猪场废水厌氧消化液好氧处理过程酸化改进及菌群结构变化
2017-05-03邓良伟姜奕圻
王 伸, 邓良伟, 徐 则 , 王 霜, 姜奕圻, 郑 丹
( 1.农业部沼气科学研究所, 成都 610041; 2.农业部农村可再生能源开发利用重点实验室, 成都 610041)
猪场废水厌氧消化液好氧处理过程酸化改进及菌群结构变化
王 伸1,2, 邓良伟1,2, 徐 则1,2, 王 霜1,2, 姜奕圻1,2, 郑 丹1,2
( 1.农业部沼气科学研究所, 成都 610041; 2.农业部农村可再生能源开发利用重点实验室, 成都 610041)

猪场废水厌氧消化液; SBR; pH值; 16SrRna; 厌氧氨氧化

1 试验材料和方法
1.1 污泥和污水
试验所用接种污泥来源于实验室培养的好氧污泥(具有硝化、反硝化活性)和厌氧氨氧化污泥。试验进水为四川邛崃某猪场废水处理沼气工程厌氧反应器出水(厌氧消化液),以及经过固液分离但未经过厌氧处理的固液分离出水,简称原水。
1.2 试验装置
试验采用SBR工艺,实验装置为直径17 cm,高度33.8 cm的刻度塑料桶,总容积6.0 L,有效容积为5.0 L。
1.3 试验方案
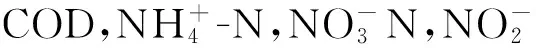
1.4 检测项目及分析方法
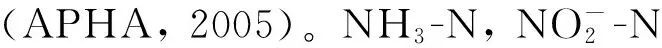
1.5 微生物高通量测序分析
取适量污泥样品,使用E.Z.N.A Soil DNA 试剂盒(Omega Bio-tek,Norcross,GA,U.S.)提取微生物总DNA。以16S rRNA V3~V4 区内338F (5'-ACTCCT ACGGGAGGCAGCA-3')和806R (5'-GGACTAC HVGGGTWTCTAA T-3')为特征引物,采用20 L混合反应体系,在GeneAmp 9700(ABI)型PCR 扩增仪上完成目标片段扩增。反应程序为95℃预变性2 min,95℃变性30s,55℃退火30 s, 72℃延伸30 s,25个循环后,72℃延伸5 min,每个样品重复3次。使用AxyPrepDNA凝胶回收试剂盒(Axygen,Union City,CA,U.S.)对PCR 扩增产物进行回收。基于Illumina Miseq PE300 平台,委托上海美吉生物医药科技有限公司完成对PCR 扩增产物的高通量测序,在多样性评估的基础上,采用Qiime 软件进行微生物分类学分析。
2 结果与讨论
2.1 反应器中混合液pH值变化
当反应器混合液pH值<6.5时,可以认为是酸化。3个反应器中混合液pH值变化列于图1。从图1中可以看出,试验期间只有CG组出现明显酸化现象,从第49天起pH值开始下降,直至稳定在5.8左右。两种改进策略AN组合RW组,只有AN组在第58~66天,出现短暂的酸化,其他时间都未出现酸化。在试验后期(第68天后),pH值比较稳定,AN组、RW组出水pH值平均值分别为7.7和7.7,说明两种改进策略能明显抑制酸化。

图1 不同反应器曝气结束时混合液pH值
2.2 SBR对COD的去除
图2和图3显示了SBR对猪场废水厌氧消化液COD去除效果。从图2和图3可知,在试验前期(低68 d前),CG组和AN组进水为厌氧消化液,其COD浓度为461 mg·L-1,COD去除率分别为7.09%和26.6%,波动较大。添加原水的RW组,进水COD为1806 mg·L-1,去除率为65.3%,随着进水COD升高,COD去除率也相应升高。在第69~91 d期间,CG组和AN组进水(厌氧消化液)COD浓度为621 mg·L-1,COD去除率分别为-13.2%和47.0%。添加原水的RW组,进水COD为3761 mg·L-1,去除率为89.4%。通过和CG组对比发现,两种酸化改进策略都能提高COD去除率,AN组和RW组分别提高了60.2%和102.6%,其中添加原水组最明显,但AN组出水COD浓度比较低。酸化后有机物的去除效果差的原因是异养细菌的最适生长pH值范围为6.5~7.5。当pH值在6.5以下时,异养细菌活性将受到抑制,在低pH值下微生物解体[18],导致出水COD大于进水COD,COD去除率为负值。添加原水后COD去除效率提高的原因是,厌氧消化液BOD5/COD 值0.26(见表1),不易生化降解;添加原水后 BOD5/COD由0.26提高到0.32改善了可生化性,为后处理中好氧微生物的生长提供了易降解的有机碳源[19],关键是pH值处于稳定在7.7左右,有利于微生物生长代谢;对于AN组,同样也是因为未出现酸化,系统处于有利于异养微生物生长的pH值之间。Bortone[20]等研究发现猪粪污水好氧生化处理出水中含有大约300 mg·L-1的难降解COD,说明AN组和RW组对COD已经达到最大程度去除。

图2 进出水 COD 浓度

图3 进出水COD 去除率
2.3 SBR对氮的去除
2.3.1 氨氮浓度和氨氮转化率

表1 运行稳定时(第69~91天)进出水主要污染物浓度和污染物去除率
2.3.2 氨氮转化产物和TIN去除率

图4 进出水浓度

图5 进出水-N去除率
2.4 微生物菌群结构的变化
利用Miseq 高通量测序平台对对照组和改进组污泥中微生物多样性进行了分析。高达99.69%以上的覆盖率表明,测序结果能真实反映样品中的菌群分布情况。CG组,AN组和RW组得到相同的24523条有效序列,平均长度为分别为443,441和441 bp,其中片段长度在421~460 bp之间旳序列占总序列数的99.89%,在97%的相似水平上可聚类产生458,502和440个OTU。从稀释度曲线可看出(图5),4组样品的曲线均趋于平坦,样品的测序数据量有效,可反映样本真实的微生物群落结构。不同反应器中样品在97%相似性上的维恩图如图8所示,显示了不同样品的OTU数目组成相似性及重叠情况。其中,有370个OTU为3组样品所共有,占各组总数的73.7%以上。不同样品的细菌群落多样性指数如表2所示。表中Chao和ACE表征菌群的丰富度,数值越大,表示样品中群落结构越丰富。Shannon和Simpson指数常用来估算样本中微生物的多样性,Shannon值越大,说明群落多样性越高,而Simpson指数值越大,说明群落多样性越低,均一性越差。表2中数据可看出,改进AN组增加了反应器中细菌群落的丰富度和多样性,同时也增加了细菌群落的均一性;而改进RW组也增加了反应器中细菌群落的丰富度和多样性,同时也增加了细菌群落的均一性,但效果不明显。对测序样品得到的序列进行比对分析,3组样品在生物分类学门的水平上进行分类,均检测到12个门,且3者门的组成类别相同,但所占比例不同,占总比例的96.1%以上。由图8可知变形菌门 (Proteobacteria) 和拟杆菌门(Bacteroidetes)为3组反应器共有的优势菌门。改进组使变形菌门 (Proteobacteria) 和拟杆菌门(Bacteroidetes)丰度显著减少,而丰度有所减小。图8显示,Proteobacteria( 变形菌门) 是各污泥样品中最丰富的门,CG,AN和RW分别占37.1%,35.1%和32.4%,改进组AN和RW组丰度分别下降2.0%和4.7%。其中,变形菌门是细菌中最大的一个门,包含多种代谢种类的细菌,变形菌门细菌根据rRNA序列被分为五类,分别以希腊字母分别以希腊字母α,β,γ,δ和ε命名,β变形菌和γ变形菌以有机物为碳源,以呼吸和发酵代谢方式进行兼性异养生长,去除废水中有机物主要参与者[27-28];δ变形菌包括严格厌氧的一些种类,同样也具有降解COD功能。另外,α-,β-和γ这3类变形菌包含了常见的氨氧化细菌(AOB),亚硝酸氧化细菌(NOB)以及反硝化细菌种属[29],是废水处理系统中含氮污染物去除的主要参与者。笔者实验中这3类细菌的大量出现,推测与本反应器中氮素的去除相关。CG,AN,和RW组第二主要门是拟杆菌门(Bacteroidetes),分别占29.2%,24.5%和20.8%。拟杆菌门细菌是反应器中的另一类优势菌。拟杆菌是化能有机营养细菌,代谢碳水化合物,能够将复杂的有机物如:纤维素、淀粉等水解为单糖,再降解为乳糖、乙酸、甲酸等;将蛋白质水解为氨基酸和有机酸等;将脂类水解为低级的脂肪酸[30]。笔者研究中检测到在拟杆菌门具有一定的优势,可能在有机物的去除中发挥了作用。
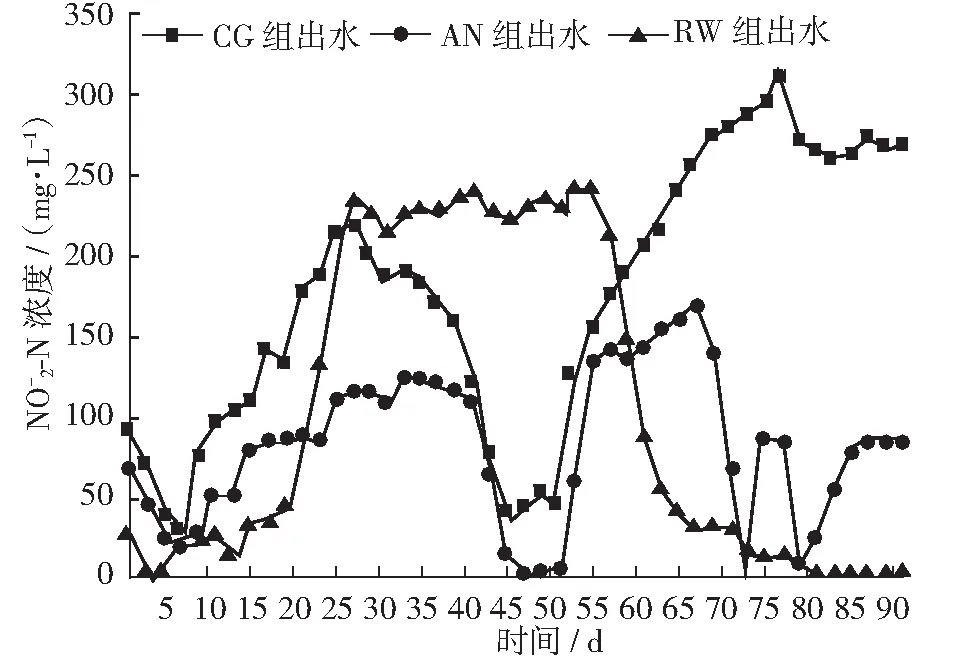
图6 各反应器出水 -N 浓度变化
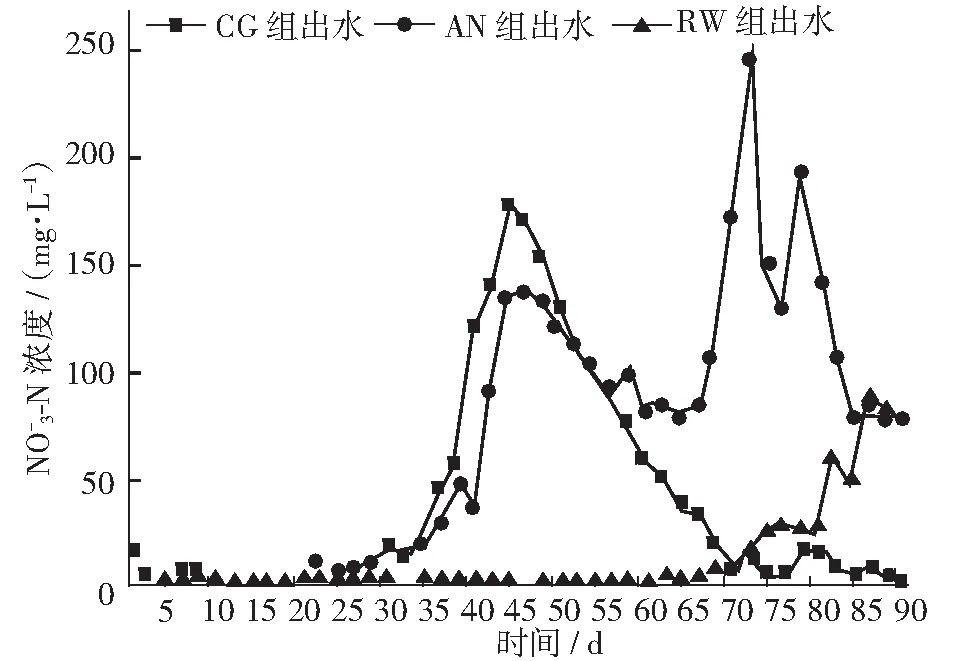
图7 各反应器出水 -N浓度变化
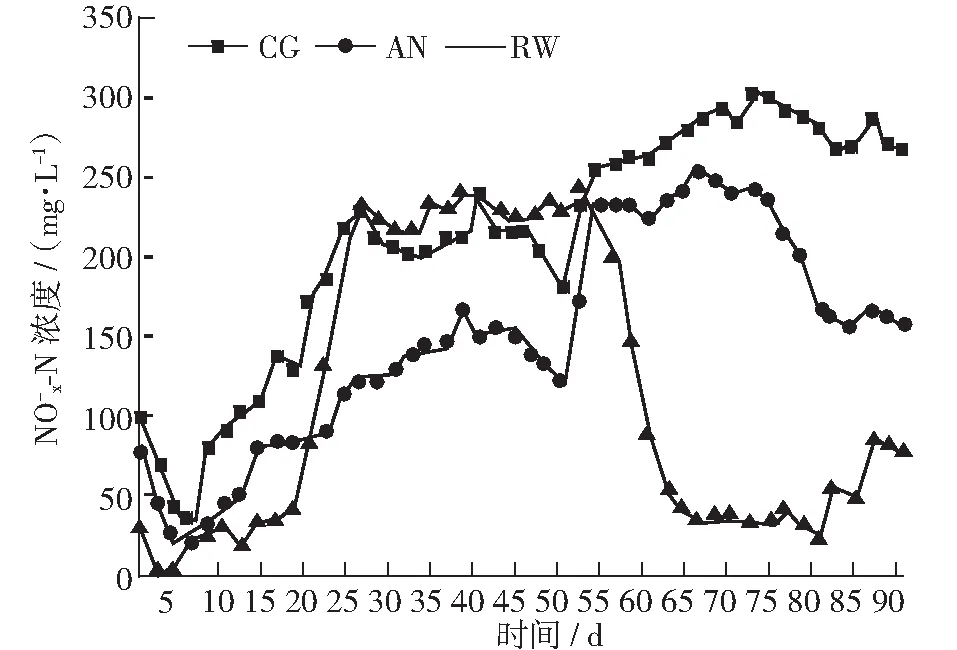
图8 各反应器出水 NOx--N 浓度变化

图9 3个反应器的TIN去除率
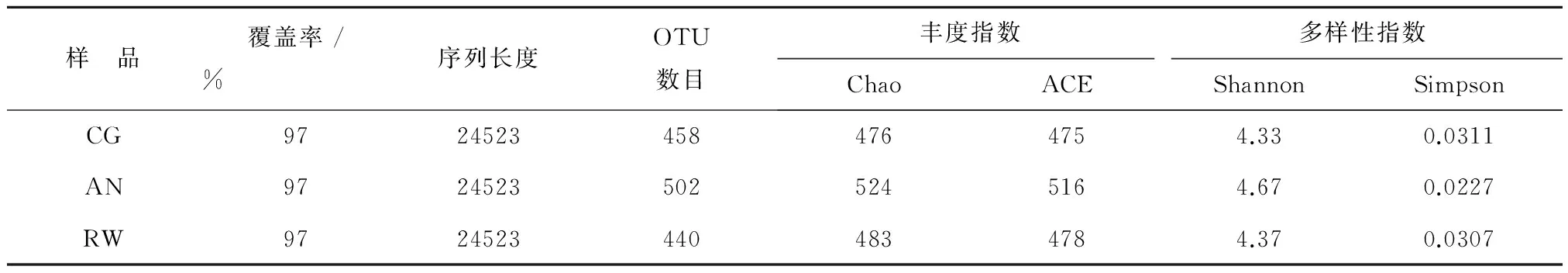
表2 微生物丰度和多样性情况
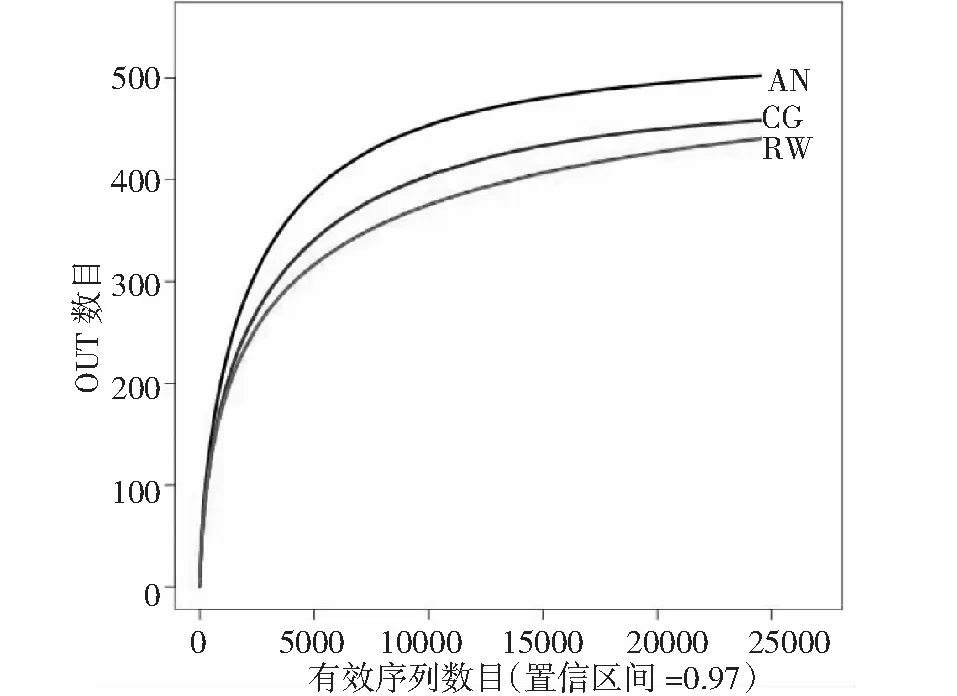
图10 3个样品稀释曲线图

图11 3个反应器各菌群的Venn图
图12显示的是4个样品在属的水平上主要菌群的分布情况。和CG组相比,AN组丰度下降最明显的3个组分别是Nitrosomonas,从7.95%下降到4.25%;OPB35_soil_group_norank从6.44%下降到3.19%;Ferruginibacter从2.90%下降到0.81%;增加最明显的三个组分别是:OPB56_norank从0.71%上升到3.67%;Deltaproteobacteria_unclassified从0.11%上升到3.11%;Candidate_division_WS6_norank从0.95%上升到2.84%。和CG组相比,RW组丰度下降最明显的三个组分别是Bacteroidetes_vadinHA17_norank从8.46%下降到1.16%;Nitrosomonas从7.95%下降到0.79%;BD1-7_clade从4.33%下降到1.16%;增加最明显的3个组分别是:Candidate_division_WS6_norank从0.95%上升到9.35%;OPB56_norank从0.71%上升到6.90%;Sterolibacterium从0.16%上升到2.82%;笔者就3个反应器出现最主要的几个属水平的细菌进行分析;在CG组,AN组和RW都发现好氧异养菌(如Saprospiraceae[31], Chitinophagaceae_uncultured[32]) ,和缺氧异养菌(如Bacteroidetes_vadinHA17_norank[33])和脱氮菌(如 Thermomonas[34], Comamonadaceae[35])出现富集,这些细菌以硝酸盐为电子受体来降解COD;同时也富集了属于疣微菌门(Verrucomicrobia)的OPB35_soil_group_norank, Verrucomicrobia是好氧甲烷氧化菌,有助于减少温室气体甲烷的排放[36];在AN组和RW组中也富集了,有助于降解高分子化合物水解和酸化细菌(如Candidate_division_WS6_norank(AN =2.84%, RW=9.35%)[37-38]和Saccharibacteria_norank. (AN =2.18%, RW=5.19%[39]),但RW组富集程度高于AN组,可能原因是RW组进水含有大量的高分子化合物,更有利于水解和酸化细菌富集。
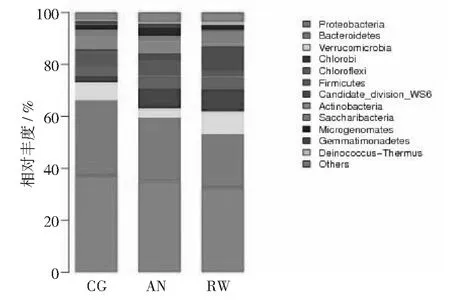
图12 3个反应器各菌群在门属水平上的相对丰度


图13 3个反应器各菌群在属水平上的相对丰度
3 结论

(2) 接种厌氧氨氧化污泥(AN组)和厌氧消化液中添加猪场废水原水(RW组)对COD去除率分别提高60.2%和102.6%,总无机氮(TIN)去除率分别为提高了11.1%和73.3%,其中RW组效果最明显。
(3) 接种厌氧氨氧化污泥(AN组)和对照(CG)组的AOB相对丰度大于NOB,出现亚硝酸积累;RW组富集的AOB相对丰度小于NOB,没出现亚硝酸积累,出水中硝酸盐以硝酸盐形式存在。
(4) 接种厌氧氨氧化污泥(AN组)富集的厌氧氨氧化细菌低于高效自养脱氮需求。
[1] Q Sui, H Dong, Z Zhu, D Guo. Ammonia stripping control parameters for improving effluent treatment effect in anaerobic digesters of piggery wastewater[J]. Transactions of the Chinese Society of Agricultural Engineering, 2012(28) :205-211.
[2] 邓良伟, 陈子爱, 袁心飞, 周文龙. 规模化猪场粪污处理工程模式与技术定位[J].养猪, 2008:21-24.
[3] G Anup, K Woo-Chang, O Sang-Eun. Removal of nitrogen from anaerobically digested swine wastewater using an anoxic/oxic (A/O) process complemented with a sulfur-packed biofilter[J]. African Journal of Biotechnology, 2013,10:9831-9838.
[4] T Pan, C M Drapcho, T Pan, C M Drapcho. Biological anoxic/aerobic treatment of swine waste for reduction of organic carbon, nitrogen, and odor[J]. Transactions of the Asae, 2001,44:1789-1796.
[5] 许振成, 谌建宇, 曾雁湘, 彭晓春, 陈 亮. 集约化猪场废水强化生化处理工艺试验研究[J].农业工程学报, 2007,10:037.
[6] Y H Song, G L Qiu, P Yuan, X Y Cui, J F Peng, P Zeng, L Duan, L C Xiang, F Qian. Nutrients removal and recovery from anaerobically digested swine wastewater by struvite crystallization without chemical additions[J]. Journal of hazardous materials, 2011,190:140-149.
[7] D Scaglione, G Tornotti, A Teli, L Lorenzoni, E Ficara, R Canziani, F Malpei. Nitrification denitrification via nitrite in a pilot-scale SBR treating the liquid fraction of co-digested piggery/poultry manure and agro-wastes[J]. Chemical Engineering Journal, 2013,228:935-943.
[8] J Meng, J Li, B Zhao, K Deng, Influence of aeration rate on shortcut nitrification in an SBR treating anaerobic-digested piggery wastewater[J]. Desalination and Water Treatment,2015:1-7.
[9] F J Cervantes, A David, J Gómez. Nitrogen removal from wastewaters at low C/N ratios with ammonium and acetate as electron donors[J]. Bioresource Technology, 2001,79:165-170.
[10] G Zhu, Y Peng, L Zhai, Y Wang, S Wang. Performance and optimization of biological nitrogen removal process enhanced by anoxic/oxic step feeding[J]. Biochemical Engineering Journal, 2009,43: 280-287.
[11] L Deng, P Zheng, Z Chen, Q Mahmood. Improvement in post-treatment of digested swine wastewater[J]. Bioresource Technology, 2008,99:3136-3145.
[12] D Yang, L Deng, D Zheng, L Wang, Y Liu. Separation of swine wastewater into different concentration fractions and its contribution to combined anaerobic-aerobic process[J]. Journal of environmental management, 2016,168:87-93.
[13] 邓良伟, 蔡昌达, 陈铬铭, 陈子爱. 猪场废水厌氧消化液后处理技术研究及工程应用[J].农业工程学报, 2002,18: 92-94.
[14] 邓良伟, 郑 平, 陈子爱. Anarwia工艺处理猪场废水节能效果的研究[J].农业工程学报, 2006,22:172-175.
[15] 郑 丹, 邓良伟, 杨 浩, 李淑兰, 张国治, 樊战辉, 陈 闯. 猪场废水厌氧消化液的厌氧氨氧化脱氮研究进展[J].中国沼气, 2011,29:3-8.
[16] I S Hwang, K S Min, E Choi, Z Yun. Nitrogen removal from piggery waste using the combined SHARON and ANAMMOX process[J]. Water Science Technology, 2005,52:487-494.
[17] 赵楠婕, 解庆林, 游少鸿, 周海妙, 史利荣. 厌氧氨氧化工艺处理猪场废水沼液的试验研究[J].四川环境, 2012,31:4-7.
[18] 王 伸, 邓良伟, 徐 则, 郑 丹, 王 兰, 王 霜. pH值对好氧处理及污泥性能的影响[J].中国沼气, 2016,34.
[19] 邓良伟, 郑 平, 李淑兰, 孙 欣, 汤玉珍. 添加原水改善 SBR 工艺处理猪场废水厌氧消化液性能[J].环境科学, 2006,26:105-109.
[20] G Bortone, S Gemelli, A Rambaldi, A Tilche. Nitrification, denitrification and biological phosphate removal in sequencing batch reactors treating piggery wastewater[J]. Water Science Technology, 1992,26:977-985.
[21] B Boiran, Y Couton, J Germon. Nitrification and denitrification of liquid lagoon piggery waste in a biofilm infiltration-percolation aerated system (BIPAS) reactor[J]. Bioresource technology, 1996,55: 63-77.
[22] W Bae, S Baek, J Chung, Y Lee. Optimal operational factors for nitrite accumulation in batch reactors[J]. Biodegradation, 2001,12:359-366.
[23] G Bitton. Wastewater Microbiology, 3ra. edic., New Jersey[J]. Editorial John Wiley Sons,2005:346-347.
[24] M H Gerardi. Nitrification and denitrification in the activated sludge process[J]. John Wiley Sons, 2003.
[25] S E Jorgensen, B Fath. Encyclopedia of Ecology[J]. Elsevier, 2008,5.
[26] T Fujii. New concepts of ammonia removal from digested swine effluents using anammox based deammonification process[J].Coastal Plain Soil,2013,3
[27] Y Miura, M N Hiraiwa, T Ito, T Itonaga, Y Watanabe, S Okabe. Bacterial community structures in MBRs treating municipal wastewater: relationship between community stability and reactor performance[J]. Water Research, 2007,41:627-637.
[28] 窦娜莎, 王 琳. 16S rDNA克隆文库法分析Biostyr曝气生物滤池处理城市污水的细菌多样性研究[J].环境科学学报, 2011,31:2117-2124.
[29] M Kumar, J G Lin. Co-existence of anammox and denitrification for simultaneous nitrogen and carbon removal—Strategies and issues[J]. Journal of Hazardous Materials, 2010,178: 1-9.
[30] A M K Vincent R Hill, Narayanan Jothikumar, Trisha B Johnson, Donghyun Hahn, Theresa L. Cromeans, Multistate Evaluation of an Ultrafiltration-Based Procedure for Simultaneous Recovery of Enteric Microbes in 100-Liter Tap Water Samples[J]. Applied Environmental Microbiology, 2007,73: 4218-4225.
[31] F Ju, T Zhang. Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant[J]. Isme Journal,2014,9: 683-695.
[32] P Kämpfer, N Lodders, E Falsen. Hydrotalea flava gen. nov, sp. nov, a new member of the phylum Bacteroidetes and allocation of the genera Chitinophaga, Sediminibacterium, Lacibacter, Flavihumibacter, Flavisolibacter, Niabella, Niastella, Segetibacter, Parasegetibacter, Terrimonas, Ferr[J]. International Journal of Systematic Evolutionary Microbiology, 2011(61): 518-523.
[33] Y Feng, X Li, Y Yu, J Qi, X Jia, J Wang, X Li. Production of unburned calcium silicon filter material (UCSFM) from oyster shell and its performance investigation in an A/O integrated biological aerated filter reactor(A/O-BAF) [J]. Rsc Advances,2016,6.
[34] S J Mcilroy, A Starnawska, P Starnawski, A M Saunders, M Nierychlo, P H Nielsen, J L Nielsen. Identification of active denitrifiers in full-scale nutrient removal wastewater treatment systems[J]. Environmental Microbiology, 2016,18:1-24.
[35] T Sadaie, A Sadaie, M Takada, K Hamano, J Ohnishi, N Ohta, K Matsumoto, Y Sadaie. Reducing sludge production and the domination of Comamonadaceae by reducing the oxygen supply in the wastewater treatment procedure of a food-processing factory[J]. Agricultural and Biological Chemistry, 2007,71:791-799.
[36] P F Dunfield, A Yuryev, P Senin, A V Smirnova, M B Stott, S Hou, B Ly, J H Saw, Z Zhou, Y Ren. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia[J]. Nature, 2007,450:879-882.
[37] M Khoshnoodi. Microbes involved in arsenic removal in passive treatment systems[M]. Vancoaver:University of British Columbia,2014.
[38] M A Dojka, J K Harris, N R Pace. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria[J]. Applied Environmental Microbiology, 2000,66: 1617-1621.
[39] M Albertsen, P Hugenholtz, A Skarshewski, K L Nielsen, G W Tyson, P H Nielsen. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes[J]. Nature Biotechnology, 2013,31:533-538.
[40] N Igarashi, H Moriyama, T Fujiwara, Y Fukumori, N Tanaka. The 2.8 A structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea[J]. Nature Structural Biology, 1997,4:276-284.
[41] Y Yue, J Liu, B Ma, L Ye, W Bo, Y Peng. Improving municipal wastewater nitrogen and phosphorous removal by feeding sludge fermentation products to sequencing batch reactor (SBR) [J]. Bioresource Technology, 2016,222:326-334.
[42] J M Regan, G W Harrington, D R Noguera. Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system[J]. Applied Environmental Microbiology, 2002,68:73-81.
[43] S Lücker, J Schwarz, C Gruberdorninger, E Spieck, M Wagner, H Daims. Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants[J]. Isme Journal, 2014,9:708-720.
[44] B Kartal, N L Van, J Rattray, V D V Jl, M C Schmid, D J Sinninghe, M S Jetten, M Strous. Candidatus ‘Brocadia fulgida’: an autofluorescent anaerobic ammonium oxidizing bacterium[J]. FEMS Microbiology Ecology, 2008,63:46-55.
Improvement in Acidification during Aerobic Treatment of Digested Swine Wastewater and Its Microbial Community Variation /
WANG Shen1,2, DENG Liang-wei1,2, XU Ze1,2, WANG Shuang1,2, JIANG Yi-qi1,2, ZHENG Dan1,2/
( 1.Biogas Institute of Ministry of Agriculture,Chengdu 610041,China; 2. Laboratory of Development and Application of Rural Renewable Energy,Ministry of Agriculture,Chengdu 610041,China)

Digested swine wastewater; SBR; pH; 16S rRNA; Anammox
2017-01-05
项目来源: 国家自然科学基金(31572450); 国家生猪技术产业体系(CARS-36-10B)
王 伸(1990-),男,安徽亳州人,在读硕士,研究方向为农村废弃物处理技术,E-mail:ws55185366@163.com
邓良伟,E-mail:dengliangwei@caas.cn
S216.4; X703
A
1000-1166(2017)02-0015-09
