Two major er1 alleles confer powdery mildew resistance in three pea cultivars bred in Yunnan Province,China
2016-10-24SuliSunYuhuHeChengDiCnxingDunZhendongZhu
Suli Sun,Yuhu He,Cheng Di,Cnxing Dun,Zhendong Zhu,*
aNational Key Facility for Crop Gene Resources and Genetic Improvement,Institute of Crop Science,Chinese Academy of Agricultural Sciences,Beijing 100081,China
bYunnan Academy of Agricultural Sciences,Kunming 650205,China
Two major er1 alleles confer powdery mildew resistance in three pea cultivars bred in Yunnan Province,China
Suli Suna,Yuhua Heb,Cheng Daib,Canxing Duana,Zhendong Zhua,*
aNational Key Facility for Crop Gene Resources and Genetic Improvement,Institute of Crop Science,Chinese Academy of Agricultural Sciences,Beijing 100081,China
bYunnan Academy of Agricultural Sciences,Kunming 650205,China
A R T I C L E I N F O
Article history:
26 April 2016
Accepted 21 May 2016
Available online 30 June 2016
Erysiphe pisi
er1
Pea
Powdery mildew
Yunnan Province
Powdery mildew,caused by Erysiphe pisi D.C.,is an important disease of pea(Pisum sativum L.). The use of cultivars carrying powdery mildew resistance alleles at the er1 locus is the most effective and economical means of controlling this disease.The objectives of this study were to screen Chinese elite pea cultivars for resistance to E.pisi and to identify the responsible gene at the er1 locus.Among the 37 pea cultivars tested,three(Yunwan 8,Yunwan 21,and Yunwan 23)were immune to E.pisi infection in phenotypic evaluations.The full-length cDNA sequences of the er1 candidate gene,PsMLO1,from the three resistant cultivars and control plants were analyzed.Comparison of the cDNA sequences of 10 clones revealed differencesamongthepowderymildew-resistantcultivars,susceptiblecontrols,and wild-type cultivar Sprinter.The observed resistance in Yunwan 8 plants resulted from a point mutation(C→G)at position 680 of PsMLO1 that introduced a stop codon,leading to premature termination of protein synthesis.The responsible resistance allele was identified as er1-1.Powdery mildew resistance in Yunwan 21 and Yunwan 23 plants was caused by identical insertions or deletions in PsMLO1.Three distinct PsMLO1 transcripts were observed in Yunwan 21 and Yunwan 23 plants.These transcripts were characterized by a 129-bp deletion and 155-and 220-bp insertions,respectively.The responsible resistance allele wasidentifiedaser1-2.Wehavecharacterizedtwoimportanter1allelesinthree
E.pisi-resistant pea cultivars bred in Yunnan Province,China.These cultivars represent important genetic resources for the breeding of powdery mildew-resistant pea cultivars.
©2016 Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.This is an open access article under the CC BY-NC-ND license
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
1.Introduction
Powdery mildew,caused by Erysiphe pisi D.C.,is one of the most serious threats to pea(Pisum sativum L.)production,causing yield losses of 25%-50%[1-3].To control this disease, the use of genetically resistant cultivars is the most efficient,economical,and environmentally friendly method[4].Researchers have focused on screening for E.pisi resistance and genetic analyses of powdery mildew resistance in pea.Three genes(er1,er2,and Er3)have been reported to be associatedwith powdery mildew(E.pisi)resistance in pea germplasm[5-7].Previous studies revealed that er1 and er2 are single recessive genes located in pea linkage groups(LGs)VI and III,respectively[8-10].Er3 is a newly identified dominant gene from a wild relative of pea(Pisum fulvum)that was recently incorporated into the genome of cultivated pea[3,7].Although Er3 has been localized between the sequence-characterized amplified region(SCAR)marker Scw4637and the random amplified polymorphic(RAPD)marker OPAG05_1240,its exact location in the pea genome is unknown[11].
The mechanisms of the three known resistance genes(er1,er2,and Er3)have been studied at the cellular level[12-14]. The er1 gene confers immunity or high-level resistance by preventing E.pisi from penetrating pea epidermal cells.In contrast,the disease resistance provided by er2 and Er3 is mediated by a post-penetration hypersensitive response[7,12,13].er2 expression is strongly influenced by temperature and leaf age.Complete disease resistance conferred by er2 occurs only at high temperatures(e.g.,25°C)or in mature leaves.Effective er2-regulated resistance to E.pisi has been observed only in specific geographic regions[6,12,15,16].
Several genetic analyses of E.pisi resistance have revealed thater1isthegeneresponsibleforconferringstableanddurable resistance in most cases[15-19].Thus,er1 has been used for decades in pea breeding programs.er2 is present only in a few E.pisi-resistant pea accessions,including SVP951,SVP952,and JI2480[15].Recently,it was reported that resistance conferred by er1 is caused by loss-of-function mutations in a powdery mildew susceptibility gene,PsMLO1,belonging to the mildew resistance locus O(MLO)gene family[20,21].
To date,nine er1 alleles(er1-1,er1-2,er1-3,er1-4,er1-5,er1-6,er1-7,er1mut1,and er1mut2)have been characterized in pea accessions resistant to E.pisi,according to differences in mutations in PsMLO1[20-27].Each er1 allele corresponds to a different PsMLO1 mutation produced by natural or artificial mutagenesis,except for er1-1 and er1mut1,which carry the identical mutation[20-22].All er1 alleles but er1-5,er1mut1,and er1mut2 were generated by natural mutations[21,25-27]. Seven alleles(er1-1,er1-3,er1-4,er1-5,er1-6,er1mut1,and er1mut2)were the result of point mutations in PsMLO1[20-22,25-27].The er1-2 allele was generated by an insertion ordeletionofaDNAfragmentofunknownsizeandidentityinto PsMLO1,resultinginabnormalPsMLO1transcription[20-24].We recentlydetecteder1-7 intheresistant pea cultivarDDR-11,and determined that this allele harbors a 10-bp deletion in exon 1 of PsMLO1[26].
In China,powdery mildew caused by E.pisi has reduced pea quality and yields since 1991[28].Thus,pea germplasm continues to be screened to detect genes conferring resistance to E.pisi[23-26,28-32].The results of these studies indicated that several Chinese pea accessions were immune or highly resistant to the Chinese E.pisi isolates EPBJ and EPYN.To identify disease resistance genes,genetic analyses and investigations of the PsMLO1 sequence were performed using the Chinese pea cultivar Xucai 1,pea line X9002,and several Chinese pea landraces resistant to E.pisi[23-25]. Wang et al.[24]and Sun.et al.[23]reported that the disease resistance in Xucai 1 and X9002 was conferred by an er1 allele,er1-2,a commonly used resistance gene in breeding programs.Recently,Sun.et al.[25]identified and characterized a novel er1 allele(er1-6)in 15 Chinese pea landraces resistant to E.pisi.Using a high-resolution melting technique,they developed and validated a functional marker specific to er1-6,SNP1121,which can be used in pea breeding by marker-assisted selection[25].These results suggest that there are various sources of resistance in Chinese pea germplasm that carry novel er1 alleles that may be useful for breeding E.pisi-resistant pea cultivars.The objectives of this study were to screen Chinese pea cultivars for resistance to E.pisi and to characterize the powdery mildew resistance alleles at the er1 locus.
2.Materials and methods
2.1.Plant materials
Thirty-six Chinese elite pea cultivars developed in eight provincesorautonomousregions(Beijing,Gansu,Hebei,Jiangsu,Qinghai,Sichuan,Tibet,and Yunnan),and preserved in the China National Genebank were evaluated for resistance to powdery mildew(E.pisi)(Table 1).Two susceptible cultivars,Bawan 6[23,24,31]and Longwan 1[23,25,26],harboring the susceptibility gene Er1,were used as susceptible controls.These two cultivars were kindly provided by Mr.Dongxu Xu of the Zhangjiakou Academy of Agricultural Sciences,China,and Dr.Xiaoming Yang of the Gansu Academy of Agricultural Sciences,China,respectively.Two resistant pea cultivars,Xucai 1[23]and YI(JI1591)[21],harboring er1-2 and er1-4,respectively,were also used as resistant controls.These cultivars were kindly provided by Prof.Fengbao Wang of the Hebei Normal University of Science&Technology,China,and Prof.Weidong Chen of Washington State University,USA,respectively.
2.2.E.pisi isolates
Two highly virulent E.pisiisolates,EPBJ(NCBI accession number: KR912079)and EPYN(NCBI accession number:KR957355),collected from Beijing and Yunnan,China,respectively,were used as inocula[23-26,31,32].The isolates were maintained on Longwan 1 seedlings.The inocula were reproduced by continuously transferring E.pisi conidia to healthy Longwan 1 seedlings by gently shaking diseased plants.The inoculated plants were incubated in a growth chamber at 10±1°C with a 12-h photoperiod.
2.3.Phenotypic evaluation
The seeds of 37 Chinese pea cultivars and susceptible(Bawan 6 and Longwan 1)and resistant(Xucai 1 and YI)controls were planted in 15-cm-diameter paper pots(five seeds per pot)filled with a mixture of vermiculite and peat moss(1:1). Twenty-five seeds of each pea cultivar and control were planted for all experiments,which were replicated five times. Seeded pots were placed in a greenhouse at 18°C-26°C. Fourteen days after planting,seedlings at the fourth or fifth leaf stage were inoculated with the two E.pisi isolates(EPBJ and EPYN),using conidia collected by gently shaking infected Longwan 1 plants[23,24].The treated plants were incubated in a growth chamber at 15±1°C with a 12-h photoperiod. Ten days later,disease severity was rated on a 0-4 scale basedon infected foliage area,macroscopic and microscopic observations of mycelial growth,and sporulation[16,23,24,32,33]. Plants with a score of 0 were considered immune,while those with scores of 1 or 2 were classified as resistant and those with 3 or 4 as susceptible.Cultivars identified as immune or resistant to E.pisi infection were retested.

Table1-PhenotypicreactionsofChineseelitepea cultivars to Erysiphe pisi isolates EPBJ and EPYN,and the er1 alleles harbored by the analyzed cultivars.
2.4.RNA extraction and PsMLO1 sequence analysis
With the aim of identifying the resistance alleles at the er1 locus in immune or resistant pea cultivars,total RNA was extracted from their young leaves using an RNAprep Pure Plant Kit(Tiangen Biotech,Beijing,China)according to the manufacturer's instructions.First-strand cDNA was synthesized using a BioRT Two-Step RT-PCR Kit(Hangzhou Bioer Technology,Hangzhou,China)with oligo(dT)primers.
To amplify the full-length cDNA of PsMLO1,a polymerase chain reaction(PCR)was performed using PsMLO1-specific primers(PsMLO1F:5′-AAAATGGCTGAAGAGGGAGTT-3′;PsMLO1R:5′-TCCACAAATCAAGCTGCTACC-3′)[21].The PCR program was as follows:95°C for 5 min;35 cycles of 94°C for 30 s,58°C for 45 s,and 72°C for 1 min;72°C for 10 min.The ampliconswere purifiedusing aPCR PurificationKit(Qiagen)and then ligated into the pEasy-T5 vector(TransGen Biotech,Beijing,China).Recombinant plasmids were cloned in Escherichia coli TOP 10 competent cells and recovered using the QIAprep Spin Miniprep kit(Qiagen).Ten clones for each cultivar were sequenced at the Beijing Genomics Institute.To confirm that the amplified PsMLO1 fragments contained the correct sequences,thesusceptible(Bawan6andLongwan1)andresistant(Xucai 1 and YI)controls were similarly analyzed.The resulting sequences were compared with PsMLO1 cDNA sequences from the controls and wild-type pea cultivar Sprinter(susceptible to E.pisi;NCBI accession number:FJ463618.1)using DNAMAN software[20].
3.Results
3.1.Phenotypic evaluation
All Bawan 6 and Longwan 1 plants(susceptible controls)were severely infected by the EPBJ and EPYN E.pisi isolates,with a disease severity of 4(Fig.1;Table 1).In contrast,no disease symptoms were observed on the resistant controls,Xucai 1 and YI.These results are consistent with those of previous studies[15,23,31].Of the 37 pea cultivars analyzed,34 were heavily infected by the two E.pisi isolates,with masses of E.pisi conidia and mycelia observed on plants(with disease ratings of 3 and 4).These 34 cultivars were considered susceptible to powdery mildew infection.Yunwan 8,Yunwan 21,and Yunwan 23,which were developed in Yunnan,China,remained healthy and symptom-free,similarly to the resistant controls(Xucai 1 and YI).These three cultivars appeared to be immune to both E.pisi isolates(with disease ratings of 0)(Fig.1;Table 1).
3.2.Analysis of PsMLO1 cDNA sequence
The PsMLO1 cDNA sequences of Bawan 6 and Longwan 1 plants were identical to that of the wild-type pea cultivar Sprinter,indicating that Bawan 6 and Longwan 1 plants carry the Er1 gene(Table 1).The PsMLO1 cDNA sequence of YI plants matched the sequence of er1-4,which harbors a single-base deletion mutation(ΔA)at position 91.[20].The PsMLO1 cDNA sequence of Xucai 1 plants was identical to that of er1-2[23]. These results confirm the accuracy of the PsMLO1 cDNA sequences of the three identified resistant cultivars,Yunwan 8,Yunwan 21,and Yunwan 23.
The PsMLO1 cDNA sequences of Yunwan 8,Yunwan 21,and Yunwan 23 plants differed from those of Bawan 6,Longwan 1,and Sprinter plants.The PsMLO1 cDNA from Yunwan 8 plants had a point mutation(C→G)at position 680,matching that in the corresponding sequence in the Mexican pea cultivar Mexique 4(JI1559),which carries er1-1[20].Thus,the resistance gene present in Yunwan 8 plantswas identified as er1-1(Fig.2;Table 1).The mutations in the Yunwan 21 and Yunwan 23 PsMLO1 cDNA sequences were identical to those in Xucai 1 PsMLO1 sequences.There are three distinct PsMLO1 transcripts in Xucai 1 plants,characterized by a 129-bp deletion between positions 1171 and 1299,and 155-bp and 220-bp insertions at position 1263 of the coding sequence[23,24](Fig.3).The sequencing results of PsMLO1 from Yunwan 21 and Yunwan 23 plants indicated that they carried er1-2(Fig.3;Table 1).Additionally,the PsMLO1 cDNA sequences in Yunwan 21 and Yunwan 23 plants contained a frame shift associated with a 5-bp deletion.
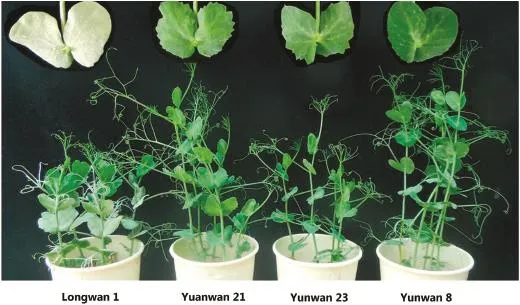
Fig.1-Phenotypic reactions of Yunwan 21,Yunwan 23,and Yunwan 8,and susceptible control Longwan 1 to Erysiphe pisi isolate EPYN.All plants of Longwan 1 were severely infected by E.pisi,with masses of E.pisi conidia and mycelia observed(disease rating 4).In contrast,there were no disease symptoms on the resistant pea cultivars Yunwan 21,Yunwan 23,and Yunwan 8.
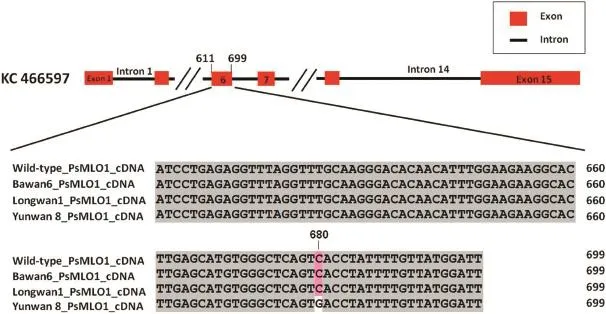
Fig.2-Comparison of PsMLO1 cDNA sequence from powdery mildew-resistant Yunwan 8 plants and susceptible Bawan 6,Longwan 1,and wild-type Sprinter(NCBI accession number:FJ463618.1)plants.The PsMLO1 cDNA sequence from Yunwan 8 plants harbors a base substitution(C→G)at position 680 in exon 6.The mutation site is indicated in the cDNA sequences.
4.Discussion
Powdery mildew of pea is an important disease that results in considerable yield and quality losses[1-3].The most efficient strategy to control this disease involves the use of resistant cultivars,given that host resistance permits sustainable pea production without potential safety risks for the environment and consumers.In the present study,the phenotypes of two susceptible and two resistant controls were consistent with those described in previous studies[20,23,24],indicating theaccuracy and reliability of the phenotypes observed for the tested pea cultivars.The cultivars Yunwan 8,Yunwan 21,and Yunwan 23 were immune to two E.pisi isolates from China. This result suggests that pea accessions from Yunnan,China,may be viable sources of resistance to E.pisi,in agreement with our previous findings[25].These three pea cultivars may be useful for the breeding of new powdery mildew-resistant cultivars in China.
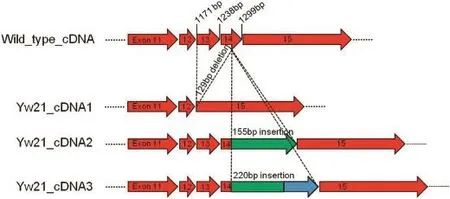
Fig.3-Three distinct PsMLO1 transcripts are present in Yunwan 21 plants.The transcripts are differentiated by a deletion(129 bp,cDNA1)or insertions(155 bp,cDNA2;220 bp,cDNA3).Yw21 refers to Yunwan 21.
The E.pisi resistance gene at the er1 locus in Yunwan 8 plants was identified as er1-1.The base substitution(C→G)at position 680 of PsMLO1 introduces a stop codon(UGA)that results in premature termination of protein synthesis.This observation is consistent with the results of previous studies by Humphry et al.[20]and Fu et al.[32].Humphry et al.[20]detected er1-1 in Mexique 4 plants,and Fu et al.[32]subsequently identified this allele in two powdery mildewresistant pea cultivars,Tara from Canada and Cooper from the Netherlands.Thus,Yunwan 8 is the fourth pea cultivar identified as carrying er1-1 but the first Chinese pea cultivar confirmed to carry this allele.
Powdery mildew resistance was conferred by er1-2(Table 1)in Yunwan 21 and Yunwan 23 plants,as in other pea cultivars/ lines including Stratagem (JI2302),Franklin,ROI3/02,Dorian,Nadir,X9002,and Xucai 1[20-24].The pea line X9002 and pea cultivar Xucai 1 were recently confirmed to carry er1-2,according to genetic mapping and sequence analyses of PsMLO1[23,24].In this study,we detected three distinct PsMLO1 transcripts in Yunwan 21 and Yunwan 23 plants,characterized by a 129-bp deletion or 155-bp and 220-bp insertions.The 129-bp deletion eliminated exons 13 and 14 from PsMLO1,resulting in the removal of 43 amino acids from the final protein(Fig.3).The 155-bp and 220-bp insertions were highly similar to five regions of a pea genomic BAC clone(accession number:CU655882).Interestingly,the 220-bp insert was also very similar(approximately 87%identical)to part of a giant retroelement that forms a major component of the pea genome(accession number:AY299395)[34,35].
The three pea cultivars identified as resistant to E.pisi were bred by researchers at the Yunnan Academy of Agricultural Sciences,China.This province is an important center of field pea diversity because it has a complex landform,which results in multiple geographical and climatic features and a variable environment[36-38].Zong et al.[36]reported that pea accessions originating in Yunnan exhibit high genetic diversity.This province is located in a low-latitude plateau region and experiences extreme biotic and abiotic stresses[38,39]. The presence of a rich stock of germplasm exhibiting disease resistance is likely the result of specific adaptations to local climates and high selection pressure.Recently,Sun.et al.[25]discovered a novel er1 allele,er1-6,in several Chinese pea landraces originating in Yunnan,suggesting that the variable climate in this province caused mutations in PsMLO1 that were specific to this geographic region.
Two single recessive genes(er1 and er2)and one single dominant gene(Er3)have been identified as influencing resistance to E.pisi[5-7].Previous studies have concluded that the resistance in most pea germplasm is controlled by er1,which is the most widely deployed gene in contemporary pea cultivars becauseofitshighlyeffectivebroad-spectrumresistancetoE.pisi[15].The er1 resistance phenotype is induced by loss-of-function mutations in PsMLO1.Nine er1 alleles have been identified as conferring resistance to E.pisi in pea germplasm[20-27].Among the reported alleles,er1-1 and er1-2 are commonly used in pea breeding programs for their ability to provide stable and durable resistance to E.pisi[20,21,23,24,32].In this study,we identified these two major er1 alleles in three powdery mildew-resistant Chinese pea cultivars,which represent valuable genetic resources for breeding E.pisi-resistant cultivars.
5.Conclusions
Three powdery mildew-resistant pea cultivars bred in Yunnan Province were confirmed to be immune to two Chinese E.pisi isolates,EPBJ and EPYN.Two(er1-1 and er1-2)of seven er1 alleles for pea powdery mildew resistance were characterized in the three resistant cultivars.These results are relevant to the breeding of E.pisi-resistant pea cultivars in China.
Acknowledgments
ThisresearchwassupportedbytheChinaAgriculture Research System (CARS-09),the Agricultural Science andTechnology Program for Innovation Team on Identification and Excavation of Elite Crop Germplasm from Chinese Academy of Agricultural Sciences(CAAS),the Special Fund for Agro-scientific Research in the Public Interest(1610092015002-01)from the Institute of Crop Science,CAAS,and the Fund(2013BB010)from Science and Technology Department of Yunnan Province.
R E F E R E N C E S
[1]R.L.Munjal,V.V.Chenulu,T.S.Hora,Assessment of losses due to powdery mildew(Erisiphe polygoni)on pea,Indian Phytopathol.19(1963)260-267.
[2]T.D.Warkentin,K.Y.Rashid,A.G.Xue,Fungicidal control of powdery mildew in field pea,Can.J.Plant Sci.76(1996)933-935.
[3]S.Fondevilla,J.I.Cubero,D.Rubiales,Confirmation that the Er3 gene,conferring resistance to Erysiphe pisi in pea,is a different gene from er1 and er2 genes,Plant Breed.130(2011)281-282.
[4]S.Fondevilla,D.Rubiales,Powdery mildew control in pea:a review,Agron.Sustain.Dev.32(2012)401-409.
[5]S.C.Harland,Inheritance of immunity to mildew in Peruvian forms of Pisum sativum,Heredity 2(1948)263-269.
[6]R.J.Heringa,A.van Norel,M.F.Tazelaar,Resistance to powdery mildew(Erysiphe polygoni D.C.)in peas(Pisum sativum L.),Euphytica 18(1969)163-169.
[7]S.Fondevilla,A.M.Torres,M.T.Moreno,D.Rubiales,Identification of a new gene for resistance to powdery mildew in Pisum fulvum,a wild relative of pea,Breed.Sci.57(2007)181-184.
[8]E.Dirlewanger,P.G.Isaac,S.Ranade,M.Belajouza,R.Cousin,D.Vienne,Restriction fragment length polymorphism analysis of loci associated with disease resistance genes and developmental traits in Pisum sativum L.Theor.Appl.Genet. 88(1994)17-27.
[9]G.M.Timmerman,T.J.Frew,N.F.Weeden,Linkage analysis of er1,a recessive Pisum sativum gene for resistance to powdery mildew fungus(Erysiphe pisi D.C.),Theor.Appl.Genet.88(1994)1050-1055.
[10]V.Katoch,S.Sharma,S.Pathania,D.K.Banayal,S.K.Sharma,R.Rathour,Molecular mapping of pea powdery mildew resistance gene er2 to pea linkage group III,Mol.Breed.25(2010)229-237.
[11]S.Fondevilla,D.Rubiales,M.T.Moreno,A.M.Torres,Identification and validation of RAPD and SCAR markers linked to the gene Er3 conferring resistance to Erysiphe pisi D.C.in pea,Mol.Breed.22(2008)193-200.
[12]S.Fondevilla,T.L.W.Carver,M.T.Moreno,D.Rubiales,Macroscopic and histological characterisation of genes er1 and er2 for powdery mildew resistance in pea,Eur.J.Plant Pathol.115(2006)309-321.
[13]S.Fondevilla,T.L.W.Carver,M.T.Moreno,D.Rubiales,Identification and characterisation of sources of resistance to Erysiphe pisi Syd.in Pisum spp.Plant Breed.126(2007)113-119.
[14]R.Iglesias-Garcia,D.Rubiales,S.Fondevilla,Penetration resistance to Erysiphe pisi in pea mediated by er1 gene is associated with protein cross-linking but not with callose apposition or hypersensitive response,Euphytica 201(2015)381-387.
[15]K.R.Tiwari,G.A.Penner,T.D.Warkentin,Inheritance of powdery mildew resistance in pea,Can.J.Plant Sci.77(1997)307-310.
[16]A.Vaid,P.D.Tyagi,Genetics of powdery mildew resistance in pea,Euphytica 96(1997)203-206.
[17]S.M.Liu,L.O′Brien,S.G.Moore,A single recessive gene confers effective resistance to powdery mildew of field pea grown in northern New South Wales,Aust.J.Exp.Agric.43(2003)373-378.
[18]B.Sharma,Identification of recessive er gene for powdery mildew resistance in a landrace of Pisum sativum,Pisum Genet.35(2003)30-31.
[19]B.Sharma,The Pisum genus has only one recessive gene for powdery mildew resistance,Pisum Genet.35(2003)22-27.
[20]M.Humphry,A.Reinstädler,S.Ivanov,T.Bisseling,R. Panstruga,Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1,Mol.Plant Pathol.12(2011)866-878.
[21]S.Pavan,A.Schiavulli,M.Appiano,A.R.Marcotrigiano,F. Cillo,R.G.F.Visser,Y.L.Bai,C.Lotti,L.Ricciardi,Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus,Theor.Appl.Genet. 123(2011)1425-1431.
[22]S.Pavan,A.Schiavulli,M.Appiano,C.Miacola,R.G.F.Visser,Y.L.Bai,C.Lotti,L.Ricciardi,Identification of a complete set of functional markers for the selection of er1 powdery mildew resistance in Pisum sativum L,Mol.Breed.31(2013)247-253.
[23]S.L.Sun,Z.Y.Wang,H.N.Fu,C.X.Duan,X.M.Wang,Z.D.Zhu,Resistance to powdery mildew in the pea cultivar Xucai 1 is conferred by the gene er1,Crop J.3(2015)489-499.
[24]Z.Y.Wang,H.N.Fu,S.L.Sun,C.X.Duan,X.F.Wu,X.M.Yang,Z. Zhu,Identification of powdery mildew resistance gene in pea line X9002,Acta Agron.Sin.41(2015)515-523(in Chinese with English abstract).
[25]S.L.Sun,H.N.Fu,Z.Y.Wang,C.X.Duan,X.X.Zong,Z.D.Zhu,Discovery of a novel er1 allele conferring powdery mildew resistance in Chinese pea(Pisum sativum L.)landraces,PLoS One 11(2016),e0147624.
[26]S.L.Sun,D.Deng,Z.Y.Wang,C.X.Duan,X.F.Wu,X.M.Wang,X.X.Zong,Z.D.Zhu,A novel er1 allele and the development and validation of its functional marker for breeding pea(Pisum sativum L.)resistance to powdery mildew,Theor.Appl. Genet.129(2016)909-919.
[27]T.Santo,M.Rashkova,C.Alabaça,J.Leitão,The ENU-induced powdery mildew resistant mutant pea(Pisum sativum L.)lines S(er1mut1)and F(er1mut2)harbour early stop codons in the PsMLO1 gene,Mol.Breed.32(2013)723-727.
[28]H.X.Peng,G.Yao,R.L.Jia,H.Y.Liang,Identification of pea germplasm resistance to powdery mildew,J.Southwest Agric.Univ.13(1991)384-386(in Chinese).
[29]X.M.Wang,Z.D.Zhu,C.X.Duan,X.X.Zong,Identification and Control Technology of Disease and Pest on Faba Bean and Pea,Chinese Agricultural Science and Technology Press,Beijing,2007(in Chinese).
[30]L.Zeng,M.Q.Li,X.M.Yang,Identification of resistance of peas resources to powdery mildew,Grass.Turf 32(2012)35-38(in Chinese with English abstract).
[31]Z.Y.Wang,S.Y.Bao,C.X.Duan,X.X.Zong,Z.D.Zhu,Screening and molecular identification of resistance to powdery mildew in pea germplasm,Acta Agron.Sin.39(2013)1030-1038(in Chinese with English abstract).
[32]H.N.Fu,S.L.Sun,Z.D.Zhu,C.X.Duan,X.M.Yang,Phenotypic and genotypic identification of powdery mildew resistance in pea cultivars or lines from Canada,J.Plant Genet.Resour.15(2014)1028-1033(in Chinese with English abstract).
[33]J.C.Rana,D.K.Banyal,K.D.Sharma,K.Sharma Manish,S.K. Gupta,S.K.Yadav,Screening of pea germplasm for resistance to powdery mildew,Euphytica 189(2013)271-282.
[34]P.Neumann,D.Pozarkova,J.Macas,Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced,Plant Mol.Biol.53(2003)399-410.
[35]T.Santo,M.Rashkova,C.Alabaca,J.Leitao,The ENU-induced powdery mildew resistant mutant pea(Pisum sativum L.)lines S(er1mut1)and F(er1mut2)harbour early stop codons in the PsMLO1 gene,Mol.Breed.32(2013)723-727.
[36]X.X.Zong,J.P.Guan,S.M.Wang,Q.C.Liu,Genetic diversity among Chinese pea(Pisum sativum L.)landraces revealed by SSR markers,Acta.Agron.Sin.34(2008)1330-1338(in Chinese with English abstract).
[37]J.G.Cheng,X.F.Wang,L.Z.Fan,X.P.Yang,P.W.Yang,Variations of Yunnan climatic zones in recent 50 years,Prog. Geogr.28(2009)18-24(in Chinese with English abstract).
[38]L.Li,R.J.Redden,X.X.Zong,J.D.Berger,S.J.Bennett,Ecogeographic analysis of pea collection sites from China to determine potential sites with abiotic stresses,Genet. Resour.Crop.Evol.60(2013)1801-1815.
[39]Z.G.Huo,L.W.Chen,W.C.Liu,C.Y.Xue,S.J.Zhao,L.W.Zhuang,Climatic zonation of wheat powdery mildew in China,Acta Ecol.Sin.22(2001)1873-1881(inChinesewithEnglishabstract).
23 February 2016
in revised form
.Tel.:+86 10 8210 9609;fax:+86 10 8210 9608.
E-mail address:zhuzhendong@caas.cn(Z.Zhu).
Peer review under responsibility of Crop Science Society of China and Institute of Crop Science,CAAS.
http://dx.doi.org/10.1016/j.cj.2016.05.010
2214-5141/©2016 Crop Science Society of China and Institute of Crop Science,CAAS.Production and hosting by Elsevier B.V.This is an open access article under the CC BY-NC-ND license(http://creativecommons.org/licenses/by-nc-nd/4.0/).
杂志排行
The Crop Journal的其它文章
- Editorial:Food Legume Diversity and Legume Research Policies
- QTL and candidate genes associated with common bacterial blight resistance in the common bean cultivar Longyundou 5 from China
- Construction of an integrated map and location of a bruchid resistance gene in mung bean
- Evaluation of common bean(Phaseolus vulgaris L.)genotypes for drought stress adaptation in Ethiopia
- Large-scale evaluation of pea(Pisum sativum L.)germplasm for cold tolerance in the field during winter in Qingdao
- Molecular cloning and characterization of a gene encoding the proline transporter protein in common bean(Phaseolus vulgaris L.)
