甘蓝型油菜polCMS育性恢复位点的全基因组关联分析
2017-10-14魏大勇谭传东崔艺馨吴道明李加纳梅家琴钱伟
魏大勇,谭传东,崔艺馨,吴道明,李加纳,梅家琴,钱伟
甘蓝型油菜CMS育性恢复位点的全基因组关联分析
魏大勇,谭传东,崔艺馨,吴道明,李加纳,梅家琴,钱伟
(西南大学农学与生物科技学院/重庆市油菜工程技术研究中心,重庆400716)
【目的】甘蓝型油菜波里马细胞质雄性不育(CMS)在中国已被广泛应用于杂交种育种,其育性恢复程度表现出受1对主效基因的控制,并受微效修饰基因的影响。通过全基因组关联分析方法挖掘育性恢复位点,并对候选基因进行比较分析。【方法】通过芸薹属60K SNP芯片对308份甘蓝型油菜自然群体进行基因型分型,并用CMS系301A作母本,与上述材料分别进行杂交得到308份F1,每份F1分别于2013年和2014年进行种植,每年2次重复,于始花期根据花粉育性和花蕊发育情况调查F1植株的育性等级,同时对测交父本自然群体进行群体结构分析和亲缘关系评估,并结合测交父本的基因型分型结果和F1的育性等级进行全基因组关联分析(GWAS)。从GWAS分析中显著的SNP左右100 kb区间或与显著SNP处于同一单体型块(2>0.5)的区间内预测候选基因,并对候选基因进行QTL比较分析和单体型或等位基因的效应分析。【结果】方差分析结果显示,两年F1的育性等级存在显著差异(<0.01),但相关分析发现,两年的育性等级存在显著的正相关(= 0.52,<0.001)。群体结构分析显示,所有测交父本被分为3个亚群(冬性、春性和半冬性),亲缘关系分析发现,任何2个材料之间平均亲缘关系值为0.072,73%的任意材料间亲缘关系值小于0.1,其中,约53%的材料亲缘关系值为0。GWAS分析共检测到13个与育性恢复程度显著关联的SNP,构成了6个候选区间,分别位于A01、A09、C03、C06和C08 5条染色体上,单个SNP解释的表型变异介于2.53%—9.96%。从中共预测到6个与育性恢复位点相关的候选基因,其中4个编码的蛋白含有恢复基因特有的PPR保守基序。共线性分析发现,4个候选基因中的2个(和)位于A09和C08染色体部分同源区间,且与已克隆的CMS育性恢复位点同源。另外2个新鉴定到的候选基因(和)连锁的SNP等位基因或单体型变化都与育性等级显著相关(<0.001)。【结论】通过GWAS分析鉴定到多个与油菜育性恢复有关的候选基因,开发基于与这些基因连锁位点或SNP的功能标记将有助于对该不育系统进行恢复系和保持系的筛选。
甘蓝型油菜;波里马细胞质雄性不育;育性恢复基因;全基因组关联分析;SNP
0 引言
【研究意义】雄性不育在植物界普遍存在,早在1763年,德国植物学家Kölreuter观察到雄性不育现象。目前,雄性不育主要包括由线粒体基因和核基因共同控制的细胞质雄性不育(cytoplasmic male sterility,CMS)和由核基因单独控制的细胞核雄性不育(genic male sterility,GMS)两种[1]。油菜是世界上继大豆和油棕之后的第三大油料作物,菜油是中国主要食用油之一。作为第一个有实用价值的油菜细胞质雄性不育类型,波里马细胞质雄性不育(Polima CMS)已被广泛应用于油菜杂交种的制种[2]。早前关于甘蓝型油菜CMS恢复基因的研究多数是基于分离群体构建的连锁图谱进行QTL定位,费时费力。随着甘蓝型油菜参考基因组的释放和高通量测序成本的不断降低,新型技术比如高通量SNP芯片的出现,加快了候选基因识别的进程。因此,本研究通过全基因组关联分析快速挖掘影响CMS的育性恢复位点,对加快杂种油菜的育种进程具有重要现实意义。【前人研究进展】由于CMS在油菜育种中的广泛应用,恢复基因()的定位越来越受到关注。前人对育性恢复性状的遗传分析说明,该性状受1对显性基因控制[3]。由于在白菜型油菜(AA)、芥菜型油菜(AABB)和甘蓝型油菜(AACC)中都发现了,因此,推测可能位于A组染色体[4]。蔡强[5]通过连续回交构建的CMS育性恢复基因近等基因系群体,将与恢复基因连锁的标记定位在N9连锁图上,位于分子标记pW123bE和CNU008之间;Li等[6]找到一个与遗传距离为0.2 cM的SSR标记KBrDP1,并与DH系遗传连锁图整合,将定位在9号连锁群上;Liu等[7]将CMS育性恢复位点定位在白菜()的A09染色体29.2 kb内,并预测区间内一个开放阅读框为候选位点;Liu等[8]验证了上述开放阅读框,并且发现是通过减少的表达来恢复油菜CMS的育性。截止到目前,已在七大作物中克隆了13个植物育性恢复基因,分别为玉米的[9-10],矮牵牛的[11],萝卜的和[12-14],水稻的()[15-18]、[18]、[19]、[20]、[21]和[22],高粱的[23],甜菜的()[24-25]以及甘蓝型油菜的[8]。除了玉米的、水稻的和甜菜的外,其他10个恢复基因的编码蛋白都含有PPR(pentatrieopeptide repeat)基序。PPR基序是由Small和Peeters于2000年发现和命名的,由35个氨基酸组成的序列单元经串联重复排列而成的一个基因家族[26]。大部分PPR蛋白N端具有线粒体和叶绿体定位序列,是研究植物核质互作的理想模型[27]。目前研究倾向于认为,恢复基因的功能是通过抑制线粒体基因组中CMS相关嵌合基因的表达,来抑制或消除雄性不育的毒害效应,但是具体机制仍不清楚。由于多个物种的恢复基因中存在PPR保守基序,因此,PPR特征可以作为鉴定植物恢复基因候选基因的有效手段。【本研究切入点】尽管油菜CMS育性恢复主效基因已被鉴定,但育性恢复仍存在微效多基因的影响,因此,本研究采用CMS系301A作母本,与308份甘蓝型油菜自然群体分别进行杂交得到308份F1,通过甘蓝型油菜自然群体(测交父本)的SNP芯片数据对308份F1的育性等级进行全基因组关联分析。【拟解决的关键问题】本研究通过GWAS期望寻找更多的甘蓝型油菜CMS育性恢复位点或基因(包括微效基因),并通过等位基因或单体型效应分析,寻找与育性相关的SNP位点,为以后的功能标记开发奠定基础,应用于该不育系统恢复系和保持系的筛选鉴定。
1 材料与方法
1.1 供试材料和表型测定
甘蓝型油菜CMS 301A作母本,与308份不同来源的甘蓝型油菜分别杂交产生308份F1,分别于2013年和2014年播种在重庆市油菜工程技术研究中心试验地(重庆北碚),3行区播种,每行10株,每年2次重复。于始花期(每个株系有一半植株至少开花3朵)观察5株长势一致F1植株的花粉育性和雄蕊/雌蕊发育情况,统计每个F1的育性等级用于后续分析。同时也记录了始花期当天和前10天的日平均温度。参考杨光圣等[4]方法,根据花粉的多少和雄蕊/雌蕊发育情况划定材料育性等级:1级,花药发育正常,大量花粉;2级,花药发育基本正常,中量花粉;3级,花药退化成三角形,雄蕊低于雌蕊,少量花粉;4级,花药退化成三角形,雄蕊明显低于雌蕊,极少花粉;5级,花药退化成三角形且呈乳白色,雄蕊明显低于雌蕊,没有花粉。其中1级和2级为可育,4级和5级为不育,3级为部分可育(图1)。
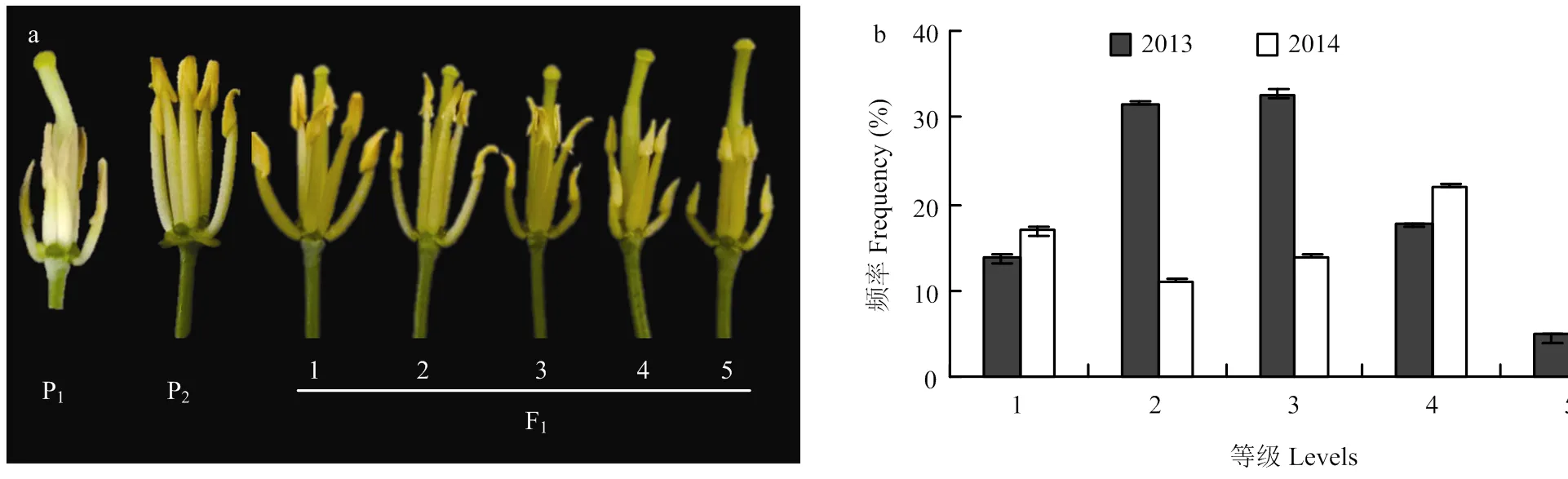
a:F1育性等级的划分依据,P1为不育母本,P2为可育父本;b:2013和2014年育性等级的频率分布
1.2 SNP基因型分型和数据过滤
于苗期选择鲜嫩叶片提取总DNA,提取及纯化使用TIANGEN®植物基因组DNA提取试剂盒(DP305),浓度最后统一稀释成100 ng·μl-1,-20℃保存备用。采用Illumina公司开发的60K SNP芯片对上述材料进行基因型分型[28]。将芯片数据得到的52 157个SNP位点与法国公布的甘蓝型油菜品种“Darmor- Bzh”的基因组v4.1(http://www.genoscope.cns.fr/brassicanapus/data/)进行本地Blastn,比对的阈值设为e-10。比对结果参考Schiessl等[29]的条件过滤:序列一致性(identity)>95%,没有gaps,比对长度(alignment length)>49 bp。
1.3 群体结构和亲缘关系评估
群体结构分析采用STRUCTURE v2.3.4[30],亚群数目k设置为1到9 ,5次模拟运算,蒙特卡罗迭代(MCMC)和模拟参数迭代(length of bum in period)都设置为1Í105次循环,在混合模型和频率相关模型下进行独立运算。最后输出的k值通过后验概率值结果LnP (D)和2个连续的后验概率值的变化速率Dk来矫正并确定群体中存在的最优类群数目[31]。数据的可视化通过基于R语言的SelectionTools包(http://www. uni-giessen.de/population-genetics/downloads)来实现。利用SPAGeDi v1.4软件[32]对甘蓝型油菜自然群体进行亲缘关系(relative kinship)评估。
1.4 全基因组关联分析和候选基因预测
利用R语言的GenABEL包进行GWAS分析[33],采用PCA + K的混合线性模型对性状和标记进行关联位点的检测,阈值设定为<4.25×10-5(1/所使用的标记,-log10= 4.37)。结果通过Manhattan图和Q-Q图显示[34]。通过R语言的p.adjusted命令计算假阳性率(false discovery rate,FDR)。将显著SNP位置左右各延伸100 kb或者与显著SNP处于同一单体型块(r2>0.5)的区间,定义为候选关联区间,在此区间参考以下条件预测候选基因:1)在甘蓝型油菜或拟南芥参考基因组上与性状相关的已知功能的基因;2)SNP直接落在基因内部;3)参考已报道QTL定位的结果。
每个SNP或单体型解释的表型变异采用SAS软件的proc glm进行计算,所用模型为y = geno,其中,y为表型观测值,geno为SNP或单体型的基因型。
2 结果
2.1 甘蓝型油菜F1的育性等级分析
前期研究发现,CMS系301A属于低温敏感型,持续低温易形成微粉,所有父本材料不受环境影响,都正常可育。301A与308份正常可育的甘蓝型油菜分别杂交所得F1,育性调查结果方差分析显示,两年的F1育性等级存在显著差异(<0.01),说明环境对育性有一定的影响。但相关分析发现,两年的育性等级存在显著的正相关(= 0.52,<0.01)(图1)。考虑到CMS系301A对温度的敏感,记录调查育性等级当天及前10天的日平均温度,相关性结果显示,育性等级与始花期前10 d的日平均温度存在显著的负相关(=-0.41—-0.20,<0.01),但和始花期当天的日平均温度没有显著的相关性(2013=-0.039,2013=0.338;2014=-0.082,2014=0.526),说明该育性恢复位点受雄蕊分化期间温度的影响但不受调查时温度的影响(电子附表1)。
2.2 SNP评价和分布
利用Illumina公司的GenomeStudio软件对308份甘蓝型油菜的60K芯片进行基因型分型。参考Schiessl等[29]方法,剔除没有定位到甘蓝型油菜参考基因组和定位在random染色体上的标记,同时删除MAF小于0.05和缺失加杂合大于25%的标记,最后剩下23 489个定位到唯一染色体上的SNP标记,用于后续分析(电子附表2,http://pan.baidu.com/s/ 1mhVbk04)。
通过SNP在基因组上的分布发现,SNP数目最多的是C04染色体,占10.3%(2 425个SNP);最少的是C09染色体,占2.80%。A和C亚基因组平均每条染色体分布1 093和1 396个SNP,结合甘蓝型油菜A、C亚基因组的大小得出,A亚基因组SNP的密度(平均每100 kb有4.6个SNP)是C亚基因组的1.5倍,说明A亚基因组发生了更多的重组交换,这可能是由于油菜从欧洲引入亚洲后导入了油菜的亲本之一白菜的遗传成分,拓宽了油菜A亚基因组的遗传多样性。
2.3 群体结构和亲缘关系
从19条染色体上均匀选取5 700个SNP(MAF>0.05)用于群体结构和亲缘关系的估测。群体结构的亚群通过独立的k值无法确定,因为LnP(D)值随着k值的增加而增大,没有出现拐点,因此,采用Evanno等[35]方法计算Δk值,Dk在k = 3时出现峰值(图2-a)。所有308份父本最后被分为3个亚群,亚群1主要由冬性材料构成,亚群2主要是春性材料,而半冬性材料主要构成了亚群3(图2-b),该结果与生态型的来源一致。
亲缘关系分析发现,任何2个材料之间平均亲缘关系值为0.072,73%的任意材料间亲缘关系值小于0.1,其中,约53%的材料亲缘关系值为0(图2-c)。以上结果表明所用材料之间的亲缘关系较远,适合GWAS的研究。
2.4 全基因组关联分析和QTL比较
为了消除年度间的环境影响,采用Merk等[36]方法对2年的表型数据进行最佳线性无偏预测(best linear unbiased prediction,BLUP),估计育性等级的BLUP值,并结合SNP基因型数据采用基于R语言GenABEL包的PCA + K混合线性模型进行GWAS分析。
GWAS分析共检测到13个SNP与育性等级显著关联(<4.26×10-5,-log10>4.37),分布在A01、A09、C03、C06和C08 5条染色体上,单个位点解释的表型变异介于2.53%—9.96%,Q-Q图显示该模型很好地控制了假阳性概率的产生(图3,表1)。根据位点间的连锁不平衡(r2>0.5),检测到的13个SNP被分为6个候选区间,预测到6个与育性相关的候选基因,其中4个候选基因的编码蛋白含有育性恢复基因特有的PPR(pentatricopeptide repeat)保守基序(表1)。
A09染色体上显著SNP(Bn-A09-p34393068和Bn-scaff_16445_1-p932699)构成的候选区间与C08染色体显著SNP(Bn-A09-p34437367)所对应的候选区间有极高的共线性(图3),且都定位在远古祖先染色体核型A block(24个祖先染色体核型block之一,拟南芥对应位置在chr1:202 136—204 189,基因从到)。同时该区间与已报道的A09染色体上1个CMS恢复基因精细定位区间重叠,候选基因都对应同一个拟南芥基因,进一步验证了该分析方法的准确性。
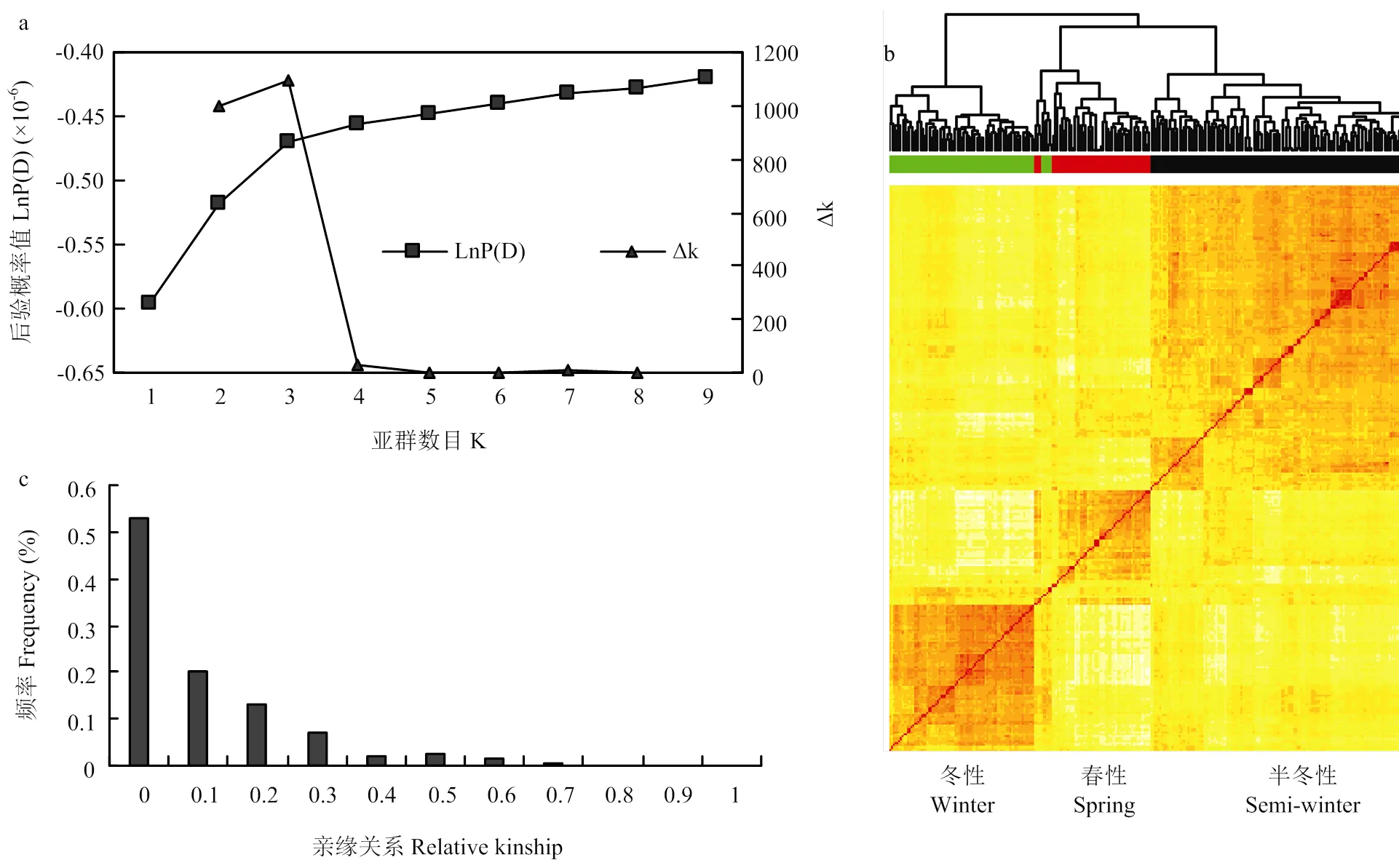
a:后验概率LnP(D)估计值和Δk值,k值取1到9;b:聚类结果;c:亲缘关系的分布
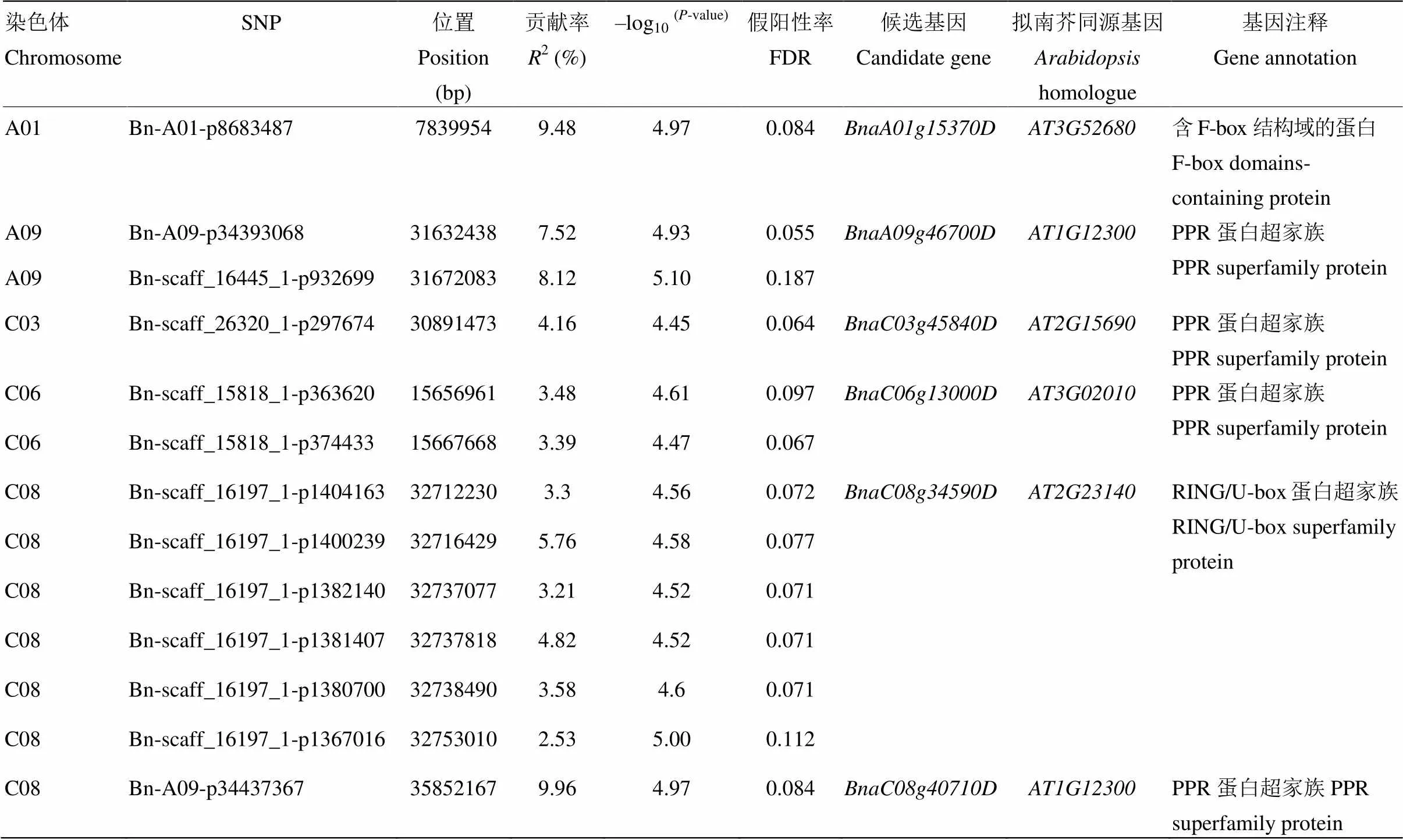
表1 GWAS结果和候选基因预测
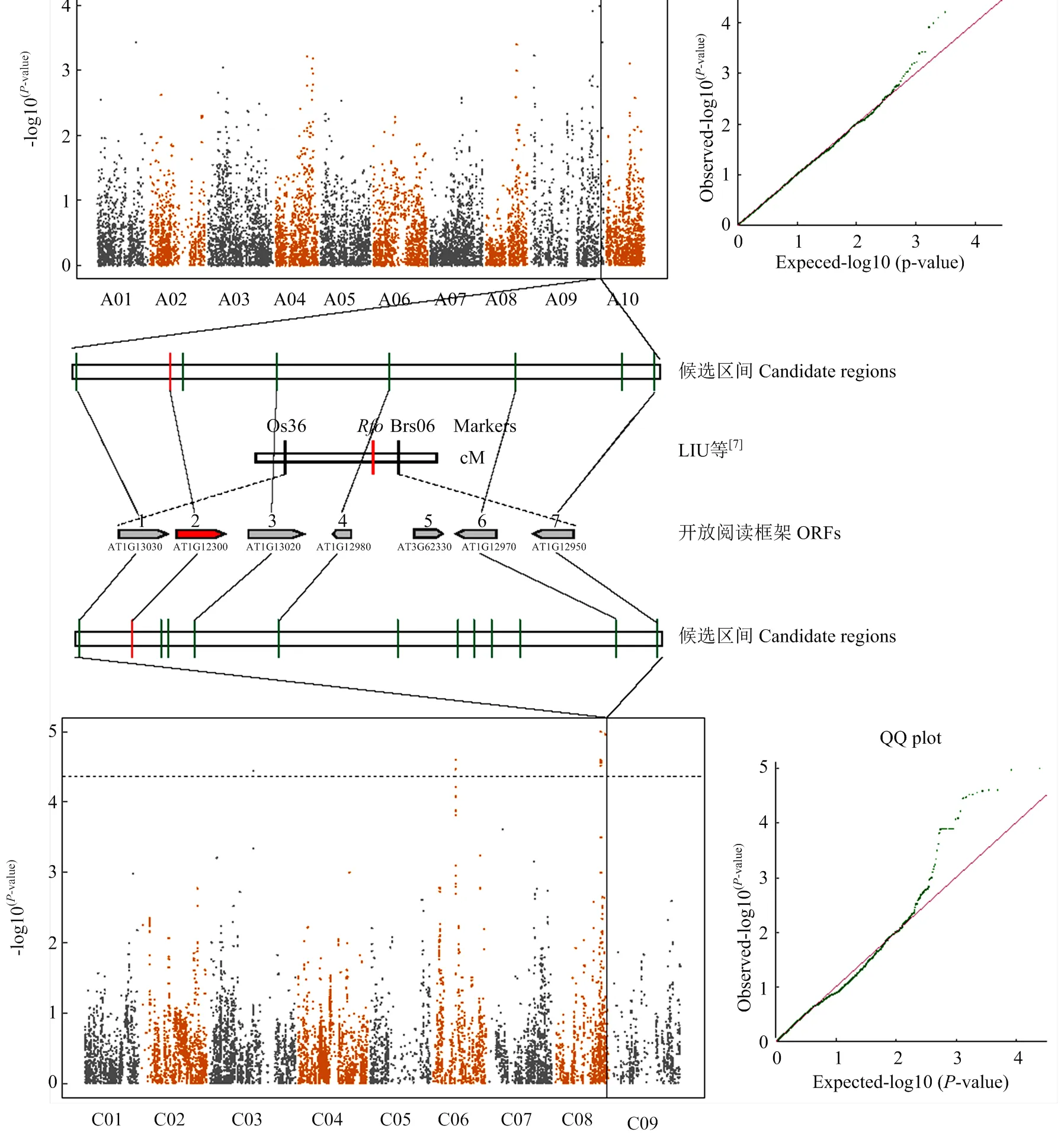
图中横的虚线代表阈值(1/23490,-log10p = 4.37),竖的虚线代表在A09和C08部分同源区间共定位的SNP。红色标记表示本研究和Liu等[7]共同预测的候选基因
2.5 单体型效应
除了上述2个候选基因与已有报道的结果一致,另外2个含有PPR保守基序的候选基因未发现相对应的QTL报道,接下来对这两个候选区间进行单体型效应分析。C03染色体上候选基因位于显著SNP(Bn-scaff_26320_1-p297674)上游45 kb处,该区域SNP平均2大于0.8,构成一个单体型块,所有7个SNP构成5种单体型,单体型效应分析发现,68.9%的材料具有单体型G-T-A-T-G-A-G,平均育性等级为3.9±1.44,20.1%的材料具有单体型A-C-C-C-N-C-A,平均育性等级为2.1±1.53,剩下3种单体型共占10.5%,2种比例最大的单体型与育性等级显著关联(<0.001)。C06染色体候选基因位于显著SNP(Bn-scaff_15818_1- p374433)下游100 kb处,编码的蛋白同样含有PPR保守基序,该100 kb区间内所有SNP处于极高的LD(r2>0.9)(图4-a),只存在2种单体型,通过一个位点的效应分析可以预测整个区间的效应。随后对候选基因上游最近的1个SNP(Bn-scaff_15818_1-p471106,上游2.9 kb)等位基因(C/T)效应分析发现,当SNP位点从CC变为TT时,可育材料(1和2级)增加了71.7%,不育材料(4和5级)降低了37.8%(图4-b),该位点与育性等级显著关联(<0.001)。
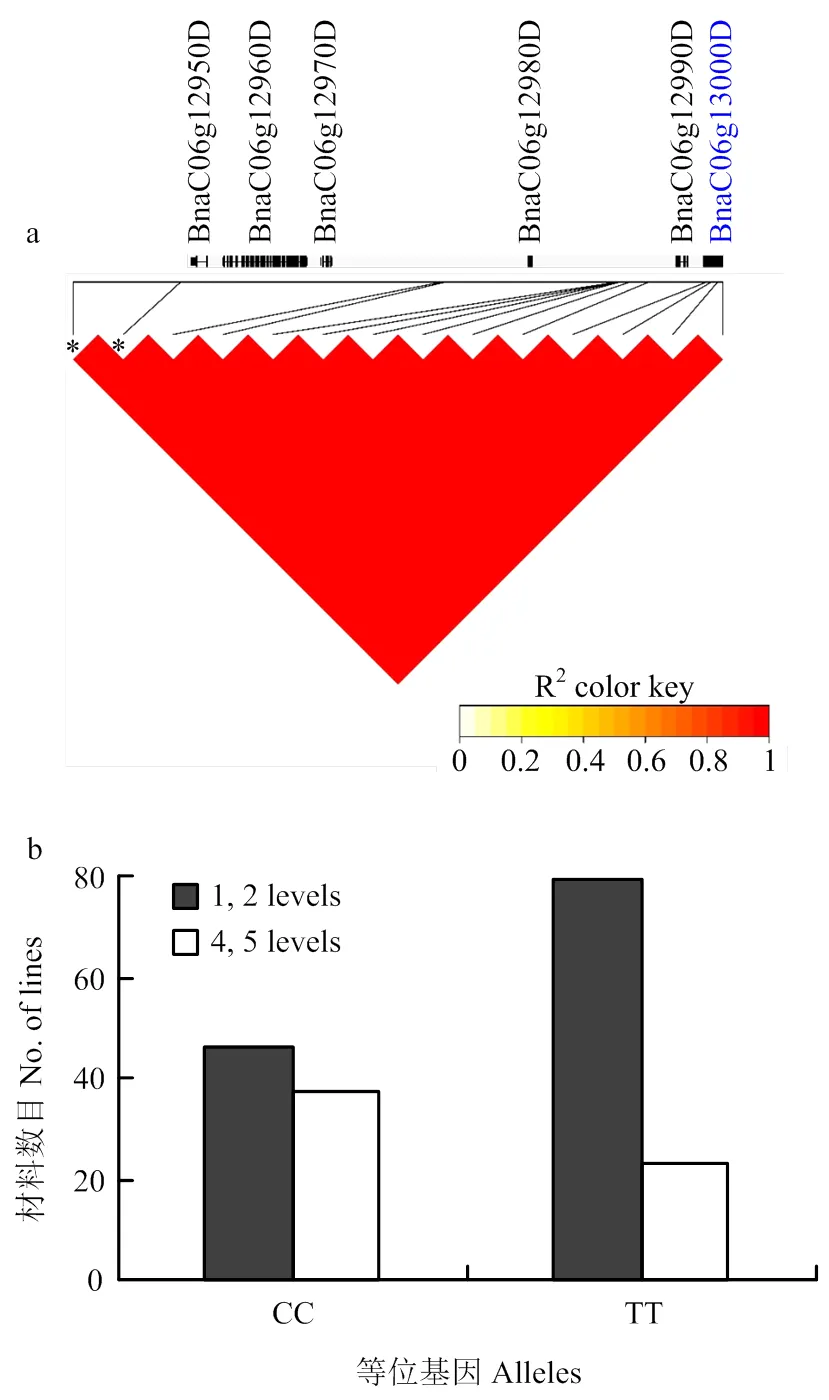
a:C06候选区间LD分析,*代表显著的SNP,蓝色字体代表预测的候选基因,基因结构显示在基因的下方。b:候选基因上游最近的SNP(Bn-scaff_15818_1-p471106)等位基因效应分析
3 讨论
本研究所使用的60K SNP芯片已被广泛应用在基于自然群体的全基因组关联分析[37]和基于分离群体的QTL定位研究中[38]。利用该SNP芯片对308份油菜F1的育性等级进行GWAS分析,最终预测了4个含PPR保守基序的候选基因,其中2个落在A09和C08染色体的部分同源区间,且与已克隆的A09上的CMS恢复基因位点()相同,对应同一个拟南芥基因。该候选区间被多个群体的QTL定位重复检测到[5-7]。另外2个候选基因对应的区间未发现相应的QTL报道,贡献率都不到5%,可能是由微效多基因控制,因而在早前基于双亲的分离群体中不容易被检测到。进一步单体型分析说明,与2个候选基因处于同一单体型块的SNP的等位基因或单体型的变化与CMS的育性显著相关,通过这些SNP的差异可以开发功能标记,应用于甘蓝型油菜CMS恢复系和保持系的筛选。
高通量测序技术的迅猛发展,将基因组学水平的研究带入了一个新的时期。芸薹属60K SNP芯片的开发和甘蓝型油菜参考基因组的释放,使我们能够快速、准确地挖掘重要农艺性状的候选基因和进一步的深入研究。本研究采用目前在植物中广泛应用的全基因组关联分析方法,成功鉴定出与甘蓝型油菜CMS育性恢复相关的位点,省时省力,同时该自然群体可以对其他农艺性状和品质性状进行定位,成为快速解码大量未知基因功能的重要途径。同时该研究将有助于其他CMS系统育性恢复候选基因的快速挖掘,促进油菜的杂种优势利用。
4 结论
通过GWAS分析,成功鉴定出13个可能影响CMS育性恢复有关的SNP位点,分布在5条染色体上。预测了4个含PPR保守基序的候选基因,其中2个候选基因在A09和C08染色体的部分同源区间,且与早前报道的一个CMS育性恢复位点一致。另外2个为新鉴定到的微效基因,与候选基因处于同一单体型块内的SNP变化与育性等级显著相关。
References
[1] Chen L, Liu Y G. Male sterility and fertility restoration in crops., 2014, 65: 579-606.
[2] Fu T D, Yang G S, Yang X N. Studies on three line Polima cytoplasmic male sterility developed in., 1990,104: 115-120.
[3] Yang G S, Fu T D, Ma C Z, Yang X N. Screening and genetic analysis of the restoring genes of polima cytoplasmic male sterility in., 1996, 29: 17-22.
[4] 杨光圣, 傅廷栋. 油菜细胞质雄性不育恢保关系的研究. 作物学报, 1991, 17(2): 151-156.
Yand G S, Fu T D. A preliminary study on the restoring-maintaining relationship in rapeseed., 1991, 17(2): 151-156. (in Chinese)
[5] 蔡强. 甘蓝型油菜波里马细胞质雄性不育恢复基因的分子标记筛选与初步定位[D]. 武汉: 华中农业大学, 2009.
Cai Q. Identification of molecular markers and perliminary mapping of fertility restorer gene () for the ‘polima’ CMS inL.[D]. Wuhan: Huazhong Agricultural University, 2009. (in Chinese)
[6] Li Y, Liu Z, Cai Q, Yang G S, He Q B, Liu P W. Identification of a microsatellite marker linked to the fertility-restoring gene for a polima cytoplasmic male-sterile line inL.., 2011, 10(47): 9563-9569.
[7] Liu Z, Liu P W, Long F R, Hong D F, He Q B, Yang G SFine mapping and candidate gene analysis of the nuclear restorer geneforCMS in rapeseed (L.)., 2012, 125(4): 773-779.
[8] Liu Z, Yang Z H, Wang X, Li K D, An H, Liu J, Yang G S, Fu T D, Yi B, Hong D FA mitochondria-targeted PPR protein restorescytoplasmic male sterility by reducingtranscript levels in oilseed rape., 2016, 9(7): 1082-1084.
[9] Cui X, Wise R P, Schnable P S. Thenuclear restorer gene of male-sterile T-cytoplasm maize., 1996, 272: 1334-1336.
[10] Liu F, Cui X Q, Horner H T, Weiner H, Schnable P S. Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize., 2001, 13(5): 1063-1078.
[11] Bentolila S, Alfonso A A, Hanson M R. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants., 2002, 99(16): 10887-10892.
[12] Brown G G, Formanova N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J F, Cheung W Y, Landry B SThe radishrestorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats., 2003, 35(2): 262-272.
[13] Desloire S, Gherbi H, Laloui W, Marhadour S, Clouet V, Cattolico L, Falentin C, Giancola S, Renard M, Budar F, Small I, Caboche M, Delourme R, Bendahmane AIdentification of the fertility restoration locus,, in radish, as a member of the pentatricopeptide-repeat protein family., 2003, 4(6): 588-594.
[14] Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura JGenetic characterization of a pentatricopeptide repeat protein gene,, that restores fertility in the cytoplasmic male-sterile Kosena radish., 2003, 34(4): 407-415.
[15] Akagi H, Nakamura A, Yokozeki-Misono Y, Inagaki A, Takahashi H, Mori K, Fujimura TPositional cloning of the ricegene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein., 2004, 108(8): 1449-1457.
[16] Hu J, Wang K, Huang W, Liu G, Gao Y, Wang J, Huang Q, Ji Y, Qin X, Wan L, Zhu R, Li S, Yang D, Zhu YThe rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162., 2012, 24(1): 109-122.
[17] Komori T, Ohta S, Murai N, Takakura Y, Kuraya Y, Suzuki S, Hiei Y, Imaseki H, Nitta NMap-based cloning of a fertility restorer gene,, in rice (L.)., 2004, 37(3): 315-325.
[18] Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D, Long Y, Zhong Y, Liu Y GCytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing., 2006, 18(3): 676-687.
[19] Itabashi E, Iwata N, Fujii S, Kazama T, Toriyama K. The fertility restorer gene,, for lead rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein., 2011, 65(3): 359-367.
[20] Fujii S, Toriyama K. Suppressed expression of retrograde- regulated male sterility restores pollen fertility in cytoplasmic male sterile rice plants., 2009, 106(23): 9513-9518.
[21] Tang H W, Luo D P, Zhou D H, Zhang Q Y, Tian D S, Zheng X M, Chen L T, Liu Y G. The rice restorerfor wild-abortive cytoplasmic male sterility encodes a mitochondrial- localized PPR protein that functions in reduction oftranscripts., 2014, 7(9): 1497-1500.
[22] Huang W C, Yu C C, Hu J, Wang, L L, Dan Z W, Zhou W, He C L, Zeng, Y F, Yao G X, Qi J Z, Zhang Z H, Zhu R S, Chen X F, Zhu Y G. Pentatricopeptide-repeat family proteinfunctions with hexokinase 6 to rescue rice cytoplasmic male sterility., 2015, 112(48): 14984-14989.
[23] Klein R R, Klein P E, Mullet J E, Minx P, Rooney W L, Schertz K FFertility restorer locus[corrected] of sorghum (L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12., 2005, 111(6): 994-1012.
[24] Hagihara E, Itchoda N, Habu Y, Iida S, Mikami T, Kubo TMolecular mapping of a fertility restorer gene for Owen cytoplasmic male sterility in sugar beet., 2005, 111(2): 250-255.
[25] Matsuhira H, Kagami H, Kurata M, Kitazaki K, Matsunaga M, Hamaguchi Y, Hagihara E, Ueda M, Harada M, Muramatsu A, Yui-Kurino R, Taguchi K, Tamagake H, Mikami T, Kubo TUnusual and typical features of a novel restorer-of-fertility gene of sugar beet (L.)., 2012, 192(4): 1347-1358.
[26] Small I D, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins., 2000, 25(2): 46-47.
[27] 丁安明, 屈旭, 李凌, 孙玉合. 植物PPR蛋白家族研究进展. 中国农学通报, 2014, 9: 218-224.
Ding A M, Qu X, Li L, Sun Y H. The progress of PPR protein family in plants., 2014, 9: 218-224. (in Chinese)
[28] Edwards D, Batley J, Snowdon R J. Accessing complex crop genomes with next-generation sequencing., 2013, 126(1): 1-11.
[29] Schiessl S, Iniguez-Luy F, Qian W, Snowdon R J. Diverse regulatory factors associate with flowering time and yield responses in winter-type., 2015, 16(1): 737-797.
[30] Pritchard J K, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data., 2000, 155(2): 945-959.
[31] Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study., 2005, 14(8): 2611-2620.
[32] Hardy O J, Vekemans X. SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels., 2002, 2(4): 618-620.
[33] Aulchenko Y S, Ripke S, Isaacs A, Van Duijn C M. GenABEL: an R library for genome-wide association analysis., 2007, 23(10): 1294-1296.
[34] Turner S D. qqman: an R package for visualizing GWAS results using QQ and manhattan plots., 2014, doi: http://dx.doi.org/ 10.1101/005165.
[35] Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study., 2005, 14: 2611-2620.
[36] Merk H L, Yarnes S C, Van Deynze A, TONG N K, MENDA N, MUELLER L A, MUTSCHLER M A, LOEWEN S A, MYERS J R, FRANCIS D M. Trait diversity and potential for selection indices based on variation among regionally adapted processing tomato germplasm., 2012, 137: 427-437.
[37] Wei L J, Jian H J, Lu K, Filardo F, Yin N W, Liu L Z, Qu C M, Li W, Du H, Li J N. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in., 2016, 14: 1368-1380.
[38] Liu L Z, Qu C M, Wittkop B, Yi B, Xiao Y, He Y, Snowdon R J, Li J N. A high-density SNP map for accurate mapping of seed fibre QTL inL.., 2013, 8(12): e83052.
(责任编辑 李莉)
附表1 供试材料及表型数据
Supplementary Table 1 List of rapeseed accessions used in this study
可育父本编号Fertility male parents accession生态型 Ecotype2013年育性等级Fertility levelsin 20132013年始花期当天的日平均温度Daily average temperature of beginning flowering in 2013 (℃)2013年始花期前10天的日平均温度Daily average temperature 10 days ages from beginning flowering in 20132014年的育性等级Fertility levles in 20142014年始花期当天的日平均温度Daily average temperature of beginning floweringin 2014 (℃)2014年始花期前10天的日平均温度Daily average temperature 10 days ages from beginning flowering in 2014 Bn-197W22015.621615.3 Bn-188W22015.611816.5 Bn-180W214.513.7115.516.8 Bn-203W314.513.7116.516.9 Bn-056W32014.151714.4 Bn-075W32014.1316.515 Bn-200W22015.6216.515 Bn-129W42014.1516.515 Bn-041W22014.1219.516 Bn-067W42014.741615.3 Bn-139W32014.1416.515 Bn-070W214.513.721615.3 Bn-141W2131141616.9 Bn-016W32014.7415.514.9 Bn-079W22015.6419.516 Bn-017W32015.621816.5 Bn-198W22014.1516.515 Bn-147W22014.741714.4 Bn-185W22015.631616.9 Bn-183W42014.1517.514.9 Bn-027W42015.641816.5 Bn-174W314.513.7218.515.5 Bn-064W21110.7219.516 Bn-191W214.513.7115.514.9 Bn-199W21315.5518.515.5 Bn-137W212.516.3416.515 Bn-160W212.516.3418.515.5 Bn-158W312.516.331616.9 Bn-213W312.516.321816.5 Bn-172W212.516.3216.515 Bn-103W22014.141616.9 Bn-133W41211.3415.514.9 Bn-121W32014.111816.5 Bn-168W21216.1215.514.9 Bn-156W32014.731816.5 Bn-119W52014.1316.515 Bn-120W32015.651814.4 Bn-134W21216.141816.5 Bn-054W217.513.7116.515 Bn-012W31712.1320.514.6 Bn-044W51311416.515 Bn-015W42010.941616.9 Bn-078W217.513.7215.514.9 Bn-028W31611.7319.516.7 Bn-040W217.513.7518.515.5 Bn-008W217.513.7516.515 Bn-072W215.512.742116.8 Bn-052W214.513.7315.514.9 Bn-059W41211.3420.514.6 Bn-085W32014.1216.515 Bn-061W215.512.741714.4 Bn-038W22419.2416.516.9 Bn-001W32015.6417.516.8 Bn-086W314.513.7316.516.9 Bn-002W32014.1517.514.9 Bn-018W32014.1316.516.9 Bn-045W22015.611816.5 Bn-053W21216.151714.4 Bn-023W21215.8317.516.9 Bn-055W312.516.3118.515.5 Bn-202W317.513.731816.5 Bn-186W22014.1219.516.7 Bn-031W314.513.731816.5 Bn-107W217.513.7119.516.7 8Q 263W21611.751614.9 8Q 265W21215.9115.514.9 8Q 264S31310.831716.8 Bn-311S31310.8518.516 Bn-395S31310.8519.514.4 Bn-273S21211.3315.514.9 Bn-320S21116.2317.516.5 Bn-238S11210.7316.516.9 Bn-364S21310.8315.514.9 Bn-245S21311.422116.8 Bn-274S21611.741716.8 Bn-279S21712.1219.516.7 Bn-343S11311.411816.5 Bn-309S31210.7517.514.9 Bn-316S21211.3516.515.3 Bn-269S3129.4416.516.9 Bn-339S412.513.5418.516 Bn-361S21211.3515.515.3 Bn-400S21210.8519.514.4 Bn-392S21210.7218.516 Bn-275S31210.851614.9 Bn-328S4129.451815.5 Bn-348S11712.112116.8 Bn-355S41210.7315.515.3 Bn-291S21110.7415.515.3 Bn-335S21211.332116.8 Bn-334S31310.8116.516.9 Bn-366S212.59419.516.7 Bn-248S4129.841815.5 Bn-313S3127.2319.516.7 Bn-330S31311316.516.9 Bn-360S21211.3119.516.7 Bn-292S412.5951815.5 Bn-340S515.512.7217.516.5 Bn-359S212.513.5416.516.9 Bn-363S31313.722116.8 Bn-365S31210.8417.516.5 Bn-376S21311219.516.7 Bn-483S21210.841815.5 Bn-299S41211.351615 Bn-342S31110.7518.516 Bn-350S3131151615 Bn-323S213.511.451714.6 Bn-314S312.5931615 Bn-290S41110.751815.5 Bn-333S21311.4120.516.9 Bn-289S21611.751615 Bn-281S21310.831616.9 Bn-240S21310.831616.9 Bn-259S42014.741716.8 Bn-283S31611.7417.516.5 Bn-393S21112.531815.5 Bn-357S2131141616.9 8Q 268S3129.841616.9 8Q 269S41210.8516.515.3 8Q 271S31310.8319.514.6 8Q 272S21611.751815.5 Bn-322S21611.741616.9 9w237SW39.55.8317.516.5 9w238SW11211.3215.515.3 9w239SW1131111716.8 9w240SW112.516215.515.3 9w241SW11212116.516.9 9w242SW11210.7116.516.7 9w243SW11210.822116.8 9w244SW31311518.516 9w245SW11311116.516.8 9w246SW11213.8117.516.5 9w247SW31211.3518.516 9w248SW4129.4519.514.6 9w249SW31611.7417.516.5 9w251SW11611.7219.516.7 9w252SW11210.8119.516.7 9w253SW21611.7515.515.3 9w255SW51110.751714.6 9w256SW11310.8114.516.7 9w257SW21310.8418.515.1 9w259SW413.511.4419.516.7 9w261SW11611.7115.515.3 9w262SW31211.3520.515.3 9w263SW31712.1520.515.3 9w264SW11311115.515.3 9w266SW11310.8116.516.8 9w267SW31611.751815.5 9w268SW31211.3518.515.3 9w269SW41712.131815.5 9w270SW11210.811815.5 9w271SW41210.8418.515.1 9w272SW21611.7516.514.9 9w273SW51210.8415.515.3 9w274SW4129.8516.514.9 9w275SW211.510.351615 9w276SW11611.721615 9w277SW311.510.351815.5 9w279SW513.511.4419.516.7 9w280SW21112.151815.5 9w281SW41311515.515.3 9w282SW41311519.514.4 9w283SW31110.7516.514.9 9w284SW313.511.4516.514.9 9w285SW41210.7315.515.3 9w286SW415.512.7316.514.9 9w287SW31311319.514.6 9w288SW1129.411615 9w289SW11210.811815.5 9w290SW41211.3515.515.3 9w292SW21311515.515.3 9w294SW21712.1117.516.5 9w295SW313.511.411815.5 9w297SW31311515.515.3 9w298SW31311519.514.6 9w299SW31611.7416.514.9 9w300SW315.512.7516.514.9 9w301SW31611.7420.517.2 9w302SW31311.4418.515.1 9w303SW2131132116.8 9w304SW513.511.4515.515.3 9w305SW11210.711616.9 9w306SW31311516.514.4 9w307SW21211.351615 9w309SW41211.3516.514.4 9w313SW212.59519.514.6 9w314SW31211.3518.515.3 9w315SW31611.7417.516.5 9w317SW41311.441615 9w318SW31211.3515.515.3 9w319SW311.510.3516.514.9 9w320SW1129.8216.514.9 9w321SW213.511.441615 9w322SW51310.8419.514.6 9w324SW41310.8519.514.6 9w325SW4129.8516.514.9 9w327SW41210.8416.514.9 9w328SW51110.7515.515.3 9w330SW31211.3416.514.9 9w333SW4127.9516.514.9 9w335SW41210.8415.515.3 9w336SW1129.411815.5 9w337SW21210.8116.514.9 9w340SW2129.4218.515.1 9w341SW21210.822116.8 9w342SW21311419.514.6 9w343SW41210.7515.515.3 9w345SW31210.741716.8 9w349SW21310.8518.515.3 9w350SW31210.8515.515.3 9w351SW41310.8516.514.9 9w354SW4127.2517.514.9 9w355SW11210.8218.515.1 9w356SW21210.751615 9w357SW465.922116.8 9w358SW211.510.351815.5 9w359SW31211.341815.5 OJ105SW31310.8519.514.6 OJ106SW411.510.3518.515.3 OJ107SW21310.8319.514.6 OJ108SW3131151615 OJ110SW21611.731615 OJ111SW51211.3518.515.3 OJ115SW3116517.514.4 OJ117SW41311.4415.515.3 OJ118SW31311515.515.3 OJ119SW410.58.4414.516.7 OJ120SW21211.3515.515.3 OJ121SW313.511.451714.6 OJ122SW31311.4516.514.9 0J125SW31311416.514.9 0Q230SW31110.7519.514.6 0Q231SW31110.7118.515.1 0Q232SW311.510.3215.515.3 0Q233SW31311515.515.3 0Q234SW21611.751815.5 0Q236SW411651615 0Q237SW11211.3416.514.9 0Q239SW51310.841815.5 0Q240SW51211.351714.6 0Q242SW21712.1518.516 0Q229SW311.510.3416.514.9 2Q 174SW41311416.514.4 2Q 175SW41210.751815.5 2Q 226SW11215.812116.8 2Q 237SW31311519.514.6 2Q 240SW41611.7516.514.9 2Q 426SW112.514.921815.5 2Q 430SW11210.8116.516.8 2Q 431SW41210.8516.514.4 2Q 433SW313.511.4216.514.4 2Q 435SW11310.8119.516.8 2Q 440SW31311419.514.6 2Q 451SW213.511.451815.5 2Q 463SW31210.841616.9 2Q 466SW41110.751615.3 2Q 467SW11210.8115.515.3 2Q 474SW31311516.514.4 2Q 476SW312.59119.516.8 2Q 477SW3129.4119.516.8 2Q 483SW4131131615 2Q 484SW21311417.516.5 2Q 494SW213.511.4318.516 2Q 495SW21211.3115.515.3 2Q 496SW31311317.516.5 2Q 511SW412.59416.514.9 2Q 516SW511.58.651615 2Q 518SW11212.5116.514.9 2Q 522SW111.510.3119.516.8 2Q 525SW11310.8115.515.3 2Q 528SW213.511.442116.8 2Q 531SW2129.4516.514.4 2Q 542SW410.58.431615 2Q 544SW11212.6116.516.8 2Q 547SW3129.8516.514.4 2Q 551SW41210.7515.515.3 2Q 553SW11210.7115.515.3 2Q 554SW31210.8515.515.3 2Q 558SW3131152114.9 2Q 560SW31110.7415.515.3 2Q 561SW31611.7316.514.4 2Q 583SW11310.8119.516.8 2Q 592SW5129.841615 2Q 594SW412.59216.514.9 2Q 605SW31611.7318.516 2Q 607SW31211.341716.8 2Q 609SW47.55.951714.6 2Q 612SW31210.7518.516 2Q 622SW111.510.311716.8 2Q 645SW21712.151714.9 2Q 706SW31210.7515.515.3 2Q 731SW21611.7216.514.9 2Q 813SW31311316.514.9 2Q 853SW2129.4518.516 2Q 002SW1131111815.5 2Q 031SW5129.4516.514.4 2Q 038SW21712.151615.3 2Q 055SW41611.751815.5 2Q 057SW21712.1418.516 2Q 071SW311.58.6518.516 2Q 073SW11311116.515 2Q 079SW31210.7516.514.4 2Q 081SW21110.751714.9 2Q 101SW41311.4515.515.3 2Q 114SW31712.1316.514.9 2Q 132SW112.5911615 2Q 135SW111.515.8116.515 2Q 151SW212.59516.514.4
Genome-wide association study of the fertility restorer loci forCMS in rapeseed (L.)
WEI DaYong, TAN ChuanDong, CUI YiXin, WU DaoMing, LI JiaNa, MEI JiaQin, QIANWei
(College of Agronomy and Biotechnology, Southwest University/Chongqing Engineering Research Center for Rapeseed, Chongqing 400716)
【Objective】 Polima system of cytoplasmic male sterility (CMS) in(rapeseed), controlled by a major gene as well as polygenes, has been widely used in China for hybrid rapeseed breeding. Genome-wide association study (GWAS) was performed in a rapeseed population to identify genetic loci and candidate genes for fertility restorer ofCMS. 【Method】 301A, a rapeseedCMS line, was chosen as the female parent to cross with a panel of 308 accessions in natural population of rapeseed which has been genotyped previously using the 60 kSNP array. The F1hybrids were grown for fertility evaluation in 2013 and 2014, respectively, with two replications each year. The fertility of F1was classified according to pollen fertility and performance of pistil and stamen, and population structure and relative kinship of 308 male accessions were analyzed. GWAS was conducted by associating the fertility of F1with the single nucleotide polymorphisms (SNPs) of males. Candidate genes was identified from the region of 100 kb each side of the peak SNP or trait-associated SNPs at LD (2>0.5). Comparative analysis of QTL and haplotype effect evaluation for candidate genes were performed. 【Result】A significant difference (<0.01) was found in the fertility of F1between two years, but a high correlation was detected in it between two years (= 0.52,<0.001). The population structure analysis classified the 308 male accessions into three genetic groups (winter, spring and semi-winter). The relative kinships analysis found that 73% of the kinship coefficients between lines were <0.1 and 53% were equal to 0. A total of 13 SNPs were detected to be with significant association with the fertility of F1, formed six genetic intervals on chromosomes A01, A09, C03, C06 and C08. Six genes related to fertility were predicted from the six intervals, and four of these could encode the PPR type proteins which is a conserve structure encoded by fertility restorer genes. Collinearity analysis revealed two PPR type candidate genes (and) detected in homoeologous regions between chromosome A09 and C08 were homology with, an open reading frame functioning as the reported rapeseed nuclear restorer gene ofCMS. The other two PPR type candidates (and) were novel candidate restorer genes forrapeseedCMS, of which the linked alleles or haplotypes of SNPs were found to significantly associated with the fertility level of the F1(<0.001).【Conclusion】The present study identified several fertility restorer genes forCMS in rapeseed from both A and C subgenomes. Developing functional markers from the alleles or SNPs linked with the candidate genes will benefit the screening of restorer and maintainer lines in theCMS system.
; polima CMS; fertility restorer gene; GWAS; SNP
2016-09-23;接受日期:2016-12-12
国家“973”计划(2015CB150201)、国家自然科学基金(31601333)
魏大勇,E-mail:dylanmay@swu.edu.cn。通信作者梅家琴,Tel:023-68250701;E-mail:jiaqinmay@163.com
