二倍体马铃薯SSR遗传图谱的构建
2017-11-04孙小磊马伟清董道峰郭宝太杨晓慧李广存
朱 云,郭 晓,孙小磊,马伟清,董道峰,郭宝太,杨 煜,杨晓慧,李广存
(1.山东省农业科学院 蔬菜花卉研究所,山东省设施蔬菜分子生物学重点实验室,山东 济南 250100;2.青岛农业大学 生命科学学院,山东 青岛 266109)
二倍体马铃薯SSR遗传图谱的构建
朱 云1,郭 晓1,孙小磊1,马伟清1,董道峰1,郭宝太2,杨 煜1,杨晓慧1,李广存1
(1.山东省农业科学院 蔬菜花卉研究所,山东省设施蔬菜分子生物学重点实验室,山东 济南 250100;2.青岛农业大学 生命科学学院,山东 青岛 266109)
为了丰富马铃薯分子标记,定位青枯病抗性基因位点,利用具有抗青枯病遗传背景的二倍体马铃薯亲本USW5337.3(C)和772102.37(E)及其123份杂交一代无性系,进行了马铃薯SSR遗传图谱的构建。根据马铃薯全基因组序列设计了864对SSR引物,其中187对在亲本间表现出差异,135对(72.2%)能够在杂交一代中扩增出清晰的条带,最终构建了一张由12个连锁群组成,包含135个SSR标记的马铃薯分子标记遗传图谱。12个连锁群总长度为948.4 cM,标记间平均间距7.03 cM,含有5个标记偏分离区域。不仅为马铃薯饱和分子遗传图谱的构建提供新的SSR标记,对于马铃薯抗青枯病QTL定位、基因克隆和分子标记辅助选择也具有重要的参考意义。
二倍体;马铃薯;SSR;连锁图谱
Potato (SolanumtuberosumL.) is the fourth most important food crop of the world behind wheat,rice,and maize and is roughly half the world′s annual output of all root and tuber crops[1].Potato exhibits a considerable amount of variability for morphological traits and for resistance to insects and diseases.However,cultivated potato is an autotetraploid (2n=4x=48) which complicates both genetic/genomic studies as well as breeding efforts to improve important traits such as disease/pest resistance,processing quality and nutritional value[2].Multiple linkage maps of potato have been developed to better understand the potato genome,develop markers for marker assisted breeding,and facilitate map-based cloning[3-5].Most potato linkage maps have been generated from diploid populations which can simplify genetic segregation and incorporate polymorphism from wild species and primitive cultivars[6-8].
Potato linkage maps have been developed from many types of markers,including isozymes,restriction fragment length polymorphisms (RFLPs),amplified fragment length polymorphisms (AFLPs),simple sequence repeats (SSRs) and single nucleotide polymorphisms (SNPs)[9-11].DNA simple sequence repeats,also known as microsatellites,are tandem repeats of 2-6 bp DNA core sequences,which are widely distributed in both non-coding and transcribed sequences.With the advantages of being PCR-based,reliable,co-dominant,multi-allelic,chromosome specific,and highly informative,SSRs are useful for many applications in plant genetics and breeding such as construction of high-density linkage maps,genetic diversity analysis,cultivar identification,and marker-assisted selection.Recent studies have shown that SSRs can detect more polymorphism in plant species than RFLP,RAPD and AFLP[12-14].There is a growing need for saturating the genetic map with SSR.Early development of SSRs was hampered by the high cost of library screening and clone sequencing.
Currently,large public SSR datasets exist for many crop species,but the number of publicly available,mapped SSRs for potato was relatively low.With the release of potato genome and advances of techniques to enrich SSRs,the development of genomic SSR markers in potato could be accelerated[15-16].In this study,a set of new SSR markers were designed and a SSR-based potato linkage map was constructed using a diploid potato population which harbored the resistance toRalstoniasolanacearum.This may be useful for further map saturation which could more facilitates the QTL mapping,identification and cloning of resistance genes,and in marker assisted breeding for resistance against disease.
1 Materials and Methods
1.1Plantmaterials
An F1(n=123) progeny developed from a cross between two diploid (2n=2x=24) potato clones USW5337.3(C) and 772102.37(E) was used as the mapping population in this study.Clone C was a hybrid betweenSolanumtuberosumL.Group Phureja PI 225696.1 and theS.tuberosumGroup Tuberosum dihaploid USW42.Clone E was obtained from a cross between clone C and theS.vernei-S.tuberosumbackcross clone VH34211[17].Previous research showed C and E clones were resistant to bacterial wilt[18].Both clones C and E were used for the polymorphism analysis of SSR markers.Genomic DNA was extracted from young leaf tissues of 123 F1progeny individuals together with C and E using CTAB method[19].
1.2SSRmarkersandpolymorphismanalysis
SSR markers were designed from the potato genome assembly using PRIMER3 online (http://www.simgene.com/Primer3).A total of 864 primer pairs were selected.All primer pairs were initially screened against two clones C and E and a small offspring screening panel for polymorphism.The polymorphic markers were then used to genotype 123 individuals of the F1population for map construction.
Assessment of genetic polymorphism was performed by separation of PCR products using non-denaturing polyacrylamide gel electrophoresis.PCR reactions were prepared in a total volume of 15 μL containing 10×Taqpolymerase Buffer (500 mmol/L KCl,200 mmol/L Tris-HCl,pH 8.4)1.5 μL ,3.0 mmol/L MgCl20.9 μL,2.5 mmol/L dNTPs 0.3 μL,5 μmol/L each of forward and reverse primers 1.0 μL,1 UTaqpolymerase (Invitrogen Corporation,USA),and 20 ng of DNA template.The final volume was adjusted with sterile distilled water.PCR was performed in 96-well plates in GeneAmp PCR 9700 thermocycler (Applied Biosystems) programmed as follows:94 ℃ for 3 min,followed by 37 cycles of 94 ℃ for 30 s,55 ℃,53 ℃,50 ℃,48 ℃ for 50 s,72 ℃ for 45 s and a final extension step at 72 ℃ for 10 min.Amplified products were separated on 6% non-denaturing polyacrylamide gels (PAGE) in 0.5 Tris-borate EDTA Buffer (TaKaRa) and visualized by silver (1 mg/mL) staining.
1.3Mapconstruction
Data were collected in the F1mapping population for all segregating marker alleles,regardless of whether segregation was from one parent or both.The segregation types were divided into biparental (genotype:ab×cd,ef×eg and hk×hk),maternal (genotype:lm×ll) and paternal (genotype:nn×np) datasets.A cross-pollinated (CP) population type was used to develop the linkage map in Joinmap 4.0[20].The goodness-of-fit between observed and expected Mendelian ratios was analyzed for each marker locus using a χ2test.Markers that deviated from the theoretical expected ratios (1∶1) were considered distorted and were marked to indicate different significance levels (P<0.05,P<0.01,P<0.001,andP<0.000 1).Markers assigned to linkage groups had a minimum LOD score of 3 and a maximum of 10.
2 Results and Analysis
2.1Identificationofpolymorphicmolecularmarkers
A total of 864 SSR primers pairs from potato were screened for polymorphisms,and 187 out of 864 SSR primers showed to be polymorphic between the parents C and E.After amplifying them on the entire map population individuals,52 gave monomorphic amplifications (no segregation) or produced unclear amplifications,resulting in difficulties in band scoring and thus were subsequently discarded.The remaining 135 SSR markers (Tab.1) were used for the linkage map construction.Four separation types were generated as nn×np,lm×ll,ef×eg,and hk×hk respectively (Fig.1).
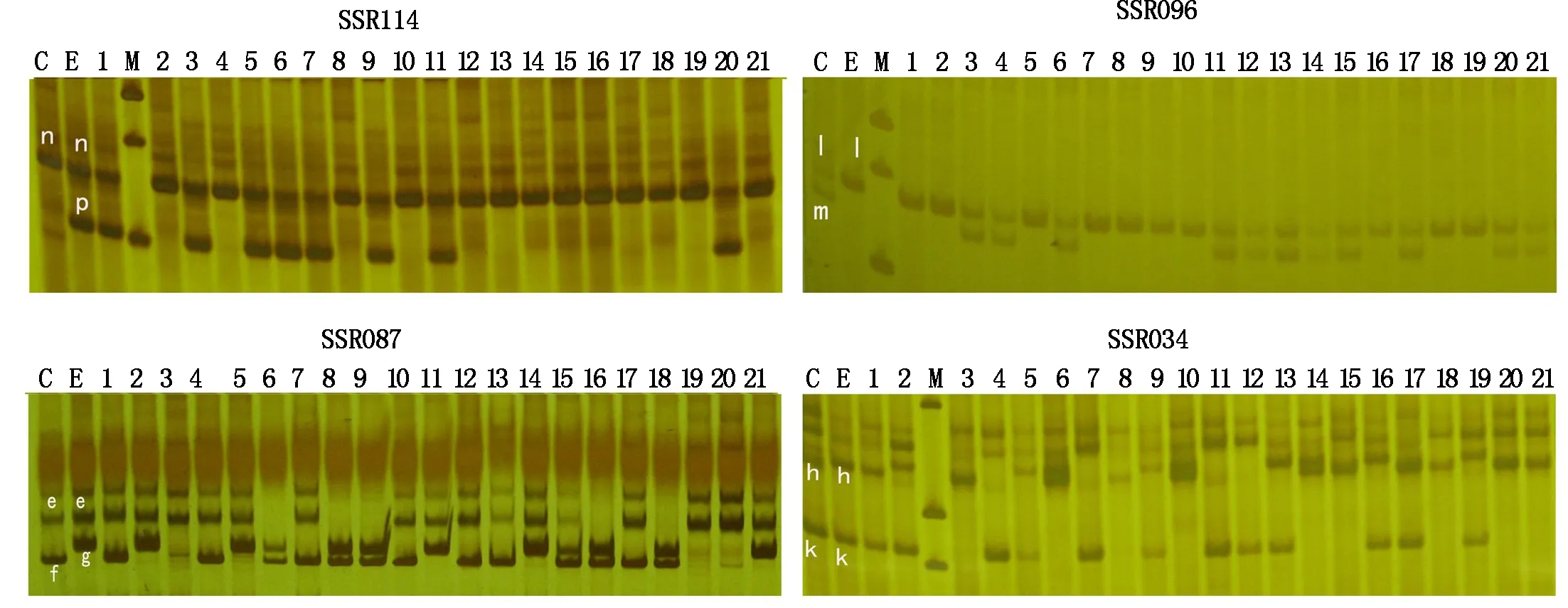
1-21.Eighteen potato individuals of F1 progeny.The different segregation types of SSR114,SSR096,SSR087 andSSR034 are nn×np,lm×ll,ef×eg,hk×hk,respectively.
2.2Mapconstruction
One hundred and thirty-five loci had been assigned to positions on 12 linkage groups (LGs),with between 6 and 20 loci per group (Tab.1).The length of each linkage group varied from 27.8 to 132.8 cM,with three linkage groups (LG1,LG9,and LG11) having a size over 100 cM.The total map distance was 948.4 cM,with an average marker interval of 7.03 cM between adjacent marker loci (Tab.1).
A total of nine gaps between two adjacent markers 20 cM existed on the maps and varied from 20.1 to 42.2 cM,with one gap on LG1 (20.4 cM between SSR069 and SSR072),LG4 (35.1 cM between SSR162 and SSR116),LG8 (20.3 cM between SSR033 and SSR090),and LG12 (42.2 cM between SSR006 and SSR005),2 gaps on LG11(32.7 cM between SSR173+and SSR163+,36.5 cM between SSR137 and SSR089),and 3 gaps on LG9 (27.7 cM between SSR049 and SSR001,20.1 cM between SSR156+and SSR134,and 20.2 cM between SSR134 and SSR154),respectively.The largest gap was 42.2 cM for SSR006-SSR005 on LG12 (Fig.2).

Tab.1 The characteristics of 12 linkage groups constructed by SSR markers in potato linkage group

Marker positions(cM)are in the left of the linkage group; The marker names are in the right.
2.3Segregationdistortion
Distorted segregation ratios of genetic markers were often observed in progeny of inter- and intra-specific hybrids and might result from competition among gametes or from abortion of the gamete or zygote.In this study,135 polymorphic markers on the linkage map were surveyed for distorted segregation ratios.Among those,40 markers (29.6%) showed a significant segregation distortion (P<0.05),favoring either the marker alleles of female parent C (52.5%) or male parent E (35.0%) or both parents (12.5%) (Tab.2).

Tab.2 Chi-square test for segregation distortion of markers in F1 population

Tab.2(Continued)
Note:*at 0.05 significant level;More than* at 0.01 extremely significant level.
Segregation distortion marker distribution along the present molecular maps of potato was far from uniform,with clusters of tightly linked loci and single marker.In this linkage map,five segregation distortion regions (SDRs,clusters of 3-7 markers) were detected on the linkage groups; LG1,LG3,and LG10 (Fig.3).Among these five SDRs,SDR-1 skewed to parent C,SDR-2 skewed to parent E,and SDR-3,SDR-4,SDR-5 skewed to C and E.The segregation distortion ratio of LG1,LG3,LG10 are 68%,100%,50%,separately.
3 Discussion
This manuscript reported the construction of an SSR-based map of diploid potato.The primary goal for the construction of this map was to develop a framework for future improvement.The application value of a genetic linkage map depends on its marker numbers,saturation,accuracy and the uniformity of marker distribution.Except for an elementary chromosome draft,the marker intervals of <20 cM are necessary for major gene location,and 1-10 cM for QTL positions and gene cloning.In this study,the total map spanned 948.4 cM,with an average marker interval of about 7.03 cM between two adjacent marker loci.This new SSR-based linkage map contained almost twice as many loci as did the previous linkage map which was constructed using 86 SSR markers[21].Moreover,the map population was derived from the cross of C and E clones conferring high resistance to bacterial wilt disease.This map will be a useful resource and tool,in which SSR markers may be used for future map saturation,QTL studies and other potato improvement.

Marker positions(cM)are in the left of the linkage group;The marker names are in the right.*Marker are declared with distorted segregation at the 0.05 level of significance and more than** at 0.01 significant level.
Allele distorted segregation is a common phenomenon in organisms.It is considered to be one of the driving forces of biological evolution.Distorted segregation ratios of genetic markers are often observed in progeny of inter- and intra-specific hybrids and might result from competition among gametes or from abortion of the gamete or zygote[22].Additionally,the mapping population,environmental factors,non-homologous chromosome recombination,gene jumping,hitchhiking,transposons,the genetic purity of parents,etc.could be the reasons of distorted segregation[23].Segregation distortion marker distribution along the present molecular maps of potato was far from uniform,with clusters of tightly linked loci and single marker.In this linkage map,five segregation distortion regions (SDRs,clusters of 3-7 markers) were detected on 3 linkage groups,indicating that the possible causes for segregation deviation of molecular markers were based on gametic selection.
Acknowledgments:This work was supported by the Natural Science Funds of Shandong Province (ZR2014YL014,ZR2016CM27),and the Youth Scientific Research Foundation of Shandong Academy of Agricultural Sciences (2014QNZ03).Special thanks to Professor Richard G F Visser (Wageningen University) for kindly providing the potato materials used in this research.
[1] Visser R F,Bachem C B,De Boer J M,et al.Sequencing the potato genome:outline and first results to come from the elucidation of the sequence of the world′s third most important food crop[J].American Journal of Potato Research,2009,86(6):417-429.
[2] Howard H W.Genetics of the potato,Solanumtuberosum[M].London: Logos Press,1970.
[3] Jacobs J ,Van Eck H J,Arens P,et al.A genetic map of potato (Solanumtuberosum) integrating molecular markers,including transposons,and classical markers[J].Theoretical and Applied Genetics,1995,91(2):289-300.
[4] Danan S,Veyrieras J B,Lefebvre V.Construction of a potato consensus map and QTL meta-analysis offer new insights into the genetic architecture of late blight resistance and plant maturity traits[J].BMC Plant Biology,2011,11:16.
[5] Prashar A,Hornyik C,Young V,et al.Construction of a dense SNP map of a highly heterozygous diploid potato population and QTL analysis of tuber shape and eye depth[J].Theoretical and Applied Genetics,2014,127(10):2159-2171.
[6] Gebhardt C,Ritter E,Salamini F.DNA-based markers in plants:RFLP map of the potato[M].Netherlands:Springer,2001:271-285.
[7] Menéndez C M,Ritter E,Schäfer-Pregl R,et al.Cold sweetening in diploid potato:mapping quantitative trait loci and candidate genes[J].Genetics,2002,162(3):1423-1434.
[8] Jin L,Liu J,Jong H D,et al.Construction of a molecular linkage map inSolanumtuberosumL.[J].Acta Horticulturae Sinica,2007,34(2):397-402.
[9] Van Os H,Andrzejewski S,Bakker E,et al.Construction of a 10,000-marker ultradense genetic recombination map of potato:providing a framework for accelerated gene isolation and a genomewide physical map[J].Genetics,2006,173(2):1075-1087.
[10] Anithakumari A M,Tang J,Van Eck H J,et al.A pipeline for high throughput detection and mapping of SNPs from EST databases[J].Molecular Breeding :New Strategies in Plant Improvement,2010,26(1):65-75.
[11] Felcher K J,Coombs J J,Massa A N,et al.Integration of two diploid potato linkage maps with the potato genome sequence[J].PLoS One,2012,7(4):e36347.
[12] He G H,Meng R H,Gao H,et al.Simple sequence repeat markers for botanical varieties of cultivated peanut (ArachishypogaeaL.) [J].Euphytica,2005,142(1/2):131-136.
[13] Barkley N A,Dean R E,Pittman R N,et al.Genetic diversity of cultivated and wild-type Peanuts evaluated with M13-tailed SSR markers and sequencing[J].Genetical Research,2007,89(2):93-106.
[14] Hong Y,Chen X,Liang X,et al.A SSR-based composite genetic linkage map for the cultivated peanut (ArachishypogaeaL.) genome[J].BMC Plant Biology,2010,10:17.
[15] Wang Y W,Samuels T D,Wu Y Q.Development of 1,030 genomic SSR markers in switchgrass[J].Theoretical and Applied Genetics,2011,122(4):677-686.
[16] Zhang H,Wei L,Miao H,et al.Development and validation of genic-SSR markers in sesame by RNA-seq[J].BMC Genomics,2012,13:316.
[17] Jacobsen E.Die chromosomen-verdopplung in der zuchtung dihaploider kartoffeln[D].Bonn:Rheinische Friedrich-Wil-helm-Universitat,1978.
[18] Qu D Y.Use of unreduced gametes of potato (SolanumtuberosumL.) for true potato seed production through 4x-2x crosses[D].Wageninger,Netherlands:Wageningen University,1996.
[19] Chen J,Hu X,Miao H,et al.Genome DNA extracted with CTAB method and its use for SSR and SRAP[J].Journal of Peanut Science,2008,37(1):29-31.
[20] Van Ooijen J.JoinMap®:software for the calculation of genetic linkage maps in experimental populations[M].The Netherlands:Kyazma BV,2006.
[21] Shan Y,Liu J,Bian C,et al.Construction of genetic map by SSR markers and QTL analysis of 3 important agronomic traits in diploid potato[J].China Vegetables,2010,18:10-14.
[22] Kozielska M,Weissing F J,Beukeboom L W,et al.Segregation distortion and the evolution of sex-determining mechanisms[J].Heredity,2010,104(1):100-112.
[23] Tang D,Guo L B,Zeng D L,et al.Genetic analysis of two extremely segregation distorted populations in rice (OryzasativaL.) [J].Hereditas,2006,28(10):1259-1264.
ConstructionofanSSR-basedGeneticLinkageMapofDiploidPotato
ZHU Yun1,GUO Xiao1,SUN Xiaolei1,MA Weiqing1,DONG Daofeng1,GUO Baotai2,YANG Yu1,YANG Xiaohui1,LI Guangcun1
(1.Vegetable and Flower Institute,Shandong Academy of Agricultural Sciences,Shandong Key Laboratory for Biology of Greenhouse Vegetables,Jinan 250100,China;2.College of Life Sciences,Qingdao Agricultural University,Qingdao 266109,China)
In order to enrich molecular markers and position the bacterial wilt resistant gene loci in potato,an SSR-based genetic linkage map was constructed from a diploid population consisting of 123 individuals derived from a cross between two potato diploid clones (C and E) which harborer resistance toRalstoniasolanacearumin this study.A total of 864 simple sequence repeat primer pairs were designed according to the potato genome sequence and used to screen parents C and E for polymorphisms.Of 187 polymorphic loci,72.2% showed clear amplifiable and scorable bands in the F1population.Then a total of 135 locus were mapped to 12 linkage groups,of which the cumulative length was 948.4 cM,with an average marker interval of 7.03 cM,and 5 segregation distortion regions in three linkage groups.This study provided a set of new SSR markers for genetic map construction and increased saturation of map in potato.The relatively high density map may provide a useful reference for further identification,mapping of quantitative trait loci (QTL) and cloning of potato bacterial wilt resistance genes and marker-assisted breeding.
Diploid;Potato;Simple sequence repeat;Linkage map
2017-08-09
山东省自然科学基金项目(ZR2014YL014;ZR2016CM27);山东省农业科学院青年科研基金项目(2014QNZ03)
朱 云(1987-),女,安徽马鞍山人,硕士,主要从事马铃薯抗病分子生物学研究。朱云、郭晓为同等贡献作者。
杨晓慧(1980-),女,山东聊城人,助理研究员,博士,主要从事马铃薯分子育种研究。
S532.03
A
1000-7091(2017)05-0117-07
10.7668/hbnxb.2017.05.018
