计算毒理学在内分泌干扰物筛选上的应用和展望
2017-10-13陈钦畅谭皓月史薇于红霞
陈钦畅,谭皓月,史薇,于红霞
污染控制与资源化研究国家重点实验室,南京大学环境学院,南京210023
计算毒理学在内分泌干扰物筛选上的应用和展望
陈钦畅,谭皓月,史薇,于红霞*
污染控制与资源化研究国家重点实验室,南京大学环境学院,南京210023
内分泌干扰物通过干扰内分泌系统导致多种疾病,如生殖疾病、肥胖症甚至癌症。然而,面对环境中大量潜在的内分泌干扰物,传统的体外、体内评估方法由于成本高、耗时长等问题,难以实现内分泌干扰物的高通量筛查。计算毒理学逐渐发展成为被美国环保局(Environmental Protection Agency, EPA)、经济合作与发展组织(Organization for Economic Co-operation and Development, OECD)等机构所推荐的内分泌干扰物筛选与预测方法。本文综述了计算毒理学在内分泌干扰物筛选上的进展,主要包括分子对接和分子动力学模拟的应用,并对有害结局路径(adverse outcome pathway, AOP)的方法进行介绍和展望。
计算毒理学;内分泌干扰;分子对接;分子动力学模拟;AOP
Received6 February 2017accepted13 March 2017
Abstract: Endocrine disrupting chemicals (EDCs) cause a variety of diseases, such as reproductive diseases, obesity and even cancer, by interfering with the endocrine system. However, in the face of a large number of potential endocrine disruptors in the environment, traditional in vitro and in vivo assays are difficult to achieve high throughput screening of endocrine disruptors due to their high cost and time consuming. Computational toxicology has been recommended as the screening and predicting method by the US Environmental Protection Agency (EPA), the Organization for Economic Co-operation and Development (OECD) and so on. Here, we discuss the application of computational toxicology methods, particularly molecular docking, molecular dynamics simulations and the developing adverse outcome pathway (AOP), in guiding the screening of EDCs.Keywords: computational toxicology; endocrine disrupting; molecular docking; molecular dynamics; AOP
1 前言(Introduction)
1.1 内分泌干扰效应与内分泌干扰物
内分泌系统指由一系列腺体分泌激素进入内循环系统运输并直接作用于目标器官的系统。内分泌系统的信号由激素传递,激素有不同的化学结构,主要包括3种:类花生酸类(eicosanoids)、甾体类(steroids)和氨基酸衍生物(胺类、肽链和蛋白质)。激素通过与目标细胞的特定受体蛋白结合,激活信号转导通路,达到调节细胞功能的作用,调节着生物体几乎所有生物学过程。已有报道表明,一些化学物质如双酚A(bisphenol A, BPA)、多环芳烃(polycyclic aromatic hydrocarbons, PAHs)及一系列杀虫剂等的暴露会干扰内分泌系统并产生有害影响,这类物质被称为内分泌干扰物(endocrine disrupting chemicals, EDCs)。内分泌干扰物的暴露会增加生殖疾病、心肺疾病、免疫系统疾病和神经系统疾病的风险,甚至导致肿瘤和癌症的发生[1]。据Attina等[2]的模型估计,2010年欧盟国家内分泌干扰物导致的疾病花费占国内生产总值(GDP)的1.28%,为2 170亿美元,而美国的达到3 400亿美元,占GDP的2.33%,内分泌干扰物筛选的研究迫在眉睫。
1.2 内分泌干扰物的实验检测手段
检测内分泌干扰物的实验手段包括体外(in vitro)和体内(in vivo)实验。体外实验包括细胞增殖实验(cell proliferation assays)[3]、报告基因实验(reporter gene assays)[4]、酵母双杂交实验(yeast two-hybrid assays)[5]、结合实验(binding assays)[6]等。体内实验多采用哺乳动物[7-8]、鸟类[9-10]、鱼类[11-12]、两栖类[13-14]等动物。过去十几年来,体外实验被合理开发应用到各种高通量测试筛选方法中,以应对数量巨大的潜在危害化合物,以及动物体内测试的巨大开销和伦理问题[15]。由美国环保局(Environmental Protection Agency, EPA)、国立卫生研究院(National Institutes of Health, NIH)和食品与药品管理局(Food and Drug Administration, FDA)等跨部门合作的21世纪毒理学(Tox21)项目,采用高通量筛选技术测试约10 000种环境化合物和药物的毒性,其中内分泌干扰是重要方面,关于雌激素干扰效应及信号通路的研究结果已于2014年发布[16]。
现阶段评估一个化合物的内分泌干扰作用往往需要体内和体外实验的结合。美国EPA内分泌干扰物筛选项目(Endocrine Disruptor Screening Program, EDSP)于2012年开展了EDSP21项目,采用2个级别的筛选测试手段(Tier 1和Tier 2)评价化合物的内分泌干扰活性。其中级别1测试包括5种体外实验和5种体内实验,级别2则是更深层次或多代体内测试。经过级别1测试具有干扰活性的化合物进入级别2测试,评估其内分泌干扰效应。目前,超过1 800种化合物内分泌相关活性的高通量筛选数据(主要为包括雌激素受体ER、雄激素受体AR结合和转录激活的级别1体外筛查数据)已可在EDSP21 Dashboard网站(http://actor.epa.gov/edsp21/)获取。
1.3 内分泌干扰物的模拟预测手段
体外和体内实验手段虽然能完整评价化合物的内分泌干扰效应,但是其成本高、耗时长,难以对全球现有超过126 000 000种化合物参考(http://www.cas.org)进行逐一筛选。因此,亟需发展化学品内分泌干扰效应筛选的计算毒理学(computational toxicology)方法[17]。计算毒理学方法指通过综合体内、体外实验和计算机模拟等不同来源的数据,开发数学或计算机模型,以更好理解或预测化合物干扰效应的方法[18]。近年来,计算毒理学得到越来越多的关注,美国EPA于2005年成立了国家计算毒理学中心(National Center for Computational Toxicology, NCCT),致力于开发新的评估化合物安全性的方法,即计算毒理方法;经济合作与发展组织(Organization for Economic Co-operation and Development, OECD)于2008年开发的计算毒理软件OECD QSAR Toolbox如今也进入3.4版本,并得到各国政府、化学工业的接受和使用。
定量结构-效应关系(quantitative structure-activity relationship, QSAR)是最早开发和发展的计算毒理学方法,将代表化合物结构、物理、化学性质的分子描述符(molecular descriptors)与特定效应终点或有害结局(adverse outcome, AO)建立联系,达到预测目的,在不同尺度的内分泌干扰效应,如核受体结合[19-20]、转录激活[21-22]、器官和个体有害结局[23-24]等,都得到广泛运用。我国陈景文教授、张爱茜教授、高士祥教授、王连生教授和于红霞教授等的团队在内分泌干扰物的QSAR研究上都做了大量工作[25-29],比如Li等[25]计算了517种有机化合物的705个分子描述符,并选取其中的13个分子描述符建立雌激素效应的QSAR模型,发现有机分子的雌激素活性主要与分子尺寸、形状特征、电负性和范德华体积等相关。如今,QSAR已经发展成较为成熟的计算毒理学方法,得到OECD等组织的认可,基于QSAR开发的毒性预测软件,如TEST、ECOSAR、OncoLogic等,也得到广泛应用。然而,QSAR往往忽略干扰物的效应机制,采用的分子描述符往往也没有直接或明确的药理学或生物学意义[30]。
激素分子与体内调节相关生理功能的大分子,如受体蛋白等之间的相互关系在内分泌系统信号传递中具有重要作用。因此对干扰物与受体作用关系的研究是内分泌干扰物筛选的重要研究手段,很多体外实验都是以干扰物与受体作用关系为对象研究化合物的内分泌干扰效应的[31-32],而分子对接(molecular docking)和分子动力学(molecular dynamics, MD)模拟方法作为基于干扰物与受体作用关系的计算毒理学研究方法也得到越来越多的应用。在此基础上,为了更加深入地理解内分泌干扰效应作用机制,OECD、美国EPA等组织开展了开发有害结局路径(adverse outcome pathway, AOP)的项目,将极大促进基于效应机制的计算毒理学的发展。因此,本文将对分子对接、MD模拟和AOP等基于效应机制的计算毒理学方法在内分泌干扰物筛选上的应用进行综述。
2 分子对接及其应用(Molecular docking and its applications)
分子对接是预测配体与受体结合成稳定复合体时配体所处的最佳位置和方向的方法[33]。内分泌系统中主要的受体蛋白(图1)是包括雌激素受体(estrogen receptor, ER)、雄激素受体(androgen receptor, AR)、甲状腺激素受体(thyroid hormone receptor, TR)、糖皮质激素受体(glucocorticoid receptor, GR)等在内的核受体(nuclear receptor, NR),它们都受激素调节并控制大量基因的表达[34]。随着晶体学和生物化学技术的发展,越来越多核受体的晶体结构被解析出来(图1)[35-39],这些晶体结构都可以通过Protein Data Bank网站(http://www.rcsb.org/pdb/home/home.do)获得,使采用对接和MD模拟等方法筛选内分泌干扰物成为可能[40]。但已有的多为人类的受体,对于其他物种,往往需要通过同源建模(homology modeling)构建受体结构[41]。
分子对接的使用有助于加深对配体受体相互作用机制的理解。Nose等[42]用对接的方法从14个酚类物质中筛选出4-(1-adamantyl)phenol为拟雌激素物质,经验证确实具有很强的雌激素活性。对对接结果的分析发现,与雌激素类似,4-(1-adamantyl)phenol的羟基也能与ERα中Glu353和Arg394氨基酸形成氢键。D'Ursi等[43]采用柔性对接的方法探索了内分泌干扰物与ER、孕酮受体(progesterone receptor, PR)和AR的相互作用,发现这些内分泌干扰物与受体的相互作用主要取决于化合物与配体结合腔(ligand binding cavity, LBC)中多个氨基酸残基之间的疏水性作用,对于亲脂性内分泌干扰物,它们有能力适应甾体受体的疏水性LBC,并呈现非特异性结合模式。

图1 部分已解析的核受体结构[35-39]Fig. 1 Some of the structures of nuclear receptors (NRs) that have been refined[35-39]
由于内分泌干扰物往往能同时作用于不同受体,研究干扰物与不同受体的相互作用能预测潜在的效应终点。Yuriev等[45]针对人体14种NR,选取了18个完整、可靠的晶体结构建立对接模型,通过对接得到化合物与各个NR的结合能,判断对哪些受体更敏感,从而判断化合物潜在的内分泌干扰效应[44],这种方法被称为反向对接法。于红霞教授团队最近也采用反向对接开发了一个针对39个人类NR的程序SPEN,该程序经过10种化合物的验证具有良好的表现[46]。
分子对接可以与QSAR结合,构建多维QSAR模型。Vedani等[47]采用柔性对接和QSAR结合的方法建立了预测ER结合能力的6维QSAR模型,并用于对106种内分泌干扰物的筛选,结果r2达到0.885,表明具有很好的筛选能力。Vedani团队将这种方法拓展到AR、TR、GR等11个核受体,并建立了基于网站的预测平台VirtualToxLab[48]。分子对接在药物开发领域也产生了一些新方法,如基于分子碎片的对接技术正在快速发展,虽然在内分泌干扰物筛选上还没有得到应用,但是不失为提高对接结果精确度的选择[49]。然而分子对接在很大程度上仍受受体柔性的制约,而对受体赋予过多的柔性会导致对计算的要求呈指数增加并且变得不切实际[50]。
在实际应用中分子对接受受体结构和配体种类的影响较大,针对相同种类、数量较少的化合物时,分子对接能得到较好的预测效果,对接的结合能可以很好预测化合物与受体的结合效力[51];而针对大量具有不同结构的化合物时,预测效果一般,甚至比普通的QSAR模型差[52]。因此,选择合适、可靠的受体模型是分子对接模型构建的重点。针对不同种类的化合物提供不同对接模型是可行的解决方法;也可以借助分子动力学模拟,产生多个受体模型进行对接[53]。
3 分子动力学模拟及其应用(Molecular dynamics simulations and the applications)
随着计算机技术的发展和计算能力的提升,分子动力学模拟逐渐成为研究生物大分子作用的标准方法[54]。分子动力学模拟是研究原子和分子物理运动的计算机模拟方法,所有分子和原子在给定的时间范围内相互作用,形成一个动态变化的系统,以此研究生物分子之间的相互作用[55]。
MD模拟有助于探索干扰物作用下受体蛋白及配体本身的构象变化。Li等[56]用MD模拟研究了内分泌干扰物的雌激素干扰效应,发现干扰物与ER在2 ns的模拟下都能达到稳定状态,并且干扰效力更强的化合物能与ER的His524氨基酸稳定形成氢键。采用MD模拟还能发现蛋白质的关键结构及活性产生的关键变化[57]。Wang等[58]通过对AR骨架变构情况的比较发现12号螺旋(Helix 12, H12)在MD模拟过程中具有最显著的位置变化,认为H12的位置变化是抗雄激素活性产生的关键。Wang等[58]还发现H12在10 ns模拟时间内达到平衡是抗雄性活性产生的重要特征,且稳定时间与活性强弱呈负相关。
干扰物与受体蛋白LBC的结合情况仍然是MD研究的重点。Martínez等[59]通过MD模拟发现了配体逃离TR-LBC的3种可能途径。有学者进一步用操纵分子动力学(steered molecular dynamics, SMD)模拟探究配体从各逃离途径逃离配体结合腔的难易程度[60-62]。Martínez等[61]发现配体逃离TR的最佳途径是位于H1、H2和H3处的通道3,而且当配体亲水部分能与受体外部的水分子接触时逃离过程会变得更轻松。另一方面,有研究表明,干扰物的诱导能使受体H12的位置发生变化,而其稳定位置正好挡住配体,使配体无法从LBC中逃离,受体形成的这种结构称为“老鼠夹(mousetrap)”结构[56, 63]。
热力学计算也是MD模拟的常用分析方法。采用MM/PBSA或MM/GBSA(molecular mechanics with Poisson-Boltzmann or generalized Born and surface area)方法计算配体-受体结合自由能ΔGbinding可用于预测配体与受体间的结合效力。van Lipzig等[64]将计算得到的雌激素干扰物与ER的结合自由能和实验测得的结合效力比较,发现两者的相关系数达到0.94。结合自由能还能区分干扰物对受体不同亚型的选择性[65-66]。Martínez等[65]分别计算了配体Triac与TRα和TRβ相互作用的结合自由能,发现Triac与TRα的结合自由能显著低于TRβ,导致其对TRα具有高度选择性。
除了干扰物与核受体的结合,核受体与其他蛋白质的相互作用,如与共调节因子作用、二聚现象等[67-69],也是影响内分泌干扰效应产生的重要过程。研究表明ER的二聚作用大大抑制了E2逃离LBC[69]。于红霞教授团队[67]最近的MD研究也表明,共调节因子在化合物甲状腺激素干扰活性产生过程具有重要作用,抗甲状腺激素干扰物与TR结合能促进共抑制因子而不是共激活因子与TR结合,从而导致抗性的产生。因此,考虑蛋白质受体与其他调剂因子的作用过程,对生物大分子间作用,如共调节因子结合、二聚作用和与DNA的结合等进行模拟,是MD模拟在内分泌干扰物筛选上的重要发展方向。另外,在MD模拟中采用量子力学/分子力学(QM/MM)耦合的方法,将配体部分用QM计算,其他部分用MM模拟的方法,有助于提高模拟的精确度,并有利于更深入探索配体受体之间的相互作用。陈景文教授团队[70]采用QM/MM的方法,探索了电中性和阴离子形态下酚类内分泌干扰物与甲状腺素运载蛋白(transthyretin, TTR)的结合,发现阴离子形态比电中性的酚类物质与TTR结合更强,认为离子形态的考虑是内分泌干扰物虚拟筛选过程不可忽视的机制。
4 AOP的发展和展望(Development and prospect of AOP)
随着对效应机制理解的不断深化,AOP概念逐渐发展起来。AOP就是描绘从分子启动事件(MIE)的开始,由一系列关键事件(KE)和之间关系(KER)连接,到有害结局(AO)之间关系的框架[71],与AOP相关的各种概念如表1所示。内分泌干扰效应的产生不只是干扰物与靶标相互作用,还包括生物大分子间、细胞层次、器官层次的变化(如图2),因此,对AOP的研究有助于获得更加精确和透彻的预测效果。然而这种预测方法是建立在对干扰效应作用通路足够明晰的基础上,这也是目前面临的最大挑战[72]。随着有害结局路径知识库(Adverse Outcome Pathway Knowledge Base, AOP-KB: http://aopkb.org/)的建立,越来越多AOP被开发并在AOP-KB平台上共享,这将大大促进AOP在计算毒理学预测上的应用。

表1 与有害结局路径(AOP)相关概念的定义[71]Table 1 Definition of concepts relevant to adverse outcome pathway (AOP)[71]
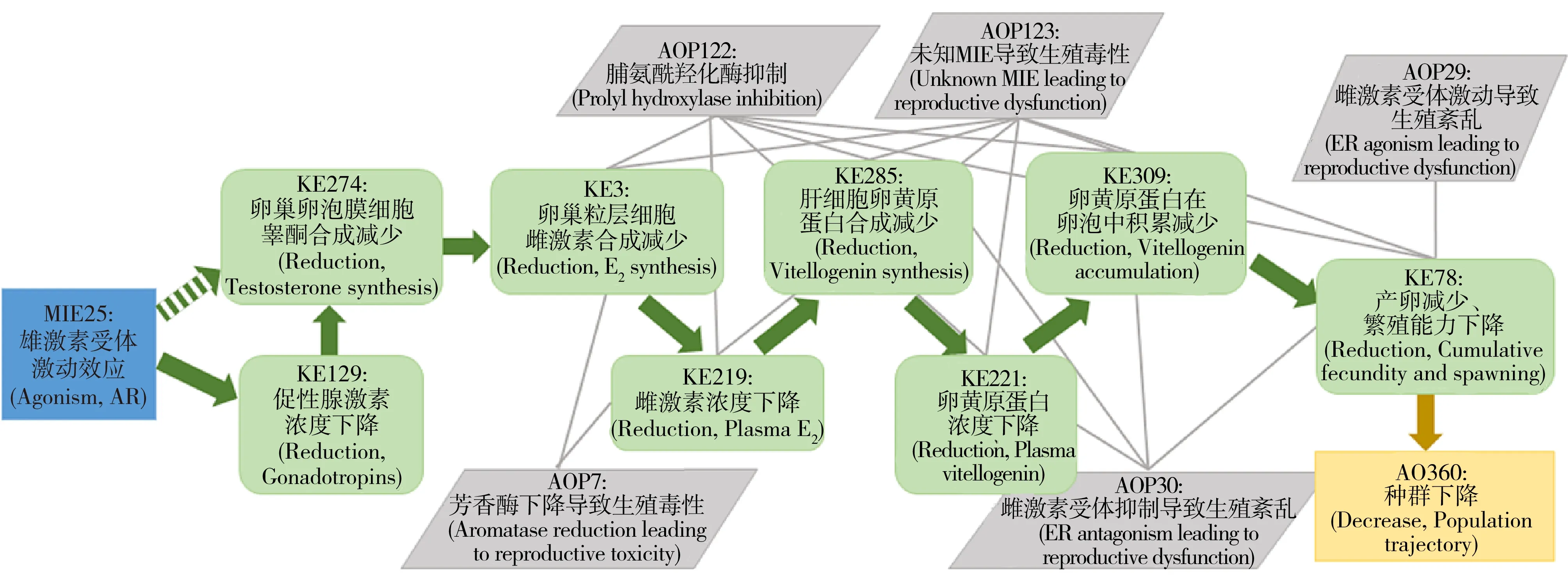
图3 雄激素受体激动效应导致生殖紊乱的AOP[73]Fig. 3 AOP: Androgen receptor agonism leading to reproductive dysfunction[73]
目前已有的AOP为20个,其中包括与雌激素、雄激素、甲状腺激素受体等相关的5个内分泌干扰AOP。Villeneuve[73]开发并发表了有关雄激素受体激动效应导致生殖功能紊乱的AOP(AOP23,如图3所示),干扰物激活雄激素受体(MIE),导致睾酮、雌激素合成下降,血液雌激素浓度降低,进而肝脏卵黄原蛋白合成下降,血液中卵黄原蛋白浓度降低,卵母细胞吸收量减少,进而使产卵下降,最终导致生殖功能紊乱的AO产生。Villeneuve等[74]还总结了AOP开发的五大原则:(1)AOP不具有化合物特异性;(2)AOP是模块化的,且构成要素可重复利用;(3)每个独立的AOP都由单一系列的KE和KER构成;(4)由具有相同KE和KER的AOP构成的网络(图2)是预测真实世界情况的基础单元;(5)AOP是可以随着新认识的形成而不断演化的。这些原则的考虑有助于对AOP的理解和应用。
MIE和KE都是AOP必不可少的组成部分。由于MIE直接与干扰物相互作用,由此开始整条通路的调节,并最终到达AO终点,干扰物的结构和性质与MIE之间的联系比其他任一节点和效应终点都强[75]。因此,正如QSAR和分子模拟所做的,大多数计算机预测模型都以MIE为研究对象。然而,MIE与AO并不是直接的相互关系,它们之间至少存在一个KE,并且不同的MIE都有可能导致相同的AO产生,形成AOP网络(图2)[76]。以前面Villeneuve[73]开发的雄激素受体激动效应导致生殖功能紊乱的AOP为例,血液雌激素浓度降低同时会与芳香酶活性降低导致生殖毒性的AOP(AOP7)产生交联,通过另一条AOP造成生殖问题,多条通路的交联从而形成AOP网络(图3)。干扰物与最终AO之间的网络关系,使针对单一靶标受体的预测方法具有更大的不确定性。
将MIE、KE、AO之间用一系列的数学模型联系起来[77],可建立定量AOP(qAOP)模型。学者们相信,通过建立qAOP可以将体外实验得到的数据,如配体受体结合效力,作为qAOP的输入信息,通过系列数学模型的模拟计算预测潜在的内分泌干扰活性,甚至模拟剂量-效应关系和时间进程行为[71]。AOP-KB平台上推出了Effectopedia模块,通过量化KE之间的关系,建立qAOP。目前,Effectopedia模块还处于发展阶段,但Beta版本的软件已发布(http://www.effectopedia.org/)。此外,还有其他系统生物学计算模拟软件,如PK-Sim和MoBi都具有很好的qAOP模型构建和模拟功能[78]。然而,qAOP需建立于明确的作用机制之下,目前还处于发展阶段,其定量预测能力还有待实验验证。
既然可以利用体外实验的数据,通过AOP预测最终的干扰效应,那么计算预测方法与AOP的结合也将成为可能。事实上,AOP概念正是来源于利用QSAR、生物标记物(biomarker)和其他机制数据提高对毒理学的认识和预测化学品暴露的潜在有害影响的想法[64]。由于对接和MD模拟的靶标往往与AOP中的MIE或KE相对应,分子模拟与AOP结合进行模拟预测将成为AOP发展中的重要研究方向。然而,限于目前AOP仍处于起步阶段,还没有较为成功的AOP与模拟预测方法结合的案例。
5 总结和展望(Conclusions and prospect)
内分泌干扰物是导致多种疾病,如生殖疾病、肥胖症和与激素相关的癌症等的重要诱因,众多化合物都具有潜在的内分泌干扰效应,使内分泌干扰问题在化合物风险评估上显得尤为突出。然而,评估一个化合物的内分泌干扰效应需要耗费大量的成本,无法对成千上万种化学品进行逐一测试。计算毒理学大大简化了这一过程,其使用也逐渐受到认可。本文基于内分泌干扰的效应机制,介绍了分子对接、MD模拟和AOP这3种计算毒理学方法及其在内分泌干扰物筛选上的应用。
分子对接与QSAR相比更有助于效应机制的理解,通过反向对接能预测化合物可能的内分泌干扰活性终点,还能与QSAR结合构建多维QSAR模型。MD模拟有助于探索配体-受体相互作用关系及两者的变化,探索重要的结构变化,并借助热力学计算预测结合效力。AOP将MIE与最终AO用一系列KE和KER连接,形成完整的、明晰的通路甚至网络,借助AOP和AOP网络将推动计算毒理学进入新的阶段。
内分泌干扰物主要通过与内分泌系统相关靶标的相互作用,正如AOP中的MIE和KE的激活,因此,分子对接、MD模拟和AOP的结合将使计算毒理学更加面向效应机制。在内分泌干扰物的筛选中,将反向对接技术与AOP模拟结合,不仅能预测干扰物潜在的内分泌干扰敏感靶标,还能进一步通过AOP网络预测可能造成的有害结局。MD模拟的发展使得通过模拟区分促进和抑制作用逐渐成为可能,甚至可以模拟生物大分子之间的相互作用,研究MIE、KE和KE、KE之间的关系。因此,将对接和分子动力学模拟技术运用到AOP和AOP网络中进行预测和筛选,将有助于计算毒理学在内分泌干扰物筛选上的发展与应用。
[1] Bergman Å, Heindel J, Jobling S, et al. State-of-the-science of endocrine disrupting chemicals, 2012 [J]. Toxicology Letters, 2012, 211(supplement): S3
[2] Attina T M, Hauser R, Sathyanarayana S, et al. Exposure to endocrine-disrupting chemicals in the USA: A population-based disease burden and cost analysis [J]. The Lancet Diabetes & Endocrinology, 2016, 4(12): 996-1003
[3] Ren X, Cao L, Yang Y, et al. In vitro assessment of thyroid hormone receptor activity of four organophosphate esters [J]. Journal of Environmental Sciences, 2016, 45: 185-190
[4] Christen V, Crettaz P, Oberli-schr Mmli A, et al. Some flame retardants and the antimicrobials triclosan and triclocarban enhance the androgenic activity in vitro [J]. Chemosphere, 2010, 81(10): 1245-1252
[5] Li F, Xie Q, Li X, et al. Hormone activity of hydroxylated polybrominated diphenyl ethers on human thyroid receptor-β: In vitro and in silico investigations [J]. Environmental Health Perspectives, 2010, 118(5): 602-606
[6] Ren X M, Guo L-H. Assessment of the binding of hydroxylated polybrominated diphenyl ethers to thyroid hormone transport proteins using a site-specific fluorescence probe [J]. Environmental Science & Technology, 2012, 46(8): 4633-4640
[7] Christiansen S, Kortenkamp A, Axelstad M, et al. Mixtures of endocrine disrupting contaminants modelled on human high end exposures: An exploratory study in rats [J]. International Journal of Andrology, 2012, 35(3): 303-316
[8] Walker D M, Kermath B A, Woller M J, et al. Disruption of reproductive aging in female and male rats by gestational exposure to estrogenic endocrine disruptors [J]. Endocrinology, 2013, 154(6): 2129-2143
[9] Porter E, Crump D, Egloff C, et al. Use of an avian hepatocyte assay and the avian toxchip polymerse chain reaction array for testing prioritization of 16 organic flame retardants [J]. Environmental Toxicology and Chemistry, 2014, 33(3): 573-582
[10] Roig B, Cadiere A, Bressieux S, et al. Environmental concentration of nonylphenol alters the development of urogenital and visceral organs in avian model [J]. Environment International, 2014, 62: 78-85
[11] Barber L B, Vajda A M, Douville C, et al. Fish endocrine disruption responses to a major wastewater treatment facility upgrade [J]. Environmental Science & Technology, 2012, 46(4): 2121-2131
[12] Liao P, Chu S, Tu T, et al. Persistent endocrine disruption effects in medaka fish with early life-stage exposure to a triazole-containing aromatase inhibitor (letrozole) [J]. Journal of Hazardous Materials, 2014, 277: 141-149
[13] Li M, Cao C, Li S, et al. Thyroid endocrine disruption of azocyclotin to Xenopus laevis during metamorphosis [J]. Environmental Toxicology and Pharmacology, 2016, 43: 61-67
[14] Scholz S, Renner P, Belanger S, et al. Alternatives to in vivo tests to detect endocrine disrupting chemicals (EDCs) in fish and amphibians—Screening for estrogen, androgen and thyroid hormone disruption [J]. Critical Reviews in Toxicology, 2013, 43(1): 45-72
[15] Abdo N, Xia M, Brown C C, et al. Population-based in vitro hazard and concentration-response assessment of chemicals: The 1000 Genomes high-throughput screening study [J]. Environmental Health Perspectives, 2015, 123(5): 458-466
[16] Huang R, Sakamuru S, Martin M T, et al. Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway [J]. Scientific Reports, 2014, 4: 5664
[17] Valerio L G. In silico toxicology for the pharmaceutical sciences [J]. Toxicology and Applied Pharmacology, 2009, 241(3): 356-370
[18] Reisfeld B, Mayeno A N. What Is Computational Toxicology? [M]// Reisfeld B, Mayeno A. (eds) Computational Toxicology. Methods in Molecular Biology (Methods and Protocols). Totowa, NJ: Humana Press, 2012: 1-7
[19] Lill M A, Winiger F, Vedani A, et al. Impact of induced fit on ligand binding to the androgen receptor: A multidimensional QSAR study to predict endocrine-disrupting effects of environmental chemicals [J]. Journal of Medicinal Chemistry, 2005, 48(18): 5666-5674
[20] Kovarich S, Papa E, Gramatica P. QSAR classification models for the prediction of endocrine disrupting activity of brominated flame retardants [J]. Journal of Hazardous Materials, 2011, 190(1): 106-112
[21] Zhang L, Sedykh A, Tripathi A, et al. Identification of putative estrogen receptor-mediated endocrine disrupting chemicals using QSAR-and structure-based virtual screening approaches [J]. Toxicology and Applied Pharmacology, 2013, 272(1): 67-76
[22] Hamers T, Kamstra J H, Sonneveld E, et al. In vitro profiling of the endocrine-disrupting potency of brominated flame retardants [J]. Toxicological Sciences, 2006, 92(1): 157-173
[23] Thomas R S, Black M, Li L, et al. A comprehensive statistical analysis of predicting in vivo hazard using high-throughput in vitro screening [J]. Toxicological Sciences, 2012, 128(2): 398-417
[24] Liu J, Mansouri K, Judson R S, et al. Predicting hepatotoxicity using ToxCast in vitro bioactivity and chemical structure [J]. Chemical Research in Toxicology, 2015, 28(4): 738-751
[25] Li F, Chen J, Qiao X, et al. Combined SVM-PLS method for predicting estrogenic activities of organic chemicals [J]. Organohalogen Compounds, 2009, 71: 1537-1541
[26] Cui S, Liu S, Yang J, et al. Quantitative structure-activity relationship of estrogen activities of bisphenol A analogs [J]. Chinese Science Bulletin, 2006, 51(3): 287-292
[27] Mao L, Colosi L M, Gao S, et al. Understanding ligninase-mediated reactions of endocrine disrupting chemicals in water: Reaction rates and quantitative structure-activity relationships [J]. Environmental Science & Technology, 2011, 45(14): 5966-5972
[28] Wu Y, Wang Y, Zhang A, et al. Three-dimensional quantitative structure-activity relationships of flavonoids and estrogen receptors based on docking [J]. Chinese Science Bulletin, 2010, 55(15): 1488-1494
[29] Yi Z, Zhang A. A QSAR study of environmental estrogens based on a novel variable selection method [J]. Molecules, 2012, 17(5): 6126-6145
[30] Fujita T, Winkler D A. Understanding the roles of the “two QSARs” [J]. Journal of Chemical Information and Modeling, 2016, 56(2): 269-274
[31] Grun F, Blumberg B. Environmental obesogens: Organotins and endocrine disruption via nuclear receptor signaling [J]. Endocrinology, 2006, 147(6): s50-s55
[32] Danzo B J. Environmental xenobiotics may disrupt normal endocrine function by interfering with the binding of physiological ligands to steroid receptors and binding proteins [J]. Environmental Health Perspectives, 1997, 105(3): 294-301
[33] Lengauer T, Rarey M. Computational methods for biomolecular docking [J]. Current Opinion in Structural Biology, 1996, 6(3): 402-406
[34] Moroy G, Martiny V Y, Vayer P, et al. Toward in silico structure-based ADMET prediction in drug discovery [J]. Drug Discovery Today, 2012, 17(1): 44-55
[35] Tanenbaum D M, Wang Y, Williams S P, et al. Crystallographic comparison of the estrogen and progesterone receptor’s ligand binding domains [J]. Proceedings of the National Academy of Sciences, 1998, 95(11): 5998-6003
[36] Sack J S, Kish K F, Wang C, et al. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone [J]. Proceedings of the National Academy of Sciences, 2001, 98(9): 4904-4909
[37] Kauppi B, Jakob C, Färnegårdh M, et al. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain RU-486 induces a transconformation that leads to active antagonism [J]. Journal of Biological Chemistry, 2003, 278(25): 22748-22754
[38] Huber B R, Desclozeaux M, West B L, et al. Thyroid hormone receptor-β mutations conferring hormone resistance and reduced corepressor release exhibit decreased stability in the N-terminal ligand-binding domain [J]. Molecular Endocrinology, 2003, 17(1): 107-116
[39] Nascimento A S, Dias S M G, Nunes F M, et al. Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function [J]. Journal of Molecular Biology, 2006, 360(3): 586-598
[40] Bongrand P. Ligand-receptor interactions [J]. Reports on Progress in Physics, 1999, 62(6): 921-968
[41] Fraccalvieri D, Soshilov A A, Karchner S I, et al. Comparative analysis of homology models of the Ah receptor ligand binding domain: Verification of structure-function predictions by site-directed mutagenesis of a nonfunctional receptor [J]. Biochemistry, 2013, 52(4): 714-725
[42] Nose T, Tokunaga T, Shimohigashi Y. Exploration of endocrine-disrupting chemicals on estrogen receptor α by the agonist/antagonist differential-docking screening (AADS) method: 4-(1-Adamantyl) phenol as a potent endocrine disruptor candidate [J]. Toxicology Letters, 2009, 191(1): 33-39
[43] D'Ursi P, Salvi E, Fossa P, et al. Modelling the interaction of steroid receptors with endocrine disrupting chemicals [J]. BMC Bioinformatics, 2005, 6(suppl 4): 1-8
[45] Yuriev E, Holien J, Ramsland P A. Improvements, trends, and new ideas in molecular docking: 2012-2013 in review [J]. Journal of Molecular Recognition, 2015, 28(10): 581-604
[46] Wang X, Zhang X, Xia P, et al. A high-throughput, computational system to predict if environmental contaminants can bind to human nuclear receptors [J]. Science of The Total Environment, 2017, 576: 609-616
[47] Vedani A, Dobler M, Lill M A. Combining protein modeling and 6D-QSAR. Simulating the binding of structurally diverse ligands to the estrogen receptor [J]. Journal of Medicinal Chemistry, 2005, 48(11): 3700-3703
[48] Vedani A, Smiesko M, Spreafico M, et al. Virtual ToxLab-in silico prediction of the toxic (endocrine-disrupting) potential of drugs, chemicals and natural products. Two years and 2,000 compounds of experience: A progress report [J]. Altex, 2009, 26(3): 167-176
[49] Yuriev E, Ramsland P A. Latest developments in molecular docking: 2010-2011 in review [J]. Journal of Molecular Recognition, 2013, 26(5): 215-239
[50] Yuriev E, Agostino M, Ramsland P A. Challenges and advances in computational docking: 2009 in review [J]. Journal of Molecular Recognition, 2011, 24(2): 149-164
[51] Rehan M, Ahmad E, Sheikh I A, et al. Androgen and progesterone receptors are targets for bisphenol A (BPA), 4-methyl-2,4-bis-(p-hydroxyphenyl) pent-1-ene—A potent metabolite of BPA, and 4-tert-octylphenol: A computational insight [J]. PloS One, 2015, 10(9): e0138438
[52] Mansouri K, Abdelaziz A, Rybacka A, et al. CERAPP: Collaborative estrogen receptor activity prediction project [J]. Journal of Environmental Health Perspectives, 2016, 124(7): 1023-1033
[53] Sivanesan D, Rajnarayanan R V, Doherty J, et al. In-silico screening using flexible ligand binding pockets: A molecular dynamics-based approach [J]. Journal of Computer-aided Molecular Design, 2005, 19(4): 213-228
[54] Hansson T, Oostenbrink C, van Gunsteren W. Molecular dynamics simulations [J]. Current Opinion in Structural Biology, 2002, 12(2): 190-196
[55] Karplus M, Mccammon J A. Molecular dynamics simulations of biomolecules [J]. Nature Structural & Molecular Biology, 2002, 9(9): 646-652
[56] Li X, Ye L, Wang X, et al. Molecular docking, molecular dynamics simulation, and structure-based 3D-QSAR studies on estrogenic activity of hydroxylated polychlorinated biphenyls [J]. Science of the Total Environment, 2012, 441: 230-238
[57] Hillisch A, von Langen J, Menzenbach B, et al. The significance of the 20-carbonyl group of progesterone in steroid receptor binding: A molecular dynamics and structure-based ligand design study [J]. Steroids, 2003, 68(10): 869-878
[58] Wang X, Yang H, Hu X, et al. Effects of HO-/MeO-PBDEs on androgen receptor: In vitro investigation and helix 12-involved MD simulation [J]. Environmental Science & Technology, 2013, 47(20): 11802-11809
[59] Martínez L, Sonoda M T, Webb P, et al. Molecular dynamics simulations reveal multiple pathways of ligand dissociation from thyroid hormone receptors [J]. Biophysical Journal, 2005, 89(3): 2011-2023
[60] Peräkylä M. Ligand unbinding pathways from the vitamin D receptor studied by molecular dynamics simulations [J]. European Biophysics Journal, 2009, 38(2): 185-198
[61] Martínez L, Webb P, Polikarpov I, et al. Molecular dynamics simulations of ligand dissociation from thyroid hormone receptors: Evidence of the likeliest escape pathway and its implications for the design of novel ligands [J]. Journal of Medicinal Chemistry, 2006, 49(1): 23-26
[62] Yang L, Zou J, Xie H, et al. Steered molecular dynamics simulations reveal the likelier dissociation pathway of imatinib from its targeting kinases c-Kit and Abl [J]. PLoS One, 2009, 4(12): e8470
[63] Celik L, Lund J D D, Schiøtt B. Conformational dynamics of the estrogen receptor α: Molecular dynamics simulations of the influence of binding site structure on protein dynamics [J]. Biochemistry, 2007, 46(7): 1743-1758
[64] van Lipzig M M, ter Laak A M, Jongejan A, et al. Prediction of ligand binding affinity and orientation of xenoestrogens to the estrogen receptor by molecular dynamics simulations and the linear interaction energy method [J]. Journal of Medicinal Chemistry, 2004, 47(4): 1018-1030
[65] Martínez L, Nascimento A S, Nunes F M, et al. Gaining ligand selectivity in thyroid hormone receptors via entropy [J]. Proceedings of the National Academy of Sciences, 2009, 106(49): 20717-20722
[66] Zeng J, Li W, Zhao Y, et al. Insights into ligand selectivity in estrogen receptor isoforms: Molecular dynamics simulations and binding free energy calculations [J]. The Journal of Physical Chemistry B, 2008, 112(9): 2719-2726
[67] Chen Q, Wang X, Shi W, et al. Identification of thyroid hormone disruptors among HO-PBDEs: In vitro investigations and coregulator involved simulations [J]. Environmental Science & Technology, 2016, 50(22): 12429-12438
[68] Sonoda M T, Martínez L, Webb P, et al. Ligand dissociation from estrogen receptor is mediated by receptor dimerization: Evidence from molecular dynamics simulations [J]. Molecular Endocrinology, 2008, 22(7): 1565-1578
[69] Costantino G, Entrena-guadix A, Macchiarulo A, et al. Molecular dynamics simulation of the ligand binding domain of farnesoid X receptor. Insights into helix-12 stability and coactivator peptide stabilization in response to agonist binding [J]. Journal of Medicinal Chemistry, 2005, 48(9): 3251-3259
[70] Yang X, Xie H, Chen J, et al. Anionic phenolic compounds bind stronger with transthyretin than their neutral forms: Nonnegligible mechanisms in virtual screening of endocrine disrupting chemicals [J]. Chemical Research in Toxicology, 2013, 26(9): 1340-1347
[71] Edwards S W, Tan Y, Villeneuve D L, et al. Adverse outcome pathways—Organizing toxicological information to improve decision making [J]. Journal of Pharmacology and Experimental Therapeutics, 2016, 356(1): 170-181
[72] Ankley G T, Bennett R S, Erickson R J, et al. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment [J]. Environmental Toxicology and Chemistry, 2010, 29(3): 730-741
[73] Villeneuve D L. Androgen receptor agonism leading to reproductive dysfunction [DB/OL]. [2017-02-06]. https://aopwiki.org/aops/23
[74] Villeneuve D L, Crump D, Garcia-reyero N, et al. Adverse outcome pathway (AOP) development I: Strategies and principles [J]. Toxicological Sciences, 2014, 142(2): 312-320
[75] Allen T E, Goodman J M, Gutsell S, et al. Defining molecular initiating events in the adverse outcome pathway framework for risk assessment [J]. Chemical Research in Toxicology, 2014, 27(12): 2100-2112
[76] Villeneuve D L, Crump D, Garcia-reyero N, et al. Adverse outcome pathway development II: Best practices [J]. Toxicological Sciences, 2014, 142(2): 321-330
[77] Chai L E, Loh S K, Low S T, et al. A review on the computational approaches for gene regulatory network construction [J]. Computers in Biology and Medicine, 2014, 48: 55-65
[78] Eissing T, Kuepfer L, Becker C, et al. A computational systems biology software platform for multiscale modeling and simulation: Integrating whole-body physiology, disease biology, and molecular reaction networks [J]. Frontiers in Physiology, 2011, 2: 1-10
◆
ApplicationandProspectofComputationalToxicologyinScreeningofEndocrineDisruptingChemicals
Chen Qinchang, Tan Haoyue, Shi Wei, Yu Hongxia*
State Key Laboratory of Pollution Control and Resource Reuse, School of the Environment, Nanjing University, Nanjing 210023, China
10.7524/AJE.1673-5897.20170206001
2017-02-06录用日期2017-03-13
1673-5897(2017)3-038-11
X171.5
A
于红霞(1963-),女,博士,教授,主要从事有机污染化学、环境监测和毒理分析等领域的研究。
国家自然科学基金(21577058);国家环保部公益性行业科研专项(201409040)
陈钦畅(1991-),男,博士研究生,研究方向为计算毒理学,E-mail:cqchang@outlook.com;
*通讯作者(Corresponding author), E-mail: yuhx@nju.edu.cn
陈钦畅, 谭皓月, 史薇, 等. 计算毒理学在内分泌干扰物筛选上的应用和展望[J]. 生态毒理学报,2017, 12(3): 38-48
Chen Q C, Tan H Y, Shi W, et al. Application and prospect of computational toxicology in screening of endocrine disrupting chemicals [J]. Asian Journal of Ecotoxicology, 2017, 12(3): 38-48 (in Chinese)
