Long non-coding RNA in liver metabolism and disease:Current status☆
2017-05-07YulnZhoJinguoWuSuthtLingpunskulLiWng
Yuln Zho,Jinguo Wu,Sutht Lingpunskul,Li Wng,e,f,g,*
aDepartment of Physiology and Neurobiology,The Institute for Systems Genomics,University of Connecticut,Storrs,CT,USA
bDivision of Gastroenterology and Hepatology,Department of Medicine,Indiana University School of Medicine,IN,USA
cDepartment of Biochemistry and Molecular Biology,Indiana University School of Medicine,Indianapolis,IN,USA
dRoudebush Veterans Administration Medical Center,Indianapolis,IN,USA
eVeterans Affairs Connecticut Healthcare System,West Haven,CT,USA
fDepartment of Internal Medicine,Section of Digestive Diseases,Yale University,New Haven,CT,USA
gSchool of Pharmaceutical Sciences,Wenzhou Medical University,Wenzhou,Zhejiang,China
1.Introduction
With the development of high-throughput RNA sequencing technology in transcriptome analysis and bioinformatics prediction,RNA transcripts,which do not code proteins but might have biological functions,constitute a new perspective in major physiological and pathological processes.It is estimated that more than 80%of the human genome is pervasively transcribed;however,protein-coding genes account for less than 2%of the human genome.1This leaves the balance of the human genome to be transcribed into thousands of non-coding RNAs(ncRNAs).According to size,the ncRNA transcripts are arbitrarily categorized into two main subgroups:short ncRNA(<200 bp)and long ncRNAs(lncRNAs)(>200 bp).Short ncRNAs include highly abundant and functionally important RNAs such as transfer RNAs(tRNAs)and ribosomal RNAs(rRNAs),as well as small regulatory RNAs such as microRNAs(miRNAs,miRs),short interfering RNAs(siRNAs),and PIWI-interacting RNAs(piRNAs).2While lncRNAs are commonly defined as transcripts of more than 200 nucleotides in length that are incapable of being translated into proteins.3Extensive studies have been conducted onmiRNAs,howeverthe functions of lncRNAs remain poorly understood.4,5Recent studies have revealed that lncRNAs significantly impact on various biological processes as X-inactivation,genomic imprinting,chromatin modification,cell fate specification,cell apoptosis,nuclear trafficking,and genome rearrangement(Table 1).6
We focus on the role of lncRNAs in liver metabolic functions and associated liverdiseases(Table 1).This new emerging research field is expected to significantly increase our understanding the fundamental mechanisms that contribute to various liver diseases with the goal of developing new diagnosis,prognosis and treatment strategies(Fig.1).
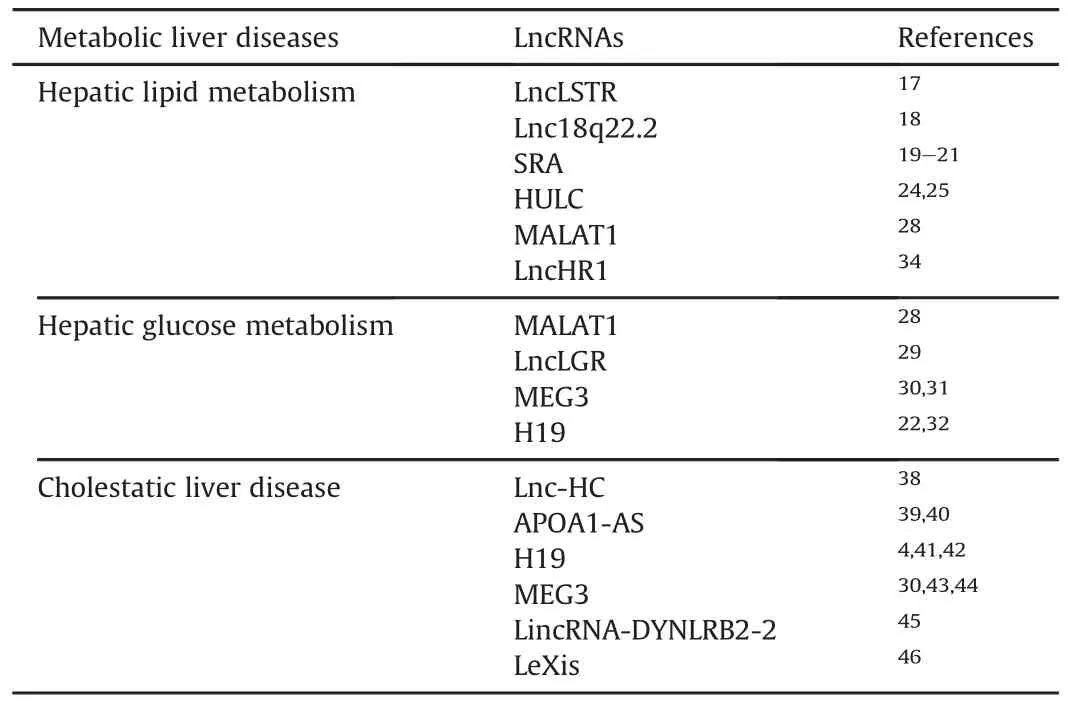
Table 1LncRNAs in metabolic liver diseases.
2.Characteristics of lncRNA
The majority of lncRNAs are transcribed from RNA polymerase II and are often capped,spliced and polyadenylated.7Unlike mRNAs,lncRNAs are poorly conserved between humans and rodents.lncRNAs are expressed in a species-and tissue-specific,and developmentally regulated manner which may contribute to their unique function.8Based on theirlocation relative to nearby proteincoding genes,lncRNAs are currently classed into five categories,namely(i)sense;(ii)antisense;(iii)bidirectional;(iv)intronic;and(v)intergenic.9As gene expression regulators,lncRNAs are involved in chromatin remodeling,epigenetic regulation,transcriptional and post-transcriptional regulation,processing of small RNAs,protein function,and other regulatory processes.10Mechanistically,molecular functions of lncRNAs are described as signals,decoys,guides,and scaffolds.11The specific expression in specific cell types and stages of development as well as the response to environmental stimuli make lncRNAs as effective signals.As decoys,lncRNAs bind and promote protein degradation.LncRNAs can also bind and recruit regulatory molecules,such as chromatin remodelers and transcription factors to specific gene targets to control gene expression.By binding different molecules with different domains,lncRNAs can also act as scaffolds,assembling a functional complex.12
3.LncRNAs in hepatic lipid and glucose metabolism
As the major site for synthesis,metabolism,storage,and redistribution of carbohydrates,proteins,and lipids,liver plays a central role in metabolic homeostasis.13Emerging studies have shown that many lncRNAs are key regulators of lipid and glucose metabolism.Lipid accumulation in liver is determined by the balance between lipogenesis and catabolic processes such as lipolysis,fatty acid boxidation and thermogenesis.14The maintenance of glucose homeostasis results from the balance of glucose production and/or storage in the liver and glucose uptake in the peripheral tissues.15Dysregulation of these processes leads to adiposity,dyslipidemia,and metabolic perturbations.16
3.1.Liver-speci fi c triglyceride regulator(lncLSTR)
Analysis of metabolic organ-enriched lncRNA identi fied that lncLSTR exhibits liver-specific expression and its expression fluctuates in responsetochanges in energylevels andmetabolic state.17LncLSTR is an intergenic lncRNA and liver-specific knockdown of lncLSTR significantly reduced plasma triglyceride (TG)via enhancing tissue TG clearance.This reduced circulating level of TG in lncLSTR knockdown mice resulted from increased apoC2 expression and LPL activities through a FXR-mediated pathway.Mechanistically,lncLSTR has been shown to directly bind toTDP43,further relieve the transcriptional suppression of Cyp8b1 and lead to substantial change in bile acid composition.The altered bile composition activate FXR and apoC2 expression,resulting in enhanced TG clearance in mice.
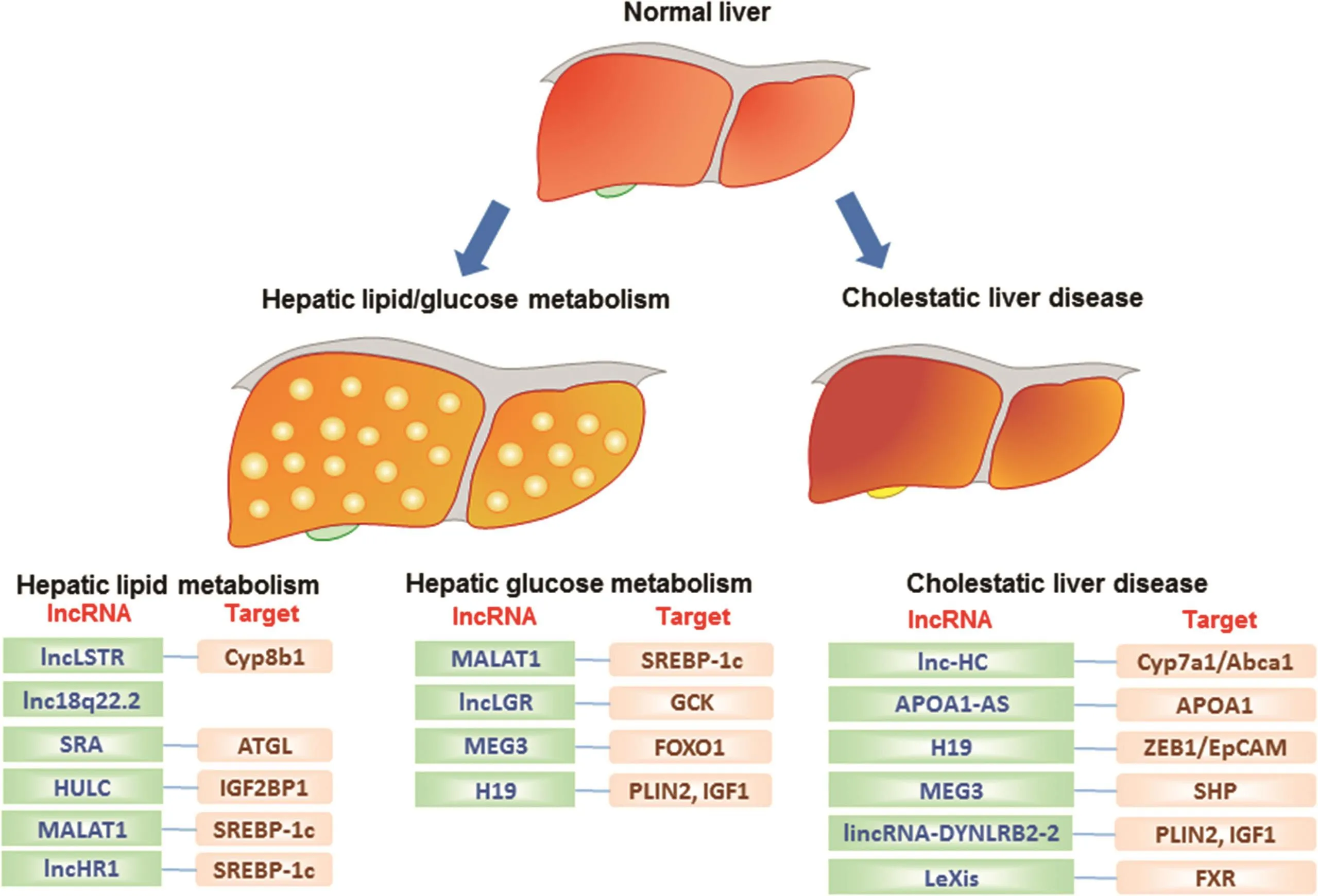
Fig.1.Diagram depicting the regulatory role of lncRNAs in liver metabolism and disease.LncRNAs(dark blue)are presented in three groups based on their function.Target genes are indicated in the right side of each lncRNA(brown).
3.2.Lnc18q22.2
Lnc18q22.2,a liver-specific lncRNA(RP11-484N16.1)on chromosome 18,has been shown to be markedly induced in the liver tissue of NASH patients,correlating with NASH grade,lobular in flammation and NAS score.18Interesting,lnc18q22.2 is signi ficantly associated with liver steatosis and steatohepatitis,but not with hepatocellular carcinoma(HCC)in 44 independent liver biopsies.Knockdown of lnc18q22.2 in hepatocyte cell lines reduced cell viability and promoted necrosis.
3.3.Steroid RNA activator(SRA)
SRA was reported existing in an SRC-1 complex and functioned as an RNA coactivator for non-steroid nuclear receptors.19A recently study showed that the expression of liver adipose triglyceride lipase(ATGL)is induced in SRA knockout mice.20SRA suppresses ATGL via inhibiting transcriptional activity of FOXO1 in an insulin-independent pathway,thus reducing FFAβ-oxidation and promoting hepatic steatosis.SRA is also the first lncRNA reported toplaya potential role in adipogenesis through regulation of PPARγand P38/JNK phosphorylation.21
3.4.Highly upregulated in liver cancer(HULC)
HULC,the first identi fied lncRNA specifically overexpressed in HCC,has been shown as an effective prognostic biomarker in many human cancers.22,23HULC can be posttranscriptional destabilized by IGF2BP1 via recruiting the CCR4-NOT deadenylase complex,a major component of the cytoplasmic RNA decay machinery.24A recently study showed that HULC induced methylation of CpG islands in the miR-9 promoter through upregulation of DNMT1.25The downregulation of miR-9 further enhanced lipogenesis and enriched intracellular triglycerides and cholesterol by activating PPARγand ACSL1 in hepatoma cells.
3.5.Metastasis associated in lung adenocarcinoma transcript 1(MALAT1)
MALAT1 is involved in the development of HCC and liver fibrosis.26,27A recently study showed that MALAT1 promoted hepatic steatosis and insulin resistance.MALAT1 induced hepatic lipid accumulation and insulin resistance by interacting with SREBP-1c and stabilizingitsprotein in hepatocytes.28Knockdown of MALAT1 effectively reversed lipid aggregation and increased insulin sensitivity inob/obmice.
3.6.Liver glucokinase repressor(lncLGR)
Glucose levels in mammals is tightly controlled through multiple mechanisms to meet systemic energy demands.As the major site of glucose regulation,the liver produces glucose through glycogenolysis and gluconeogenesis during fasting,while promoting glucose uptake and glycogen storage during feeding.lncLGR,a fasting-induced lncRNA in the liver,has been shown to suppress glucokinase(GCK)transcription and glycogen storage in fasted mice.29Mechanistically,lncLGR specifically binds to heterogenous nuclear ribonucleoprotein L(hnRNPL),an RNA-binding protein which is further con firmed to be recruited to the promoter of GCK and suppress its transcription.
3.7.Maternally expressed gene 3(MEG3)
MEG3 is upregulated in high-fat diet andob/obmice through histone acetylation.30Knockdown of MEG3 remarkably abolishes hepatic TG accumulation,up-regulates glycogen content,and promotes glucose tolerance in high-fat diet mice andob/obmice.Another study showed that MEG3 was highly expressed in mouse islets and was down-regulated during the pathogenesis of diabetes.31Moreover,the expression of MEG3 was modulated by glucose bothin vitroandin vivo.Knockdown of MEG3 reduced insulin synthesis,secretion,and increased beta cell apoptosis,leading to impaired glucose tolerance.
3.8.H19
H19 was one of the first discovered lncRNAs highly expressed in fetal mouse and human liver,but repressed in adult liver by zinc fingers and homeoboxes 2(Zhx2).32H19 is significantly upregulated in mouse model of CCl4-induced tumors and in human fibrotic/cirrhotic liver.33H19 plays a role in affecting transcription through chromatin remodeling.One study related H19 to steatosis by a lipid droplet protein PLIN2 which was significantly induced in fatty liver.22H19 was up-regulated in the liver of high fat diet fed PLIN2 knockdown mice with decreased hepatic TG level.
3.9.LncHR1
LncHR1 was recently identi fied as a novel human-specific lncRNA that lower hepatic and plasma TG levels and down regulates sterol regulatory element binding protein(SREBP)1c,fatty acid synthase(FAS),and ACCαin high-fat diet model.In Huh7 cells,lncHR1 abolished oleic acid induced TG accumulation and cytosolic lipid droplets.Mechanistically,lncHR1 regulates hepatic lipid metabolism by suppressing SREBP1c promoter activity and negatively regulating SREBP1c and FAS expression.34
4.LncRNA in cholestatic liver disease
In hepatocytes,cholesterol homeostasis is maintained by a complex network involving cholesterol uptake,synthesis,intracellular transport,and excretion.35As the end-products of cholesterol catabolism in the liver,bile acids(BAs)aresecreted bythe liver to aid in the digestion of fats and are reported as signaling molecules that regulate glucose,lipid,and energy metabolism.13,36Cholestasis is a pathological condition that BAs are blocked to flow out of liver,resulting intrahepatic accumulation.High levels of toxic BAs cause bile duct epithelium damage,hepatocytes injury and in flammation.37Chronic cholestasis then leads to liver fibrosis,cirrhosis,liver failure,HCC,and cholangiocarcinoma.
4.1.Lnc-HC
A novel lncRNA named lnc-HC is up-regulated in HFD MetS rat liver to negatively regulate cholesterol metabolism by physically interactingwith hnRNPA2B1in hepatocytes.38Thelnc-HC-hnRNPA2B1 complex further binds to two target message RNAs Cyp7a1 or Abca1;both are critical for the conversion of cholesterol into BAs and for facilitating cholesterol ef flux.Knockdown of lnc-HC significantly recovers glucose tolerance disorder,and improves total cholesterol,TG,and HDL-cholesterol in the HCD rat model.
4.2.Apolipoprotein A1 antisense transcript(APOA1-AS)
Another potential group of lncRNAs that consist of 70%of total lncRNAs is natural antisense transcripts(NATs),which regulate sense gene expression in a positive or negative manner.APOA1-AS is transcribed from the opposed DNA strand to APO gene cluster and negatively regulates APOA1 expressionin vitroandin vivo.As the major protein component of high-density lipoprotein in plasma and cofactor for the lecithin cholesterol acyltransferase,APOA1 plays a signi ficant role in reverse cholesterol transport,promoting cholesterol ef flux from tissues.39APOA1-AS epigenetically inhibits APOA1 expression and multiple neighboring genes,though LSD1 and suppressor of Zeste 12 homolog(SUZ12)recruitment.40
4.3.H19
We reported that hepatic H19 expressionwas strikingly induced in Bcl2-induced cholestatic liver injury,which was alleviated by knockdown of H19.41Hepatic overexpression of H19RNA facilitated the development of obstructive cholestatic liver fibrosis.42Mechanistically,H19 downregulated hepatic zinc finger E-box-binding homeobox 1(ZEB1),and impeded its suppression of epithelial cell adhesion molecule(EpCAM),which in turn augmented bile duct ligation(BDL)-induced liver fibrosis.In addition,H19RNA is upregulated in both PBC and PSC patients.Another group reported a similar role of H19 in cholestasis.H19 was shown to be representing a key factor that causes the gender disparity of cholestatic liver injury not only in Mdr2 KO mice,and probably in human PSC patients.4
4.4.MEG3
MEG3,which was shown to promote hepatic insulin resistance,was down-regulated in the liver of mice fed with DDC.30,43Recently,MEG3 was identi fied as a guide RNA scaffold to recruit PTBP1 to destabilize nuclear receptor Shp mRNA in cholestatic liver injury.44Overexpression of MEG3 RNA induced a rapid Shp mRNA degradation and elevation of liver injury enzymes such as alanine aminotransferase(ALT)and aspartate aminotransferase(AST)in mouse liver,and disrupted bile acids homeostasis.Moreover,the expression of MEG3 was also inhibited by SHP via a CREB-dependent feedback regulation.
4.5.DYNLRB2-2
lincRNA-DYNLRB2-2 was reported to be significantly induced in THP-1 macrophage-derived foam cells by Ox-LDL in a dose and time-dependentmanner.45Up-regulated lincRNA-DYNLRB2-2 promoted ABCA1 expression,which plays an essential role in cholesterolmetabolism and in flammation,viainducingthe expression of G protein-coupled receptor 119(GPR119)and activating GLP-1R pathway.
4.6.Liver-expressed LXR-induced sequence(LeXis)
LeXis,an lncRNA robustly induced in primary mouse hepatocytes treated with LXR agonist GW3965,decreased serum cholesterol,but not triglycerides in a mouse model.46LeXis has been shown to inhibit cholesterol biosynthesis by associating with and affecting the DNA interaction of Raly which is required for the maximal expression of cholesterologenic genes in mouse liver.
5.Conclusions and future perspectives
LncRNAs control several important aspects of liver function involved in the pathophysiology of human liver injuryand disease.5The miRNAs comprise a class of short single-stranded non-coding RNAsthatregulategeneexpression mainly through posttranscriptional regulation by binding to the 3′UTRs of target mRNAs.For example,a new study demonstrated that miR-127 modulates the pluripotency of liver tumor initiating cells by targeting HMGB2.47Compared to miRNAs,lncRNAs can exert their diverse functions through interaction with proteins,miRNAs and mRNAs.48HCC is one of the most common cancers in the world.49Recent studies have also identi fied the critical roles of lncRNAs in the pathogenesis and metastasis of HCC through regulating tumor growth and proliferation,metastasis and invasion,and apoptosis.48Novel concepts related to lncRNAs and liver physiology have been successfully integrated into the drug development process to develop effective therapies.Continued efforts are still needed to expand the current knowledge in order to develop lncRNA-based therapeutics for the treatment of chronic liver diseases(Fig.1).
Con flict of interest
The authors declare that they have no con flict of interest.
Authors'contributions
Y.Zhao,J.Wu and S.Liangpunsakul prepared the manuscript.L.Wang supervised the work and wrote the manuscript.
This work was supported by the National Institutes of Health(NIH) grants (R01DK104656,R01DK080440,R01ES025909,R21AA022482,R21AA024935,R01AA026322 to L.Wang);VA Merit Award(1I01BX002634 to L.Wang);the National Natural Scienti fic Foundation of China(Grant No.81572443 to L.Wang),VA Merit Award(1I01CX000361 to S.Liangpunsakul),NIH(U01AA021840,R01DK107682,R01AA025208,R21AA024935 to S.Liangpunsakul),US DOD(W81XWH-12-1-0497 to S.Liangpunsakul).
1.ENCODE Project Consortium.An integrated encyclopedia of DNA elements in the human genome.Nature.2012;489:57-74.
2.Gomes AQ,Nolasco S,Soares H.Non-coding RNAs:multi-tasking molecules in the cell.Int J Mol Sci.2013;14:16010-16039.
3.He Y,Meng XM,Huang C,et al.Long noncoding RNAs:novel insights into hepatocelluar carcinoma.Cancer Lett.2014;344:20-27.
4.Li X,Liu R,Yang J,et al.The role of LncRNA H19 in gender disparity of cholestatic liver injury in multidrug resistance 2 gene knockout mice.Hepatology.2017;66:869-884.
5.Yang Z,Ross RA,Zhao S,Tu W,Liangpunsakul S,Wang L.LncRNA AK054921 and AK128652 are potential serum biomarkers and predictors of patient survival with alcoholic cirrhosis.Hepatol Commun.2017;1:513-523.
6.Sun J,Lin Y,Wu J.Long non-coding RNA expression pro filing of mouse testis during postnatal development.PloS one.2013;8,e75750.
7.Danko CG,Hah N,Luo X,et al.Signaling pathways differentially affect RNA polymerase II initiation,pausing,and elongation rate in cells.Mol Cell.2013;50:212-222.
8.Backofen R,Vogel T.Biological and bioinformatical approaches to study crosstalk of long-non-coding RNAs and chromatin-modifying proteins.Cell tissue Res.2014;356:507-526.
9.Ma L,Bajic VB,Zhang Z.On the classi fication of long non-coding RNAs.RNA Biol.2013;10:925-933.
10.Moran VA,Perera RJ,Khalil AM.Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs.Nucleic acids Res.2012;40:6391-6400.
11.Wang KC,Chang HY.Molecular mechanisms of long noncoding RNAs.Mol Cell.2011;43:904-914.
12.Chen Z.Progress and prospects of long noncoding RNAs in lipid homeostasis.Mol Metab.2016;5:164-170.
13.Rudraiah S,Zhang X,Wang L.Nuclear receptors as therapeutic targets in liver disease:are we there yet?Annu Rev Pharmacol Toxicol.2016;56:605-626.
14.Lee SM,Zhang Y,Tsuchiya H,Smalling R,Jetten AM,Wang L.Small heterodimer partner/neuronal PAS domain protein 2 axis regulates the oscillation of liver lipid metabolism.Hepatology.2015;61:497-505.
15.Huang J,Iqbal J,Saha PK,et al.Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver.Hepatology.2007;46:147-157.
16.Tabbi-Anneni I,Cooksey R,Gunda V,et al.Overexpression of nuclear receptor SHP in adipose tissues affects diet-induced obesity and adaptive thermogenesis.Am J Physiol Endocrinol Metab.2010;298:E961-E970.
17.Li P,Ruan X,Yang L,et al.A liver-enriched long non-coding RNA,lncLSTR,regulates systemic lipid metabolism in mice.Cell Metab.2015;21:455-467.
18.Atanasovska B,Rensen SS,van der Sijde MR,et al.A liver-specific long noncoding RNA with a role in cell viability is elevated in human non alcoholic steatohepatitis.Hepatology.2017;66:794-888.
19.Zhao X,Patton JR,Davis SL,Florence B,Ames SJ,Spanjaard RA.Regulation of nuclearreceptoractivity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator.Mol Cell.2004;15:549-558.
20.Chen G,Yu D,Nian X,et al.LncRNA SRA promotes hepatic steatosis through repressing the expression of adipose triglyceride lipase(ATGL).Sci Rep.2016;6,35531.
21.Liu S,Xu R,Gerin I,et al.SRA regulates adipogenesis by modulating p38/JNK phosphorylation and stimulatinginsulin receptorgeneexpression and downstream signaling.PLoS One.2014;9:e95416.
22.Imai Y,Boyle S,Varela GM,et al.Effects of perilipin 2 antisense oligonucleotide treatment on hepatic lipid metabolism and gene expression.Physiol Genomics.2012;44:1125-1131.
23.Fan YH,Wu MJ,Jiang Y,et al.Long non-coding RNA HULC as a potential prognostic biomarker in human cancers:a meta-analysis.Oncotarget.2017;8:21410-21417.
24.H¨ammerle M,Gutschner T,Uckelmann H,et al.Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1(IGF2BP1).Hepatology.2013;58:1703-1712.
25.Cui M,Xiao Z,Wang Y,et al.Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway.Cancer Res.2015;75:846-857.
26.Hou Z,Xu X,Fu X,et al.HBx-related long non-coding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma.Am J Cancer Res.2017;7:845-856.
27.Yu F,Lu Z,Cai J,et al.MALAT1 functions as a competing endogenous RNA to mediate Rac1 expression by sequestering miR-101b in liver fibrosis.Cell Cycle.2015;14:3885-3896.
28.Yan C,Chen J,Chen N.Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability.Sci Rep.2016;6,22640.
29.Ruan X,Li P,Cangelosi A,Yang L,Cao HA.Long non-coding RNA,lncLGR,regulates hepatic glucokinase expression and glycogen storage during fasting.Cell Rep.2016;14:1867-1875.
30.Zhu X,Wu YB,Zhou J,Kang DM.Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression.Biochem Biophys Res Commun.2016;469:319-325.
31.You L,Wang N,Yin D,et al.Downregulation of long noncoding RNA Meg3 affects insulin synthesis and secretion in mouse pancreatic beta cells.J Cell Physiol.2016;231:852-862.
32.Perincheri S,Dingle RW,Peterson ML,Spear BT.Hereditary persistence of alpha-fetoprotein and H19 expression in liver of BALB/cJ mice is due to a retrovirus insertion in the Zhx2 gene.Proc Natl Acad Sci U S A.2005;102:396-401.
33.Chen X,Yamamoto M,Fujii K,et al.Differential reactivation of fetal/neonatal genes in mouse liver tumors induced in cirrhotic and non-cirrhotic conditions.Cancer Sci.2015;106:972-981.
34.Li D,Cheng M,Niu Y,et al.Identi fication of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c.Int J Biol Sci.2017;13:349-357.
35.Datta S,Wang L,Moore DD,Osborne TF.Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter by nuclear receptors liver receptor homologue-1 and small heterodimer partner:a mechanism for differential regulation of cholesterol synthesis and uptake.J Biol Chem.2006;281:807-812.
36.Chiang JYL.Bile acid metabolism and signaling in liver disease and therapy.Liver Res.2017;1:3-9.
37.Jung D,York JP,Wang L,et al.FXR-induced secretion of FGF15/19 inhibits CYP27 expression in cholangiocytes through p38 kinase pathway.Pflugers Arch.2014;466:1011-1019.
38.Lan X,Yan J,Ren J,et al.A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism.Hepatology.2016;64:58-72.
39.Glomset JA.The plasma lecithins:cholesterol acyltransferase reaction.J lipid Res.1968;9:155-167.
40.Halley P,Kadakkuzha BM,Faghihi MA,et al.Regulation of the apolipoprotein gene cluster by a long noncoding RNA.Cell Rep.2014;6:222-230.
41.Zhang Y,Liu C,Barbier O,et al.Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function.Sci Rep.2016;6,20559.
42.Song Y,Liu C,Liu X,et al.H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule.Hepatology.2017;66:1183-1196.
43.Oliva J,Bardag-Gorce F,French BA,Li J,French SW.The regulation of noncoding RNA expression in the liver of mice fed DDC.Exp Mol Pathol.2009;87:12-19.
44.Zhang L,Yang Z,Trottier J,Barbier O,Wang L.Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay.Hepatology.2017;65:604-615.
45.Hu YW,Yang JY,Ma X,et al.A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis.J lipid Res.2014;55:681-697.
46.Sallam T,Jones MC,Gilliland T,et al.Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis.Nature.2016;534:124-128.
47.Zhao Y,Yang Z,Wu J,et al.High-mobility-group protein 2 regulated by microRNA-127 and small heterodimer partner modulates pluripotency of mouse embryonic stem cells and liver tumor initiating cells.Hepatol Commun.2017;1:816-830.
48.Sun J,Bie B,Zhang S,Yang J,Li Z.Long non-coding RNAs:critical players in hepatocellular carcinoma.Int J Mol Sci.2014;15:20434-20448.
49.Yang Z,Koehler AN,Wang L.A novel small molecule activator of nuclear receptor SHP inhibits HCC cell migration via suppressing Ccl2.Mol Cancer Ther.2016;15:2294-2301.
杂志排行
Liver Research的其它文章
- Interleukin-22 in the pathogenesis and potential treatment of liver diseases☆
- Recent development and gene therapy for glycogen storage disease type Ia☆
- FOXO transcription factors in non-alcoholic fatty liver disease☆
- Interaction between stress responses and circadian metabolism in metabolic disease☆
- Decoding the role of extracellular vesicles in liver diseases☆
- Guide for Authors
