PTGER4基因多态性与炎症性肠病易感性的Meta分析①
2017-04-10杨春贵马立兴侯海峰
郭 政 杨春贵 孙 涛 马立兴 侯海峰
(泰山医学院基础医学院,泰安271000)
·临床免疫学·
PTGER4基因多态性与炎症性肠病易感性的Meta分析①
郭 政 杨春贵 孙 涛②马立兴③侯海峰②
(泰山医学院基础医学院,泰安271000)
目的:系统评价人群中PTGER4基因多态性与炎症性肠病(Inflammatory bowel disease,IBD)的关系。方法:检索PubMed、Embase、Web of Science中2016年2月29日以前发表的相关病例对照研究文献,选择符合质量要求的研究。用STATA12.0软件进行Meta分析,计算合并OR值及其95%可信区间(Confidence interval,CI),并通过敏感性分析判断结果的稳定性,通过Egger′s 检验分析发表偏倚。结果:共纳入20篇文献中发表的44项病例对照研究,包括25 179例克罗恩病(Crohn′s disease,CD)病例、5 261例溃疡性结肠炎(Ulcerative colitis,UC)病例和44 652名对照。Meta分析结果表明,rs4613763T/C多态性与CD关系的等位基因频率、共显性模型、显性模型、隐性模型分析的合并OR值(95%CI)分别是1.24(1.06~1.45)、1.32(1.06~1.64)、1.25(1.06~1.48)和1.28(1.03~1.59);rs17234657T/G多态性与CD关系四种模型分析合并OR值(95%CI) 为:1.43(1.34~1.52)、2.12(1.70~2.63)、1.46(1.36~1.57)和1.93(1.56~2.40);rs4495224A/C多态性与CD关系的四种模型分析合并OR值(95%CI) 为:1.05(0.79~1.41)、1.08(0.62~1.88)、1.12(0.75~1.65)和1.00(0.67~1.49);rs9292777G/T多态性与CD关系的四种模型分析合并OR值(95%CI)为:0.77(0.67~0.88)、0.59(0.51~0.69)、0.73(0.61~0.87)和0.68(0.59~0.79);rs1373692T/G多态性与CD关系的四种模型分析合并OR值(95%CI)为:1.23(0.96~1.57)、1.39(0.74~2.59)、1.26(0.74~2.13)和1.31(1.00~1.72)。rs4613763T/C多态性与UC易感性分析的四种模型分析合并OR值(95%CI)为:1.30(1.17~1.44)、1.73(1.16~2.59)、1.32(1.17~1.48)和1.64(1.10~2.45)。结论:rs17234657T/G、rs4613763T/C和rs9292777G/T多态性与CD易感性相关;rs4613763T/C多态性与UC的易感性相关。
炎症性肠病;PTGER4;基因多态性;Meta分析
炎症性肠病(Inflammatory bowel disease,IBD)是一类多种病因引起的、异常的自身免疫介导的肠道慢性和复发性炎症,包括溃疡性结肠炎(Ulcerative colitis,UC)和克罗恩病(Crohn′s disease,CD)[1]。IBD的患病水平有广泛的地域差异性,且近来发病率有所增加[2]。虽然发病机制尚不完全清楚,目前认为该病是微生物感染、环境因素等引发的肠道免疫功能混乱[3],且与遗传因素有密切关系[4]。Hugot等[5]发现了人类第一个CD易感基因CARD15,Libioulle等[6]首次发现PTGER4基因是CD的易感基因。临床研究表明,肠黏膜屏障功能异常与IBD的发病关系密切[7],破坏上皮屏障的完整性在IBD的进展中起着至关重要的作用[8],而PTGER4编码产生的前列腺素受体EP4在维护上皮屏障的完整性中是必不可少的[9]。但是当前报道的人群中关于PTGER4与IBD关系的研究结果却不尽相同[6,10-28]。为明确PTGER4基因多态性与IBD的关系,本研究搜集了现有已发表相关病例对照研究,并进行Meta分析,以期更为全面地认识PTGER4基因多态性与IBD的关系。
1 材料与方法
1.1 材料 从Pubmed、Embase、Web of Science中检索2016年2月29日以前发表的人群中IBD与PTGER4基因多态性关系的病例对照研究。中文检索词为炎症性肠病、克罗恩病、溃疡性结肠炎、PTGER4基因、多态性、突变;英文检索词为Inflammatory bowel disease、IBD、Crohn′s disease、Ulcerative colitis、CD、UC、PTGER4、Polymorphism、Variant、Mutation。
1.2 方法
1.2.1 纳入标准和排除标准 纳入标准:①炎症性肠病的诊断明确;②文献为病例对照研究,病例组是经过临床诊断证实的IBD病患者,对照组为健康人或非患有IBD病的其他患者,且两组具有可比性;③对照组基因型符合哈迪-温伯格遗传平衡性(Hardy-Weinberg equilibrium,HWE);④文献研内容为PTGER4基因多态性与IBD(CD和/或UC)的相关性;⑤有充足的计算OR值和95%可信区间(Confidence interval,CI)的数据信息。 排除标准:①对同一组研究数据的重复报道;②非病例对照研究;③研究包括重叠数据;④发表类型为细胞学和动物学的研究;⑤计算OR值和95%CI的信息不充足,且通过与作者联系无法获得。
1.2.2 数据提取 由两名流行病专业研究人员共同对文献质量进行评价,对检索出的相关文献严格执行纳入和排除标准,对纳入文献提取的资料包括第一作者、研究对象的国别、发表时间、研究对象类型、研究对象的种族、样本量及基因型等。
1.3 统计学处理 采用STATA12.0软件进行统计学分析。分别计算等位基因频率、共显性基因模型、显性基因模型、隐性基因模型的合并OR值及其95%可信区间(CI)。通过Q检验和I2检验进行异质性检验,若P>0.10和/或I2<50%则认为各原始研究间无明显的异质性,并采用固定效应模型进行合并计算,否则认为存在异质性,并采用随机效应模型计算。通过逐一剔除每项原始研究进行敏感性分析。采用Egger′s检验分析发表偏倚。
2 结果
2.1 资料检索结果 初步检索到相关文献104篇,依照纳入标准和剔除标准逐层筛选,最终20篇文献纳入Meta分析[6,10-28],均为英文文献,其中研究对象包括25 179例CD病例、5 261例UC病例和44 652例对照。文献筛选流程图见图1,纳入研究的基本情况见表1。
2.2 合并分析结果
2.2.1 rs4613763多态性与CD关系 rs4613763等位基因频率、共显性模型、显性模型、隐性模型比较的合并OR值(95%CI)分别是:1.24(1.06~1.45)、1.32(1.06~1.64)、1.25(1.06~1.48)、1.28(1.03~1.59)。异质性检验结果分别为:I2=83.0%(P<0.10)、I2=48.2%(P>0.10)、I2=81.7%(P<0.10);I2=34.6%(P>0.10 ),见表2和图2。
表1 纳入研究的基本资料
Tab.1 Basic information of included studies
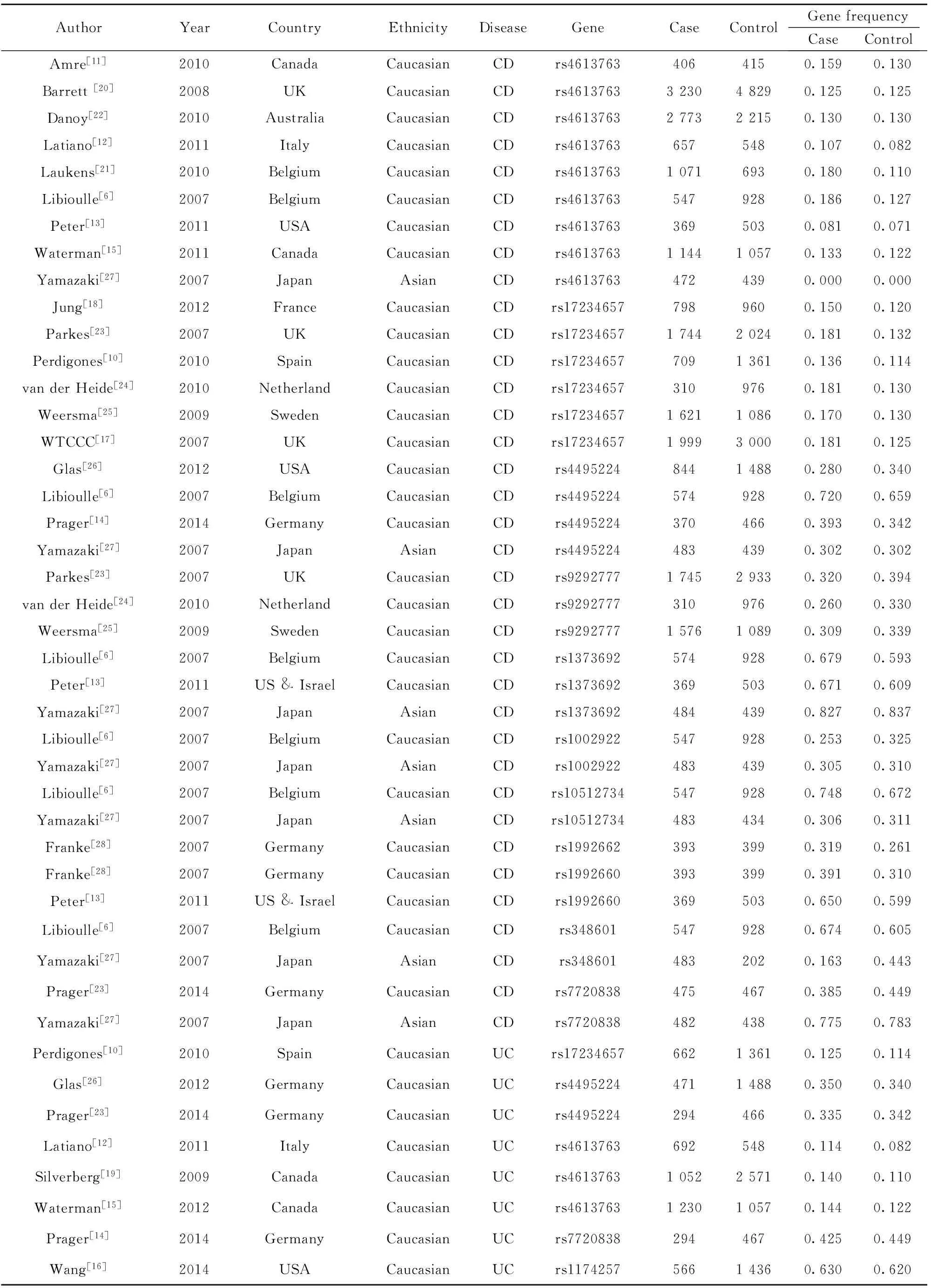
AuthorYearCountryEthnicityDiseaseGeneCaseControlGenefrequencyCaseControlAmre[11]2010CanadaCaucasianCDrs46137634064150.1590.130Barrett[20]2008UKCaucasianCDrs4613763323048290.1250.125Danoy[22]2010AustraliaCaucasianCDrs4613763277322150.1300.130Latiano[12]2011ItalyCaucasianCDrs46137636575480.1070.082Laukens[21]2010BelgiumCaucasianCDrs461376310716930.1800.110Libioulle[6]2007BelgiumCaucasianCDrs46137635479280.1860.127Peter[13]2011USACaucasianCDrs46137633695030.0810.071Waterman[15]2011CanadaCaucasianCDrs4613763114410570.1330.122Yamazaki[27]2007JapanAsianCDrs46137634724390.0000.000Jung[18]2012FranceCaucasianCDrs172346577989600.1500.120Parkes[23]2007UKCaucasianCDrs17234657174420240.1810.132Perdigones[10]2010SpainCaucasianCDrs1723465770913610.1360.114vanderHeide[24]2010NetherlandCaucasianCDrs172346573109760.1810.130Weersma[25]2009SwedenCaucasianCDrs17234657162110860.1700.130WTCCC[17]2007UKCaucasianCDrs17234657199930000.1810.125Glas[26]2012USACaucasianCDrs449522484414880.2800.340Libioulle[6]2007BelgiumCaucasianCDrs44952245749280.7200.659Prager[14]2014GermanyCaucasianCDrs44952243704660.3930.342Yamazaki[27]2007JapanAsianCDrs44952244834390.3020.302Parkes[23]2007UKCaucasianCDrs9292777174529330.3200.394vanderHeide[24]2010NetherlandCaucasianCDrs92927773109760.2600.330Weersma[25]2009SwedenCaucasianCDrs9292777157610890.3090.339Libioulle[6]2007BelgiumCaucasianCDrs13736925749280.6790.593Peter[13]2011US&IsraelCaucasianCDrs13736923695030.6710.609Yamazaki[27]2007JapanAsianCDrs13736924844390.8270.837Libioulle[6]2007BelgiumCaucasianCDrs10029225479280.2530.325Yamazaki[27]2007JapanAsianCDrs10029224834390.3050.310Libioulle[6]2007BelgiumCaucasianCDrs105127345479280.7480.672Yamazaki[27]2007JapanAsianCDrs105127344834340.3060.311Franke[28]2007GermanyCaucasianCDrs19926623933990.3190.261Franke[28]2007GermanyCaucasianCDrs19926603933990.3910.310Peter[13]2011US&IsraelCaucasianCDrs19926603695030.6500.599Libioulle[6]2007BelgiumCaucasianCDrs3486015479280.6740.605Yamazaki[27]2007JapanAsianCDrs3486014832020.1630.443Prager[23]2014GermanyCaucasianCDrs77208384754670.3850.449Yamazaki[27]2007JapanAsianCDrs77208384824380.7750.783Perdigones[10]2010SpainCaucasianUCrs1723465766213610.1250.114Glas[26]2012GermanyCaucasianUCrs449522447114880.3500.340Prager[23]2014GermanyCaucasianUCrs44952242944660.3350.342Latiano[12]2011ItalyCaucasianUCrs46137636925480.1140.082Silverberg[19]2009CanadaCaucasianUCrs4613763105225710.1400.110Waterman[15]2012CanadaCaucasianUCrs4613763123010570.1440.122Prager[14]2014GermanyCaucasianUCrs77208382944670.4250.449Wang[16]2014USACaucasianUCrs117425756614360.6300.620

图1 文献检索和筛选流程图Fig.1 Flow chart of study selection for this Meta-analysis
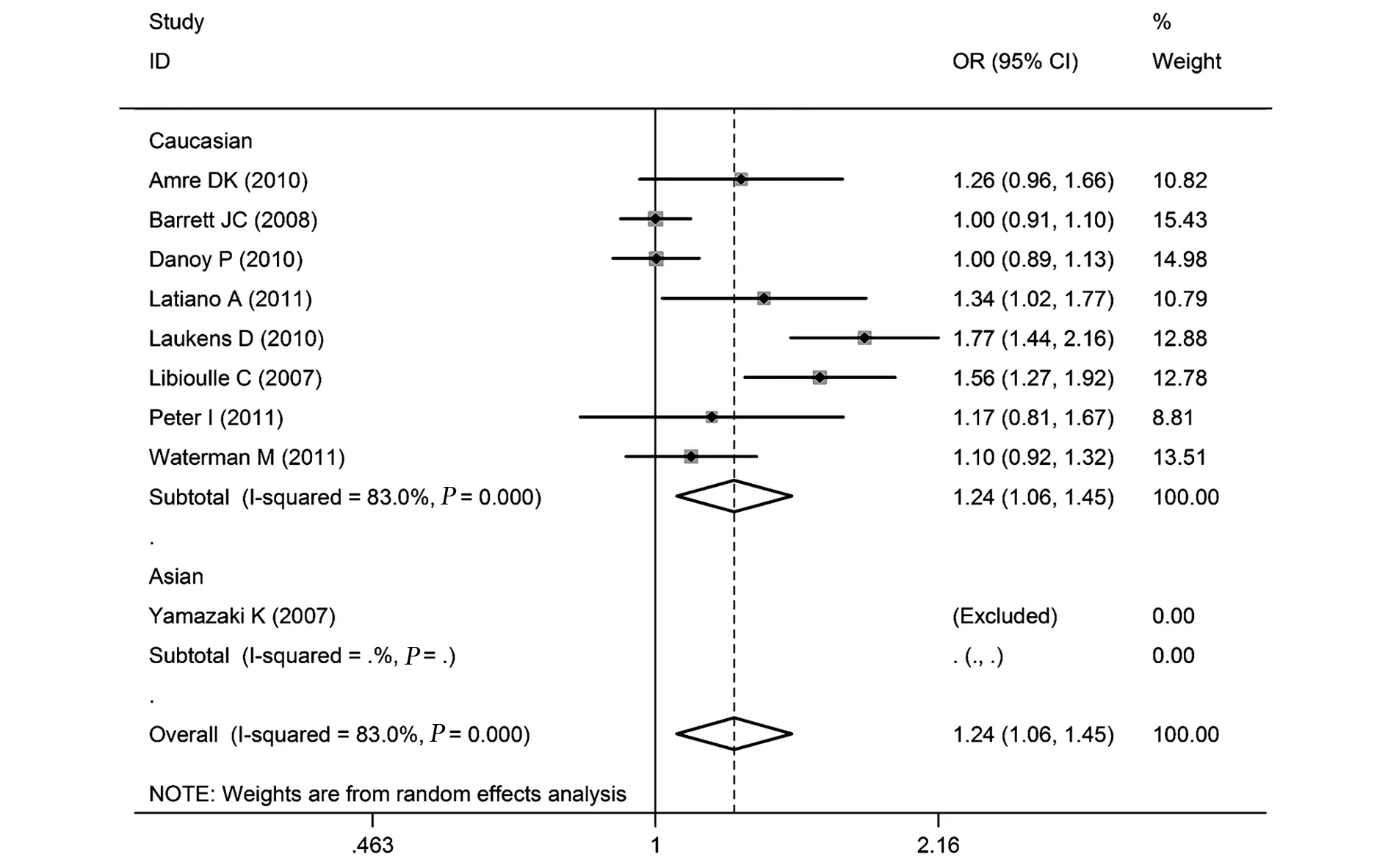
图2 rs4613763的等位基因频率与CD关系的森林图(C vs T)Fig.2 Forest plot of rs4673163 and CD in allelic frequency(C vs T)
表2 PTGER4基因多态性与炎症性肠病(IBD)的关系Meta分析结果
Tab.2 Results of Meta-ananlysis for relationship between PTGER4 genetic polymorphisms and inflammatory bowel disease(IBD)
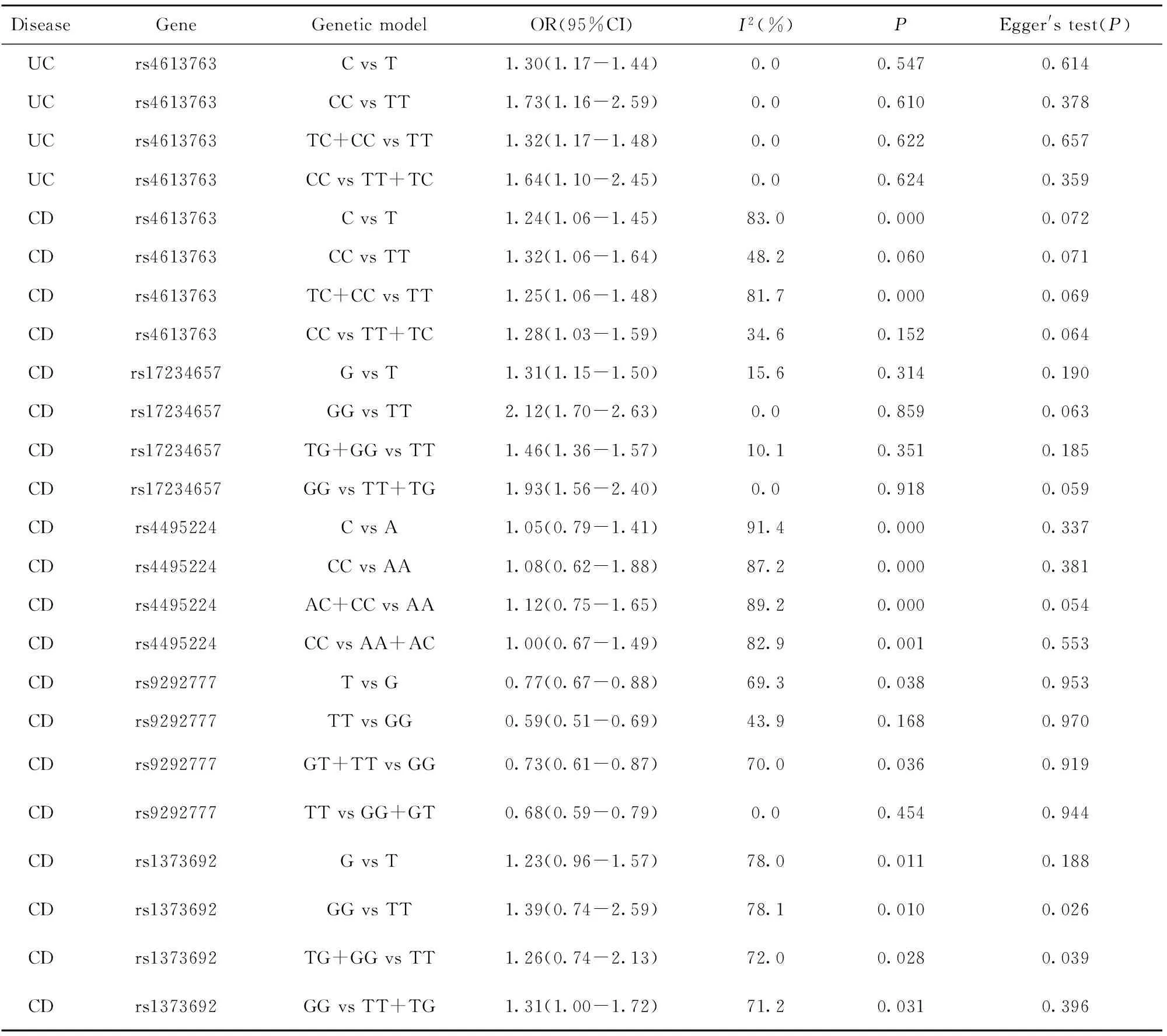
DiseaseGeneGeneticmodelOR(95%CI)I2(%)PEgger'stest(P)UCrs4613763CvsT1.30(1.17-1.44)0.00.5470.614UCrs4613763CCvsTT1.73(1.16-2.59)0.00.6100.378UCrs4613763TC+CCvsTT1.32(1.17-1.48)0.00.6220.657UCrs4613763CCvsTT+TC1.64(1.10-2.45)0.00.6240.359CDrs4613763CvsT1.24(1.06-1.45)83.00.0000.072CDrs4613763CCvsTT1.32(1.06-1.64)48.20.0600.071CDrs4613763TC+CCvsTT1.25(1.06-1.48)81.70.0000.069CDrs4613763CCvsTT+TC1.28(1.03-1.59)34.60.1520.064CDrs17234657GvsT1.31(1.15-1.50)15.60.3140.190CDrs17234657GGvsTT2.12(1.70-2.63)0.00.8590.063CDrs17234657TG+GGvsTT1.46(1.36-1.57)10.10.3510.185CDrs17234657GGvsTT+TG1.93(1.56-2.40)0.00.9180.059CDrs4495224CvsA1.05(0.79-1.41)91.40.0000.337CDrs4495224CCvsAA1.08(0.62-1.88)87.20.0000.381CDrs4495224AC+CCvsAA1.12(0.75-1.65)89.20.0000.054CDrs4495224CCvsAA+AC1.00(0.67-1.49)82.90.0010.553CDrs9292777TvsG0.77(0.67-0.88)69.30.0380.953CDrs9292777TTvsGG0.59(0.51-0.69)43.90.1680.970CDrs9292777GT+TTvsGG0.73(0.61-0.87)70.00.0360.919CDrs9292777TTvsGG+GT0.68(0.59-0.79)0.00.4540.944CDrs1373692GvsT1.23(0.96-1.57)78.00.0110.188CDrs1373692GGvsTT1.39(0.74-2.59)78.10.0100.026CDrs1373692TG+GGvsTT1.26(0.74-2.13)72.00.0280.039CDrs1373692GGvsTT+TG1.31(1.00-1.72)71.20.0310.396
2.2.2 rs17234657多态性与CD关系 rs17234657等位基因频率、共显性模型、显性模型、隐性模型分析的合并OR值(95%CI)分别是:1.31(1.15~1.50)、2.12(1.70~2.63)、1.46(1.36~1.57)、1.93(1.56~2.40)。异质性检验结果分别为:I2=80.0%(P<0.10)、I2=75.7%(P<0.10)、I2=69.7%(P<0.10)、I2=74.0%(P<0.10),见表2和图3。
2.2.3 rs4495224多态性与CD关系 rs4495224等位基因频率、共显性模型、显性模型、隐性模型分析的合并OR值(95%CI)分别是:1.05(0.79~1.41);1.08(0.62~1.88)、1.12(0.75~1.65)、1.00(0.67~1.49)。异质性检验的结果分别为:I2=91.4%(P<0.10)、I2=87.2%(P<0.10)、I2=89.2%(P<0.10)、I2=82.9%(P<0.10),见表2。
2.2.4 rs9292777多态性与CD关系 rs9292777等位基因频率、共显性模型、显性模型、隐性模型分析的合并OR值(95%CI)分别是:0.77(0.67~0.88)、0.59(0.51~0.69)、0.73(0.61~0.87)、1.23(0.96~1.57)。异质性检验结果分别为:I2=69.3%(P<0.10)、I2=43.9%(P>0.10)、I2=70.0%(P<0.10)、I2=0.0%(P>0.10),见表2和图4。
2.2.5 rs1373692多态性与CD关系 rs1373692等位基因频率、共显性模型、显性模型、隐性模型分析的合并OR值(95%CI)分别是:1.23(0.96~1.57)、1.39(0.74~2.59)、1.26(0.74~2.13)、1.31(1.00~1.72)。异质性检验结果分别为:I2=78%(P<0.10)、I2=78.1%(P<0.10)、I2=72.0%(P<0.10)、I2=71.2%(P<0.10),见表2和图5。
2.2.6 rs4613763多态性与UC关系 rs4613763等位基因频率、共显性模型、显性模型、隐性模型分析的合并OR值(95%CI)分别是:1.30(1.17~1.44)、1.73(1.16~2.59)、1.32(1.17~1.48)、1.64(1.10~2.45)。异质性检验结果分别为:I2=0.0%(P>0.10)、I2=48.2%(P>0.10);I2=0.0%(P>0.10)、I2=0.0%(P>0.10),见表2和图6。
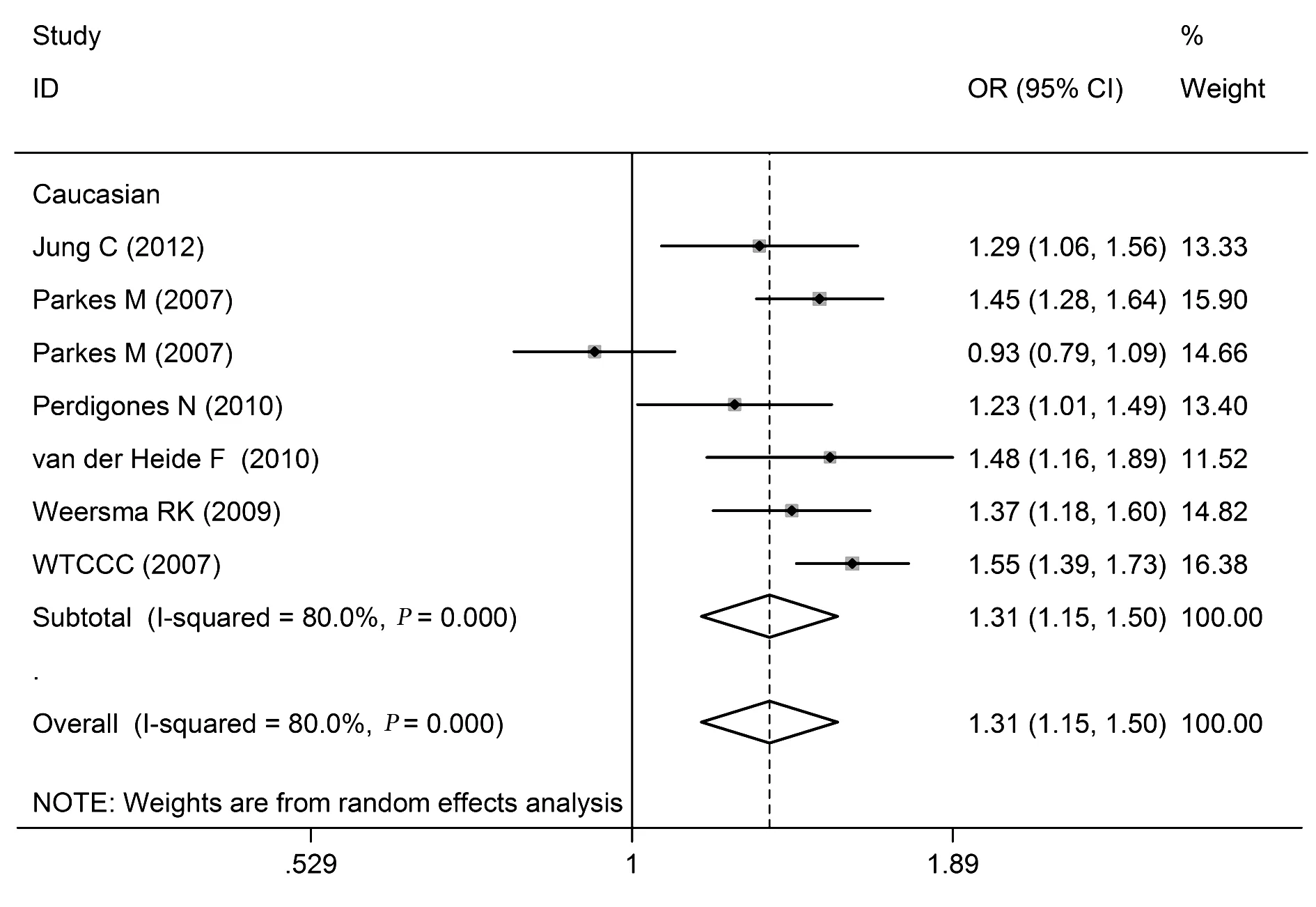
图3 rs17234657的等位基因频率(G vs T)模型与CD关系的森林图Fig.3 Forest plot of rs17234657 and CD in allelic frequency(G vs T)
2.2.7 rs17234657、rs4495224、rs7720838、rs11742 57基因多态性 由于相关文献在2篇以下,故本次研究未对rs17234657、rs4495224、rs7720838、rs1174-257基因IBD的关系进行Meta分析。
2.3 敏感性分析 我们通过依次剔除各项原始研究进行敏感性分析,计算了新的合并OR值和95%CI。分析结果显示:在rs1373692 与CD关系的分析中,在剔除Libioulle[6]和 Peter[13]的研究后,rs1373 692基因隐性模型的合并OR值与原结果有较大改变;rs9292777与CD关系的分析中,在剔除Parkes[23]、van der Heide[24],Weersma[25]的研究后,rs9292777基因显性模型的合并OR值与原结果有较大改变;rs17234657与CD关系的分析中,在剔除Jung[18]、Parkes[23]、Perdigones[10]、van der Heide[24]、Weersma[25]、WTCCC[17]的研究数据后rs17234657共显性和隐性模型的合并OR值与原结果有较大改变;在rs4613763与UC关系的分析中,在剔除Silverberg MS的研究数据后rs4613763共显性和隐性模型的合并OR值与原结果有较大改变。而其他16项合并OR值和95%CI均为有明显改变。可见本次Meta分析的结果具有较好的稳定性。
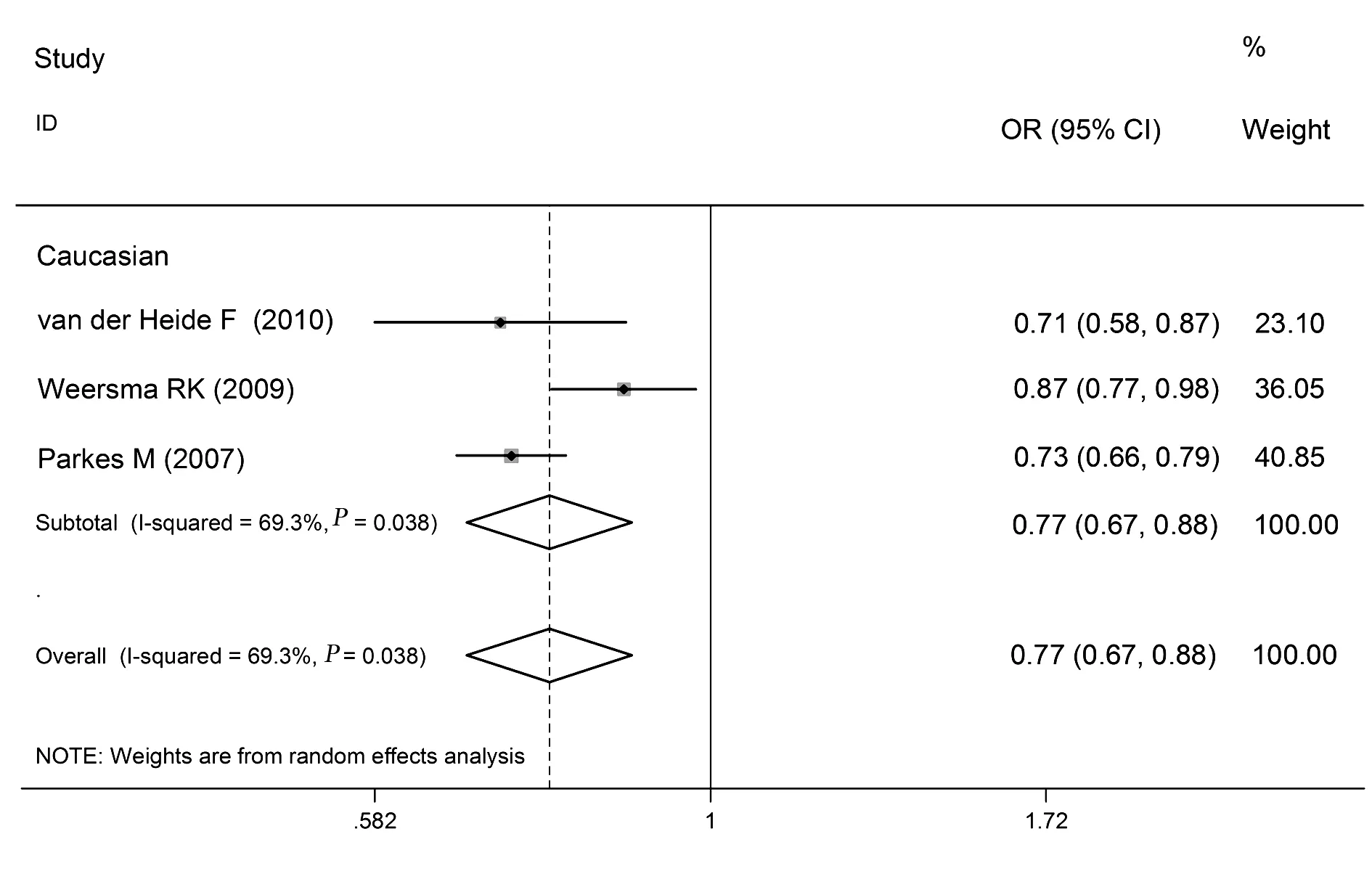
图4 rs9292777的等位基因频率(T vs G)与CD关系的森林图Fig.4 Forest plot of rs9292777 and CD in allelic frequency(T vs G)
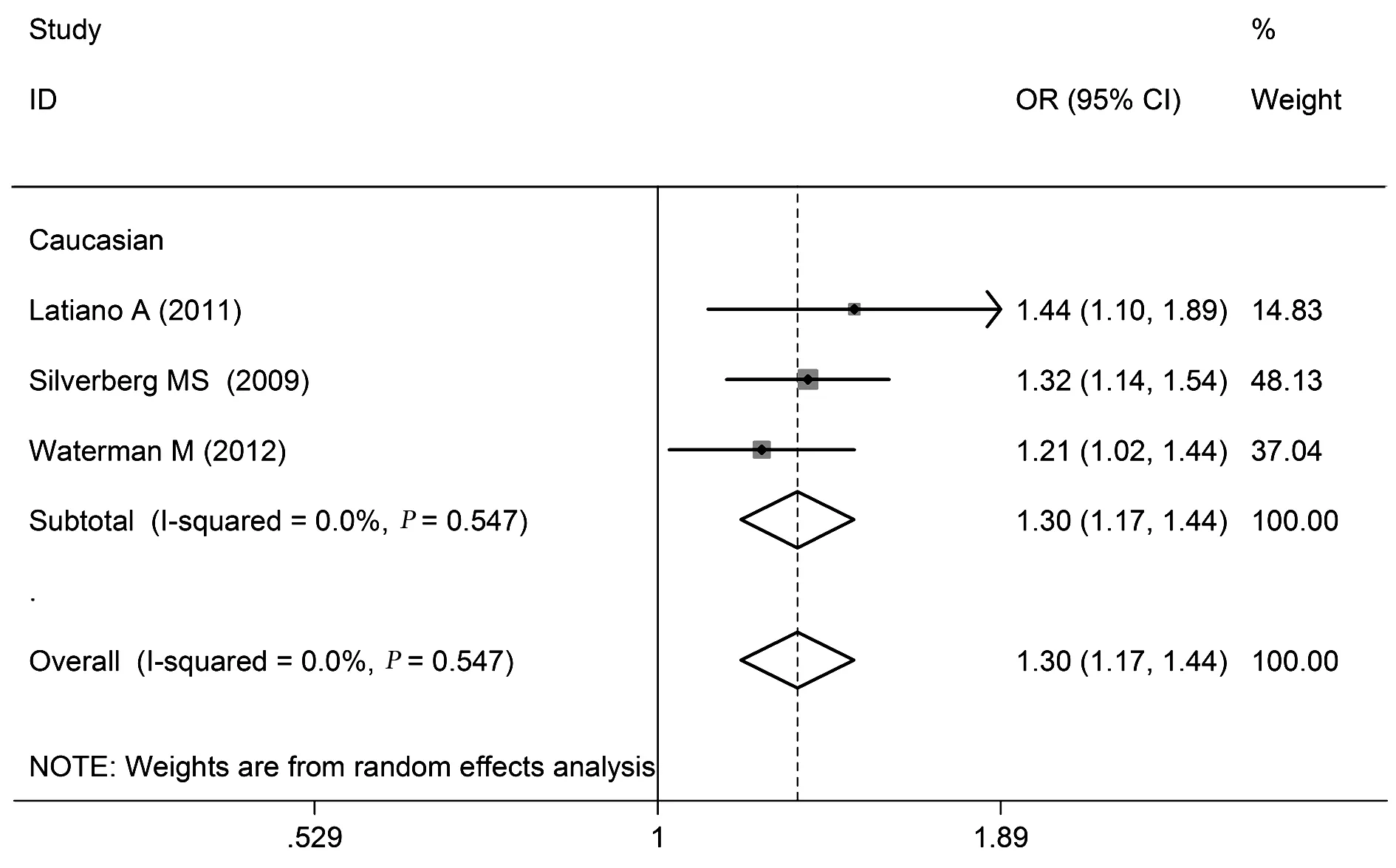
图6 rs4613763的等位基因频率(C vs T)与UC相关性的森林图Fig.6 Forest plot of rs4613763 and UC in allelic frequency(C vs T)
2.4 发表偏倚分析 对于PTGER4基因多态性与IBD关系的研究,用STATA12.0软件进行Egger线性回归分析和漏斗图分析,表明rs1373692、rs17234657等位基因频率和显性基因模型均存在发表偏倚,尚需更多文献来评价发表偏倚;其他相关位点与IBD关系的研究发表偏倚均不显著(P>0.05),表2。
3 讨论
本研究利用系统评价的方法回顾了PTGER4基因与IBD关系的研究成果。通过对20篇文献中的44项病例对照研究进行Meta分析发现,rs1373692T/G、rs4495224A/C基因多态性与CD发病无明显关联;rs17234657T/G、rs4613763T/C和rs9292777G/T基因多态性与CD发病有关;rs4613763T/C基因多态性与UC发病有关。
前列腺素E2(PGE2)在胃肠道内发挥特殊的生理和病理作用,正常情况下PGE2调节胃肠道内的胃酸、盐类和黏液的分泌,在胃肠肠道蠕动、黏膜完整性和血液循环方面也发挥着作用[29,30]。然而在IBD发病时PGE2又可发挥炎症介质的作用,引起病灶部位发生充血、水肿和炎细胞浸润,导致肠道组织破坏及溃疡形成[31]。位于染色体5p13.1上的PTGER4基因,约有270 kb,编码产生PGE2受体的EP4亚基,其基因多态性影响了EP4的表达水平,也间接发挥着影响PGE2 功能的作用[5,13,32],使得PTGER4基因成为IBD的易感基因。另有文献介绍采用 EP4选择性激动剂治疗结肠炎,可增强上皮细胞存活和再生而起到治疗作用[33,34]。本研究结果表明,rs4613763基因的C等位基因变异是UC发病的危险因素,证明了上述联系的合理性。研究表明,活化的前列腺受体EP4可以促进Th17细胞的分化和Th1细胞的增殖[35,36],这两种炎性细胞亚群在CD的发病中起了非常重要的作用[37]。本研究证实,EP4亚基的编码基因rs4613763、rs17234657和rs9292777的基因多态性与CD的发病有显著联系。
此外,针对PTGER4基因与IBD发病的相关研究也发现,被诱导剂激活的核转录因子NF-kB可促进炎症因子的表达,并调控PTGER4基因表达[38,39],而XBP1是内质网应激的重要组成部分[40],内质网应激会引起炎症的发生,也可调控PTGER4基因的表达,并影响IBD的发病[26,41]。
本研究通过Meta分析来评估PTGER4基因多态性与IBD易感性的联系,能客观、系统地对多个研究进行综合评价和分析,从而避免了单个研究的随机误差、样本量小对研究结果影响较大的缺陷,提供更为可信的危险因素的评估[27],使我们能够更为确切地评价PTGER4基因多态性与IBD的关联,为今后相关的研究提供了参考。本次Meta分析也存在以下的局限性:①部分基因分析中纳入研究存在异质性,且存在一定的发病偏倚;②仅考虑到IBD与基因多态性相关,未涉及抽烟、喝酒、年龄、性别、种族、地区等因素对发病的影响;③纳入的rs4495224、rs9292777与CD发病以及rs4613763与UC发病关系的文献数量偏少,尤其是2015~2016年没有相关文献报道。
尽管PTGER4基因与IBD关系已经取得了一定的进展,但是有关前列腺素、前列腺素受体EP4及PTGER4基因表达调控在IBD发病中的调节机制还不是很清楚。本文研究结果将为进一步探讨IBD与遗传因素的关系,以及前列腺素及其受体在IBD发病中的调节机制提供新的证据。
[1] Podolsky DK.Inflammatory bowel disease [J].N Engl J Med,2002,347:417-429.
[2] Burisch J,Munkholm P.The epidemiology of inflammatory bowel disease[J].Scand J Gastroenterol,2015,50:942-951.
[3] Gabbani T,Deiana S,Annese AL,etal.The genetic burden of inflammatory bowel diseases:implications for the clinic[J].Exp Rev Gastroenterol Hepatol,2016,9:1-9.
[4] Baumgart DC,Carding SR.Inflammatory bowel disease:cause and immunobiology [J].Lancet,2007,369:1627-1640.
[5] Hugot JP,Chamaillard M,Zouali H,etal.Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn′s disease [J].Nature,2001,411:599-603.
[6] Libioulle C,Louis E,Hansoul S,etal.Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4 [J].PLoS Genet,2007,3:e58.
[7] Xavier RJ,Podolsky DK.Unravelling the pathogenesis of inflammatory bowel disease [J].Nature,2007,448:427-434.
[8] Ogura Y,Bonen DK,Inohara N,etal.A frameshift mutation in NOD2 associated with susceptibility to Crohn′s disease [J].Nature,2001,411:603-606.
[9] Lejeune M,Leung P,Chadee K.Role of EP4 receptor and prostaglandin transporter in prostaglandin E2-induced alteration in colonic epithelial barrier integrity [J].Am J Physiol Gastrointest Liver Physiol,2010,299:1097-1105.
[10] Perdigones N,Martín E,Robledo G,etal.Study of chromosomal region 5p13.1 in Crohn′s disease,ulcerative colitis,and rheumatoid arthritis [J].Hum Immunol,2010,71:826-828.
[11] Amre DK,Mack DR,Morgan K,etal.Susceptibility loci reported in genome-wide association studies are associated with Crohn′s disease in Canadian children [J].Aliment Pharmacol Ther,2010,31:1186-1191.
[12] Latiano A,Palmieri O,Latiano T,etal.Investigation of multiple susceptibility loci for inflammatory bowel disease in an Italian cohort of patients [J].PloS One,2011,6:e22688.
[13] Peter I,Mitchell AA,Ozelius L,etal.Evaluation of 22 genetic variants with Crohn′s disease risk in the Ashkenazi Jewish population:a case-control study [J].BMC Med Genet,2011,12:63.
[14] Prager M,Buttner J,Buning C.PTGER4 modulating variants in Crohn′s disease [J].Int J Colorectal Dis,2014,29:909-915.
[15] Waterman M,Xu W,Stempak JM,etal.Distinct and overlapping genetic loci in Crohn′s disease and ulcerative colitis:correlations with pathogenesis[J].Inflamm Bowel Dis,2011,17:1936-1942.
[16] Wang MH,Fiocchi C,Zhu X,etal.Gene-gene and gene-environment interactions in ulcerative colitis[J].Hum Genet,2014,133:547-558.
[17] Wellcome Trust Case Control Consortium.Genome-wide association study of 14 000 cases of seven common diseases and 3 000 shared controls [J].Nature,2007,447:661-678.
[18] Jung C,Colombel JF,Lemann M,etal.Genotype/phenotype analyses for 53 Crohn′s disease associated genetic polymorphisms [J].PLoS One,2012,7:e52223.
[19] Silverberg MS,Cho JH,Rioux JD,etal.Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study [J].Nat Genet,2009,41:216-220.
[20] Barrett JC,Hansoul S,Nicolae DL,etal.Genome-wide association defines more than 30 distinct susceptibility loci for Crohn′s disease [J].Nat Genet,2008,40:955-962.
[21] Laukens D,Georges M,Libioulle C,etal.Evidence for significant overlap between common risk variants for Crohn′s disease and ankylosing spondylitis [J].PloS one,2010,5:e13795.
[22] Danoy P,Pryce K,Hadler J,etal.Association of variants at 1q32 and STAT3 with ankylosing spondylitis suggests genetic overlap with Crohn′s disease [J].PLoS Genet,2010,6:e1001195.
[23] Parkes M,Barrett JC,Prescott NJ,etal.Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn′s disease susceptibility [J].Nat Genet,2007,39:830-832.
[24] van der Heide F,Nolte IM,Kleibeuker JH,etal.Differences in genetic background between active smokers,passive smokers,and non-smokers with Crohn′s disease[J].Am J Gastroenterol,2010,105:1165-1172.
[25] Weersma RK,Stokkers PC,Cleynen I,etal.Confirmation of multiple Crohn′s disease susceptibility loci in a large Dutch-Belgian cohort [J].Am J Gastroenterol,2009,104:630-638.
[26] Glas J,Seiderer J,Czamara D,etal.PTGER4 expression-modulating polymorphisms in the 5p13.1 region predispose to Crohn′s disease and affect NF-kappaB and XBP1 binding sites [J].PloS one,2012,7:e52873.
[27] Yamazaki K,Onouchi Y,Takazoe M,etal.Association analysis of genetic variants in IL23R,ATG16L1and 5p13.1 loci with Crohn′s disease in Japanese patients[J].J Hum Genet,2007,52:575-583.
[28] Franke A,Hampe J,Rosenstiel P,etal.Systematic association mapping identifies NELL1 as a novel IBD disease gene[J].PLoS One,2007,2:e691.
[29] Dai L,King DW,Perera DS,etal.Inverse expression of prostaglandin E2-related enzymes highlights differences between diverticulitis and inflammatory bowel disease[J].Dig Dis Sci,2015,60:1236-1246.
[30] Huang C,Haritunians T,Okou DT,etal.Characterization of genetic loci that affect susceptibility to inflammatory bowel diseases in African Americans[J].Gastroenterology,2015,149:b1575-1586.
[31] Nitta M,Hirata I,Toshina K,etal.Expression of the EP4 prostaglandin E2 receptor subtype with rat dextran sodium sulphate colitis:colitis suppression by a selective agonist,ONO-AE1-329[J].Scand J Immunol,2002,56:66-75.
[32] Kabashima K,Saji T,Murata T,etal.The prostaglandin receptor EP4 suppresses colitis,mucosal damage and CD4 cell activation in the gut [J].J Clin Invest,2002,109:883-893.
[33] Jiang GL,Nieves A,Im WB,etal.The prevention of colitis by E Prostanoid receptor 4 agonist through enhancement of epithelium survival and regeneration [J].J Pharmacol Exp Ther,2007,320:22-28.
[34] Brand S.Crohn′s disease:Th1,Th17 or both? The change of a paradigm:new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn′s disease [J].Gut,2009,58:1152-1167.
[35] Yao C,Sakata D,Esaki Y,etal.Prostaglandin E2-EP4 signaling promotes immune inflammation through Th1 cell differentiation and Th17 cell expansion[J].Nat Med,2009,15:633-640.
[36] Chen Q,Muramoto K,Masaaki N,etal.A novelantagonist of the prostaglandin E(2) EP(4) receptor inhibits Th1 differentiationand Th17 expansion and is orally active in arthritis models[J].Br J Pharmacol,2010,160:292-310.
[37] Atreya I,Atreya R,Neurath MF.NF-kappaB in inflammatory bowel disease [J].J Intern Med,2008,263:591-596.
[38] Neurath MF,Pettersson S,Meyer zum Büschenfelde KH,etal.Local administration of antisense phosphorothioate oligo-nucleotides to the p65 subunit of NF-kappa B abrogates established experimental colitis in mice [J].Nat Med,1996,2:998-1004.
[39] Kaser A,Lee AH,Franke A,etal.XBP1 linksER stress to intestinal inflammation and confers genetic risk for humaninflammatory bowel disease [J].Cell,2008,134:743-756.
[40] Ellson CD,Davidson K,Ferguson GJ,etal.Neutrophils from p40phox-/-mice exhibit severe defects in NADPH oxidase regulation and oxidant-dependent bacterial killing [J].J Exp Med,2006,203:1927-1937.
[41] Noble JH Jr.Meta-analysis:Methods,strengths,weaknesses,and political uses [J].J Lab Clin Med,2006,147:7-20.
[收稿2016-04-20 修回2016-06-28]
(编辑 倪 鹏)
Meta-analysis of relationship between PTGER4 genetic polymorphisms and inflammatory bowel disease
GUOZheng,YANGChun-Gui,SUNTao,MALI-Xing,HUOHai-Feng.
SchoolofBasicMedicalScience,TaishanMedicalUniversity,Taian271000,China
Objective:To systematically review the relationship between PTGER4 genetic polymorphisms and the risk of inflammatory bowel disease (IBD).Methods: All eligible case-control studies which published up to February 29,2016 were searched by PubMed,Embase,Web of Science.The studies in accordance with high quality were included in this study.We synthesized pooled odds ratio (OR) and its 95% confidence interval (CI) using STATA12.0.The sensitivity analysis was used to determine the stability of results in meta-analysis.The Egger′s analysis was performed to evaluate the publication bias.Results: Twenty original publications involving 44 case-control studies were included in this study,in which 25 179 patients with Crohn′s disease (CD),5 261 patients with ulcerative colitis (UC) and 44 652 control subjects were detected.The allelic frequency,additive model,dominant model and recessive model were used to analyze the association of rs4613763T/C polymorphism and CD,and the pooled OR and 95%CIwere as followings:1.24 (1.06-1.45),1.32 (1.06-1.64),1.25 (1.06-1.48),1.28 (1.03-1.59).For rs17234657T/G polymorphism and CD,the pooled OR (95%CI) of four genetic models were 1.35 (1.28-1.47),2.12 (1.70-2.63),1.46 (1.36-1.57) and 1.90 (1.54-2.35),respectively.For rs4495224A/C polymorphism and CD,the pooled OR(95%CI) were 1.05 (0.79-1.41),1.08 (0.62-1.88),1.12 (0.75-1.65) and 1.00 (0.67-1.49).And the pooled OR (95%CI) were 0.77 (0.67-0.88),0.59 (0.51-0.69),0.73 (0.61-0.87),0.68 (0.59-0.79) for rs9292777G/T polymorphism and CD.For rs1373692T/G polymorphism and CD,the pooled OR (95%CI) were 1.23 (0.96-1.57),1.39 (0.74-2.59),1.26 (0.74-2.13),1.31 (1.00-1.72).The pooled OR (95%CI) of allelic frequency,additive model,dominant model and recessive model for the association of rs4613763T/C polymorphism with UC were 1.30 (1.17-1.44),1.73 (1.16-2.59),1.32 (1.17-1.48),1.64 (1.10-2.45),respectively.Conclusion: Polymorphisms of rs17234657T/G,rs4613763T/C and rs9292777G/T are associated with CD.Polymorphism of rs4613763T/C is associated with UC susceptibility.
Inflammatory bowel disease;PTGER4;Genetic polymorphism;Meta-analysis
10.3969/j.issn.1000-484X.2017.03.019
①本文为国家自然科学基金资助项目(81202170)和国家级大学生创新创业训练项目(201510439202,201610439032)。
郭 政(1992年-),男,主要从事循证医学研究,E-mail:935133632@qq.com。
及指导教师:侯海峰(1979年-),男,医学博士,副教授,硕士生导师,主要从事病因流行病学研究,E-mail:hfhou@163.com。
R574
A
1000-484X(2017)03-0407-08
②泰山医学院公共卫生学院,泰安271000。
③泰山医学院附属医院消化内科,泰安271000。
