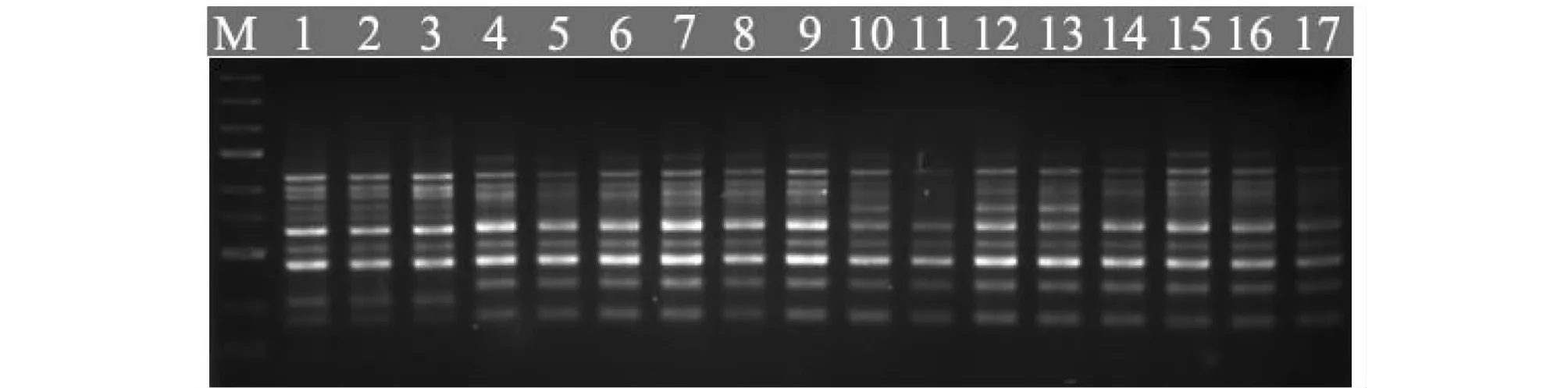
Fig.1 Electrophoresis of B. fimbristipulata Hance DNA samples
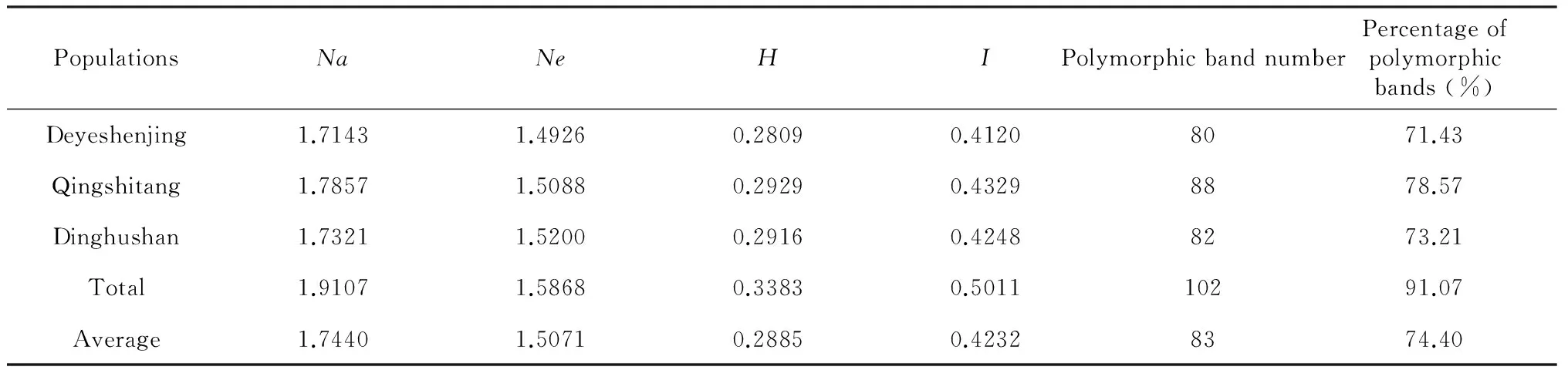
PopulationsNaNeHIPolymorphicbandnumberPercentageofpolymorphicbands(%)Deyeshenjing1.71431.49260.28090.41208071.43Qingshitang1.78571.50880.29290.43298878.57Dinghushan1.73211.52000.29160.42488273.21Total1.91071.58680.33830.501110291.07Average1.74401.50710.28850.42328374.40
Notes:Abbreviation:Na=Observed number of alleles,Ne=Effective number of alleles,H=Nei's gene diversity (1973),I=Shannon's Information index.

Table 4 Genetic similarity coefficient (above diagonal) and genetic distanced (below diagonal) of B. fimbristipulata Hance
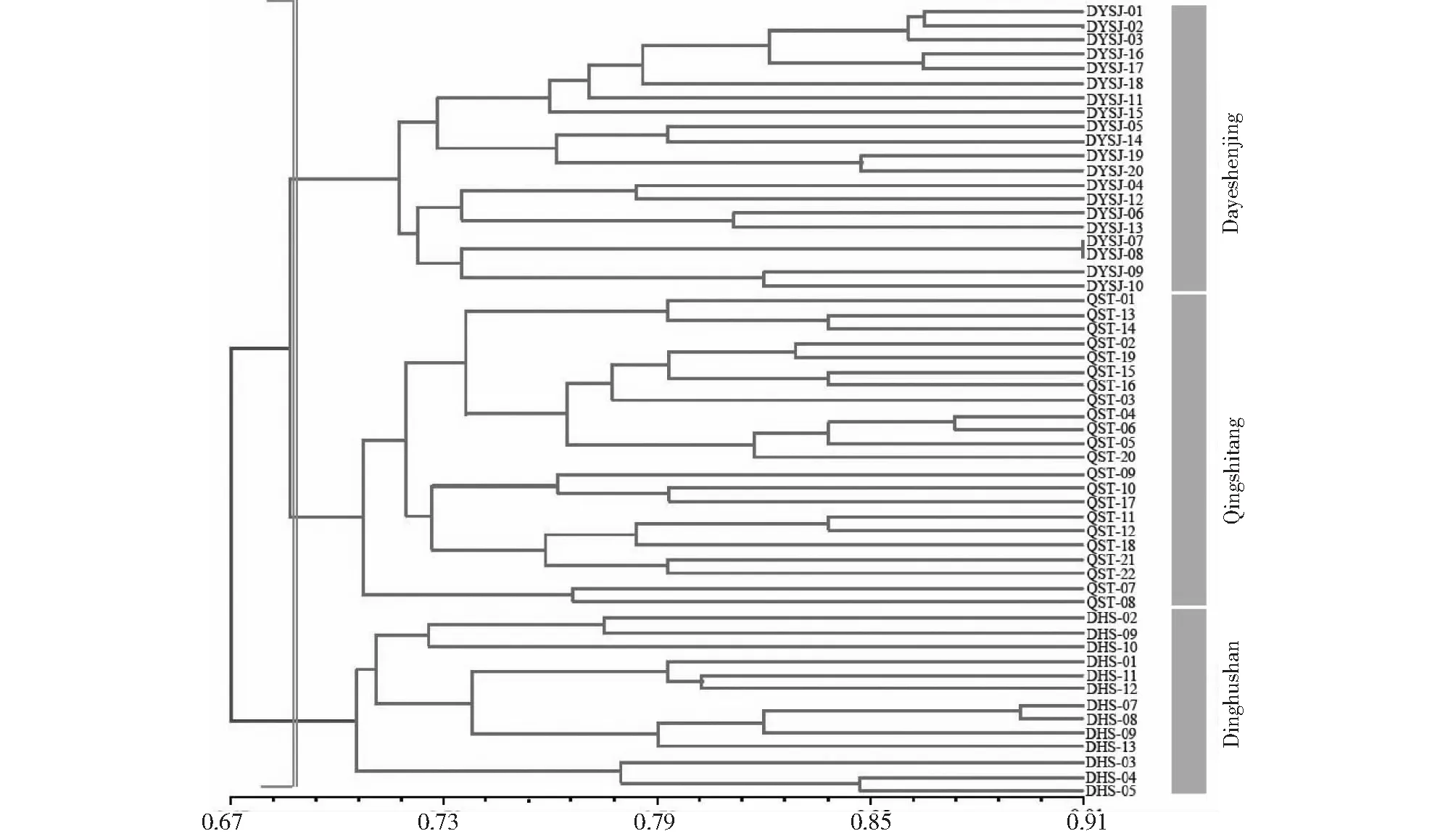
Fig.3 UPGMA Dendrogram of 3 pupulations of B. fimbristipulata Hance
3 Discussion
Assessment of the genetic variability within a wild population has important consequences in plant breeding and the conservation of genetic resources, and ISSR markers could offer an approach to reveal the genetic diversity among samples based on genetic polymorphisms. In this study, ISSR marks were applied for the first time to assess the level and pattern of genetic diversity of 55B.fimbristipulataHance samples from three different populations. Wide genetic variability among different population ofB.fimbristipulataHance was observed in the study (Table 3). A high percentage of polymorphic bands (up to 91.07 %), and the highest Shannon information index (0.4329) were found in the samples from Qingshitang. Meanwhile, the lowest values were recorded in the populations from Dayeshenjing (I=0.4120) (Table 3). These data suggest thatB.fimbristipulataHance samples from Qingshitang possess relatively higher genetic variation. The high genetic diversity could be attributed to a special geomorphic type, which has been shown to strongly correlate with the variation levels within a population[25]. Samples from Qingshitang mainly grew in the plain field, human activities have strongly contributed to the genetic differentiation. While theB.fimbristipulataHance plants from Dayeshenjing and Dinghushan grew in the mountain with an isolated topographic conditions. This geographical isolation might have a limit to seed and pollen transmission[26].
In a sense, the genetic structure of a population results from the environmental heterogeneity. The present study revealed molecular differentiation mainly occured within population and the results of cluster analyses corresponding to the latitude and longitude of the three populations. A low level of genetic differentiation among populations (Gst=0.1507), indirect estimates suggest that gene flow (Nm=1.4089) betweenB.fimbristipulataHance populations is limited. The limited gene flow detected in this study has been an important factor shaping the observed population structure and will influence future changes in these populations[27]. Therefore, efforts should be made to increase the levels of gene flow among populations to maintain the natural genetic variation. The richness of genetic diversity determines the ability to adapt and evolve, and it also provides very important information on the status of a species and conservation value[28].
4 Conclusion
Currently there are very limited resources of naturalB.fimbristipulataHance populations in China, and the most important aspect in protecting these resources is maintaining their genetic diversity. Hence, the information gained from the levels and distribution of ISSR variation in the threatenedB.fimbristipulataHance can be used to devise appropriate management strategies. Although the genetic diversity of wildB.fimbristipulataHance was high, the genetic variation mainly occurred intra-population due to the current status of habitat loss and fragmentation. When selecting the provenance for core conservation, we should paid special attention to the populations and individuals that come from special habitats or demonstrate irreplaceable uniqueness. In order to broaden the genetic bases, population with greater genetic differences from Qingshitang should be a priority to protect or as parent strain for cross-breeding, and these combinations might have potential for improving their genetic diversity.
[1]Hughes M,Hollingsworth P M. Population genetic divergence corresponds with species level biodiversity patterns in the large genusBegonia[J]. Molecular Ecology, 2008, 17(11): 2643-2651.
[2]Ku S M, Liu Y, Peng C I. Four new species ofbegoniasect.Coelocentrum(Begoniaceae) from limestone areas in Guangxi, China[J]. Bot. Stud. (Taipei, Taiwan), 2006, 47: 207-222.
[3]De Wilde J J F E, Van Valkenburg J L C H. Novitates Gabonenses 57.Begoniasosefiana(Begoniaceae): a new species in section Loasibegonia from Gabon[J]. Blumea, 2005, 50: 467-471.
[4]Thomas D C, Hughes M, Phutthai T, et al. A non-coding plastid DNA phylogeny of AsianBegonia(Begoniaceae): Evidence for morphological homoplasy and sectional polyphyly [J]. Molecular Phylogenetics and Evolution, 2011, 60(3): 428-444.
[5]Wang Y, Chen X W, Shao L, et al. Research Advances on the Ecological and Biological Characteristics of a Rare and Endangered PlantBegoniafimbristipulataHance[J]. Chinese Wild Resources, 2014, 33(6): 26-32.
[6]Wang H S, Cao Y M, Li G H, et al. Effects of extract ofBegoniafimbristipulataon the development of diabetic nephrophathy rats [J]. Chinese Journal of Biochemical Pharmaceutics, 2012, 33(3): 272-274.
[7]Shao L, Liang X. Biological Characteristics of BegoniaceaeBegoniafimbristipulataHance[J]. Journal of Agriculture, 2012, 2(8): 49-52.
[8]Wang Y, Yi H L, Shao L, et al. Survival and eco-biological characteristics ofBegoniafimbristipulataHance in the process of reintroduction[J]. Ecological Science, 2017, 36(2):32-41
[9]Zhao P J, Xiao J Z. Science of Chinese Wild Vegetable Resourses[M]. Beijing: China Environmental Science Press, 2006: 103.
[10]Guo Z, Xu L. Procyanidine: A wide development prospect botanical drug[J]. World Notes on Botanical drug, 1996, 11(5): 196-204.
[11]Tan X S, Zhou B H, Tang T X. Research for Preparation Technique ofBegoniafimbristipulaEffervescent Tablets[J]. Chinese Journal of Experimental Traditional Medical Formulae, 2011, 17(3): 53-55.
[12]XingF W. Rare plants of China [M]. Changsha: Hunan Education Press, 2005: 107.
[13]Li Y. Studies of the Surfactant Assisted Microwave Extraction of the Pigment fromBegoniafimbristipulataHance[J]. Journal of Instrumental Analysis, 2005, 24(4): 95-97.
[14]Cai D J, He Y, Yang J H. The extraction of red pigment frombegoniafimbristipulahance and the stability of the red pigment[J]. Food Sci. Technol., 2005(2): 48-51.
[15]Shao L, Liang G J, Lianglian,et al. Artificial Cultivation of Endangered PlantBegoniafimbristipulaHance on Dinghu Mountain[J]. Plant Physiology Journal Plant Physiology Journal, 2012, 48(10): 979-985.
[16]Shao L, Liang X. Biological Characteristics of BegoniaceaeBegoniafimbristipulataHance[J]. Journal of Agriculture, 2012, 2(08):49-52.
[17]Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR) - anchored polymerase chain reaction amplification[J]. Genomics, 1994, 20: 176-183.
[18]Sun Z Y, Yao H. ISSR analysis and identification onIsatisindigoticafrom different habitats[J]. Chinese Traditional and Herbal Drugs, 2014, 45(22): 3323-3326.
[19]Wei X Y, Tian Y X, Zhao Z L,et al. RAPD and ISSR analyses of genetic diversity ofAmericanginsenggermplasm from different habitats in China [J]. Chinese Traditional and Herbal Drugs, 2014, 45(21): 3153-3158.
[20]LiM, Wang P, Sun J K, et al. Study on Genetic Diversity of Twelve NaturalZanthoxylumdissitumPopulations[J]. Journal of Chinese Medicinal Materials, 2014, 37(12):2159-2163.
[21]Li X L, Yangjin, Zhang X, et al. Genetic Variation within Ten Accessions ofDipsacusin China by ISSR Analysis[J]. Bull Bot Res, 2015, 35(3):450-456.
[22]Doyle J J, Doyle J L, 1990. Isolation of plant DNA from fresh tissue. Focus. 1990, 12: 13-15.
[23]Yeh F C, Yang R C, Boyle T B J, et al. POPGENE, the user-friendly shareware for population genetic analysis[J]. Molecular Biology and Biotechnology Centre. University of Alberta, Canada. 1997.
[24]Rohlf F J. NTSYS-pc version 2.1, Numerical Taxonomy and Multivariate Analysis System[M]. Exeter Software, Setauket, New York. 2000.
[25]Hamrick J, Godt M. Effects of life history traits on genetic diversity in plant species[J]. Philos. Trans. Roy. Soc. B. 1996, 351: 1291-1298.
[26]Chen X L, Shao L, Liang G J, et al. A Study on Floral Syndrome and Breeding System ofBegoniafimbristipulaHance[J]. Acta Horticulturae Sinica, 2013, 40(2): 363-372.
[27]Wright S. The genetical structure of populations[M]. Galton Laboratory. Annals of Eugenics. London, England: Eugenics Society, 1951: 323.
[28]Ellstrand N C, Elam D R. Population genetics consequences of small population size: Im plications for plant conservation[J]. Ann. Review Ecol. Systemat, 1993, 34: 217.
1001-4829(2017)10-2224-06
2016-10-23
广西秋海棠属(秋海棠科)植物DNA条形码技术应用(2012GXNSFBA053075)
赵 博(1981-),女,河南西峡人,副研究员,主要从事植物细胞学与分子系统发育研究,E-mail:2052886016@qq.com,*为通讯作者:钟树华,(1958-),男,高级工程师,从事植物分类及保育研究,E-mail:zsh@gxib.cn。
紫背天葵遗传多样性的ISSR分析
赵 博1,2,毛世忠1,李景剑1,黄仕训1,钟树华1*
(1. 中国科学院广西植物研究所,广西 桂林 541006;2. 中国中医科学院中药研究所,北京 100700)
【目的】对野生紫背天葵资源的遗传多样性研究可为其育种、利用和保护提供参考依据。【方法】应用ISSR分子标记技术对广东肇庆鼎湖山、广西桂林大野神境、青狮潭3个紫背天葵野生居群的遗传多样性进行分析。采用 POPGEN32 进行数据分析,UPGMA 绘制聚类图。【结果】12条ISSR引物共检测到112个清晰的扩增位点,多态性位点102个,多态位点百分率为91. 07 %;Nei' s基因多样性指数(He)为0.3383,Shannon多样性指数(I)为0.5011,基因分化系数(Gst)为0.1507。居群间的遗传相似系数为0.8949~0.9162,平均为0.9026。聚类分析可知,各居群个体的聚类与其地理位置一致。【结论】紫背天葵具有较高的遗传多样性,但居群间的遗传分化水平较低,建议可将各种群后代种子或幼苗相互交叉移栽,以提高群体间的基因交流,增加遗传多样性水平。
紫背天葵;ISSR分子检测;遗传多样性
Q948
A
10.16213/j.cnki.scjas.2017.10.011
(Edited by LI Jie)
