Fosmid library construction and screening for the maize mutant gene Vestigial glume 1
2016-04-05ChaoxianLiuXiaoliLiuLeiLeiHaiyingGuanYilinCai
Chaoxian Liu, Xiaoli Liu, Lei Lei, Haiying Guan, Yilin Cai*
aMaize Research Institute, Southwest University, Chongqing 400715, ChinabNational Maize Improvement Center of China, China Agricultural University, Beijing 100193, ChinacMaize Institute, Shandong Academy of Agricultural Sciences, Jinan 250100, China
Fosmid library construction and screening for the maize mutant gene Vestigial glume 1
Chaoxian Liua,1, Xiaoli Liua,1, Lei Leib, Haiying Guanc, Yilin Caia,*
aMaize Research Institute, Southwest University, Chongqing 400715, China
bNational Maize Improvement Center of China, China Agricultural University, Beijing 100193, China
cMaize Institute, Shandong Academy of Agricultural Sciences, Jinan 250100, China
A R T I C L E I N F O
Article history:
Received 2 June 2015
Received in revised form
27 August 2015
Accepted 27 November 2015
Available online 4 December 2015
Keywords:
Vestigial glume 1
Fosmid
Library construction
Library screening
A B S T R A C T
The maize mutant gene Vestigial glume 1 (Vg1) has been fine-mapped to a narrow region by map-based cloning and the candidate gene for Vg1 spanned 19.5 kb. Here we report Vg1 genomic fosmid library construction and screening. The fosmid library of Vg1 consisted of 574,000cloneswithanaverageinsertsizeof36.4 kb,representing7.9-foldcoverageofthemaize genome. Fosmid stability assays indicated that clones were stable during propagation in the fosmid system. Using Vg1 candidate gene-specific primers, a positive clone was successfully identified. This discovery will pave the way for identifying the function of Vg1 in maize development.
©2015 Crop Science Society of China and Institute of Crop Science, CAAS. Production and hosting by Elsevier B.V. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
* Corresponding author.
E-mail address: caiyilin1789@163.com (Y. Cai).
Peer review under responsibility of Crop Science Society of China and Institute of Crop Science, CAAS.
1Chaoxian Liu and Xiaoli Liu contributed equally to this work.
http://dx.doi.org/10.1016/j.cj.2015.09.003
2214-5141/©2015 Crop Science Society of China and Institute of Crop Science, CAAS. Production and hosting by Elsevier B.V. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
1. Introduction
Bacterial artificial chromosome (BAC) and fosmid systems are frequently used methods for generating large and stable genomic clones for genome sequencing, physical mapping, positional cloning, and analysis of gene structure and function. The BAC genomic library is a system for cloning larger DNA inserts. Given that the insert size of a BAC library generally ranges from 80 to 200 kb, with maximum sizes up to 340 kb [1], BAClibrariesarewidelyusedasgenomiclibrariesforcropswith larger genomes [2–6]. But for smaller insert sizes of about 40 kb, fosmids are a powerful complementary tool for BACs. A fosmid cloning system employs a low-copy number cosmid vector based on the Escherichia coli F-factor replicon and provides a method for preparing mini-BACs with an average insert size of 40 kb with high efficiency [7]. The fosmid vector offers many advantages over other genomic library systems, including ease of handling and propagation of clones, stability of the insert DNA, and unbiased cloning.
Recently, fosmid libraries have been developed for many species including a plant pathogenic bacterium [8],rice [9], sugar beet [10], Taxus [11], and human [12]. Fosmid libraries have been usedforgenomegapclosing.Forexample,thecompletegenome of Bacillus thuringiensis strain HD521 was sequenced with anIllumina HiSeq 2000 platform, and assembly of the sequence reads resulted in 77 scaffolds [13]. The gaps between scaffolds were closed with 186 fosmid clones to yield a single gapless chromosome. The human genome sequence had been finished to very high standards in 2004. However, 341 gaps remained when the finished genome was published [12]. Using fosmid resources generated from multiple individuals, Bovee et al. [14] filled gaps in the euchromatic part of the human genome. Fosmid libraries have been widely used in physical map construction [11,15].Magrini etal. [16] generated a fosmid library representing 10-fold coverage of the Histoplasma capsulatum G217B genome. A total of 9551 fosmid clones were analyzed for physical mapping, which resulted in joining of existing physical map contigs into 84 clusters. Park et al. [17] reported the development of a fosmid library for Chinese cabbage consisting of 97,536 clones with an average insert size of approximately 40 kb, corresponding to seven genome equivalents. Among many uses of fosmid libraries, one of our interests is to develop resources for positional cloning of genes with large size or complex structure. Cadavid et al. [18] positionally cloned alr2 using a tightly linked molecular marker, constructed fosmid genomic libraries from a homozygous laboratory strain and identified a clone containing the f allele at both alr1 and alr2 [19].
However, to date, there are few reports of identification candidate genes using fosmid libraries for crops with large genomes. Vg1 is a dominant mutation resulting in severely diminished size of ligules and greatly reduced tassel and ear glumes [20]. Vg1 has been fine-mapped to an interval containing a single candidate gene of 11.5 kb according to the B73 reference genome, and a large DNA fragment about 8 kb inserted in the candidate gene has been confirmed by Southern blot analysis (unpublished data). The difficulty of amplifying a mutant gene of 19.5 kb from the complex maize genome by PCR prompted the construction of a genomic library to screen for the mutant gene. We describe fosmid library construction and screening for Vg1.
2. Materials and methods
2.1. Plant material and DNA extraction
Vg1 mutants were grown in Sanya, Hainan province, in November 2011. Ear bracts of Vg1 mutants were collected and ground in liquid nitrogen. Genomic DNA extracted by the CTAB method was dissolved in 800 μL TE buffer (pH 8.0), and then the extracted DNA was purified with chloroform–isoamyl alcohol and precipitated with isopropanol for two times. DNA quality was assessed by pulsed-field gel electrophoresis (PFGE) and NanoDrop 2000 (Thermo).
2.2. Randomly sheared Vg1 genomic DNA
The CopyControl HTP Fosmid Library Production Kit with pCC2FOS vector (Epicentre) was used for fosmid library construction. According to the protocol, at least 10% of genomic DNA fragments should be of length 40 kb. The DNA was divided into three equal parts and each was subjected to shearing via repetitive pipetting 200, 400 and 600 times using a 200-μL pipette tip, until 10% or more of the genomic DNA was of a length of about 40 kb.
2.3. End repair and recovery of insert DNA
The randomly sheared DNA was end-repaired using a blunt end repair kit to generate blunt-ended, 5′-phosphorylated DNA and was separated on a 1% low-melting point agarose gel using PFGE for 15 h for 1–12 s at 120°and 6 V cm−1in 0.5×TBE buffer. DNA fragments ranging from 25 to 48 kb in size were excised from the gel and recovered using GELase (Epicentre) according to the instruction manual.
2.4. Fosmid library titer determination
The recovered DNA was ligated to vector pCC2FOS at 16°C overnight. The ligation mixture was subsequently packaged using MaxPlax Lambda Packaging Extracts, and then the number of phage particles was determined by serial dilution followed by infection of EPI300-T1 cells. The titer of the genomic library was caculated by the following formula:
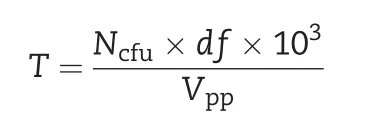
where T denotes the titer of the fosmid library (cfu mL−1), Ncfuthe number of colonies, df the dilution factor, and Vppthe volume of phage plated (μL). Finally, the infected bacterial cells supplemented with 20% glycerol were transferred to 2 mL sterile tubes and stored at−80°C.
2.5. Average size estimation of fosmid library
To determine theaveragesizeofinsertfragments,115randomly picked clones were cultured overnight in 15 mL vials with 3 mL LB liquid media containing 12.5 μg mL−1chloramphenicol and 30 μL auto-induction solution. Fosmid clones were then completely digested with Not I (NEB) and the size of insert fragments was estimated using PFGE under the following conditions: 1–12 s, 10 h, 120°, and 6 v cm−1in 0.5×TBE buffer.
2.6. Stability assays of fosmid library
Ten randomly picked fosmid clones were used for stability assays.Following Kimetal.[21],thecloneswereculturedin2 mL LB with 12.5 μg mL−1chloramphenicol overnight at 37°C and 200 r min−1. On day 1, these clones were considered to be generation 0 and on day 6 they had propagated to generation 100. Plasmids prepared from generations 0 to 100 were digested with Hind III and the digestion patterns were compared by PFGE.
2.7. Vg1 candidate gene screening Theinfectedbacterialcellsstoredat−80°Cwerethawedatroom temperature and then shaken at 37°C, 200 r min−1for 1 h. Then, infected bacterial cells (22 μL) were transferred to 1.5 mL sterile tubes containing 350 μL LB and an appropriate antibiotic and incubated at 37°C and 220 r min−1for 4 to 6 h.
Gene-specific primers were designed for screening for Vg1-positive clones (forward primer: CAGGCGATGCAAAGTTG CCGAG, reverse primer: CACGCTGGTGCTACAAAGGGCT). A 5-μLaliquot of culture was used as template for PCR screening. PCR was performed as follows: one cycle of 94°C for 10 min followed by 35 cycles of denaturation at 94°C for 50 s, annealing at 64°C for 50 s, and elongation at 72°C for 50 s, with a final extension at 72°C for 5 min. PCR products were separated on a 1% agarose gel and visualized by ethidium bromide staining. When a positive clone was identified, the positive pool diluted 2×106times with LB liquid media was plated on LB agar plates, and 100 colonies were picked and subjected to colony PCR.
3. Results
3.1. Extraction of high-quality Vg1 genomic DNA
DNA was prepared from bracts of immature ears of Vg1 plants to construct a fosmid library for map-based cloning of Vg1. The size, concentration and quality of Vg1 genomic DNA were determined by PFGE and Thermo NanoDrop 2000. The majority of the genomic DNA ranged from 48.5 kb to 97 kb (Fig. 1). The concentration was 342.5 ng μL−1and A260/A280was 1.79. The concentration and quality of genomic DNA could thus satisfy the needs of fosmid genomic library construction.
3.2. Randomly sheared Vg1 genomic DNA
Because the insert DNA size should be about 40 kb, the prepared DNA could not be used directly. The prepared DNA in fragment sizes ranging from 23 kb to 97 kb was divided into three tubes and randomly sheared 200, 400 and 600 times with a 200 μL small-bore pipette tip. A small amount (about 5 μL) of DNA was subjected to PFGE. The results showed that at least 20% of DNA fragment sheared 600 times migrated with the 48.5 kb DNA band (Fig. 2). Accordingly, 2–4 cm of the gel below the 48.5 kb band was excised for insert DNA recovery.
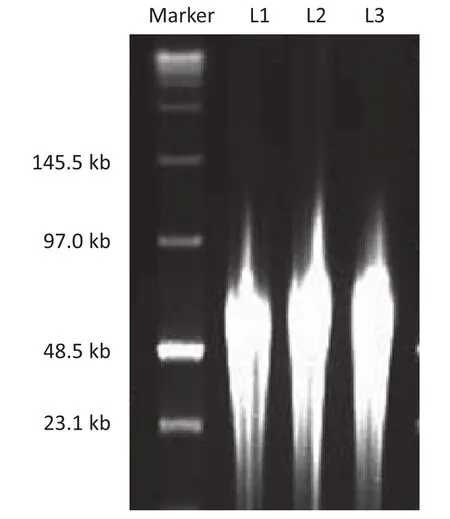
Fig. 2–Vg1 DNA randomly sheared 200, 400, and 600 times. L1: DNA randomly sheared 600 times; L2: DNA randomly sheared 400 times; L3: DNA randomly sheared 200 times.
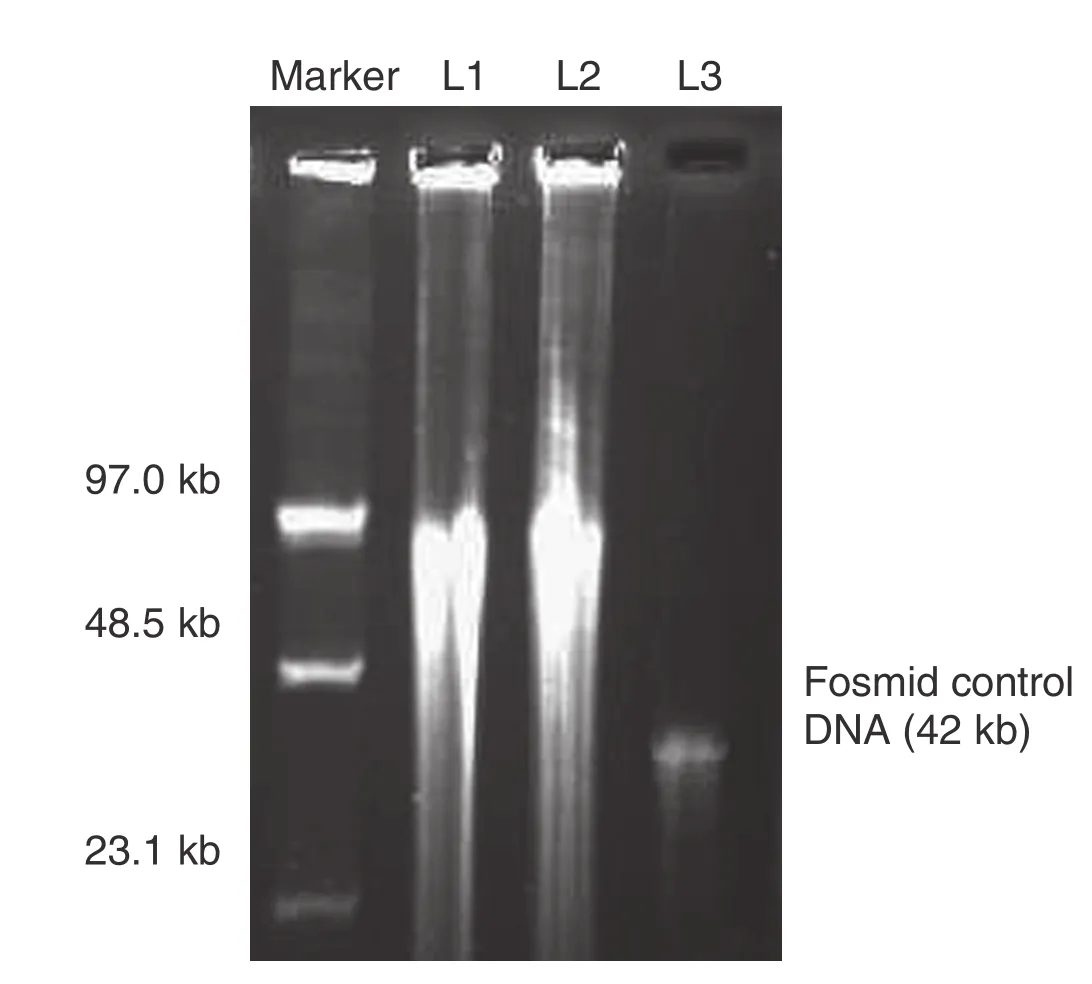
Fig. 1–Vg1 genomic DNA Preparation. L1: lane 1, L2: lane 2, L3: lane 3.
3.3. Determination of library titer
The recovered DNA was subjected to end repair and ligated into the pCC2FOS vector. The ligation product was packed using MaxPlax Lambda Packaging Extracts according to the manufacturer's protocol. Packaged phage particles (10 μL) were serially diluted to 1:10, 1:100, and 1:1000 with phage dilution buffer and 10 μL of each dilution was added to 100 μL of prepared EPI300-T1 host cells for determination of the library titer. The results indicated that the numbers ofcolonies from serial dilutions 1:10, 1:100, and 1:1000 were 471, 65, and 6, respectively. The library titers were thus 4.71×105cfu mL−1, 6.5×105cfu mL−1, and 6×105cfu mL−1, with an average of 5.74×105cfu mL−1, meaning that the library contained 5.74×105fosmid clones.
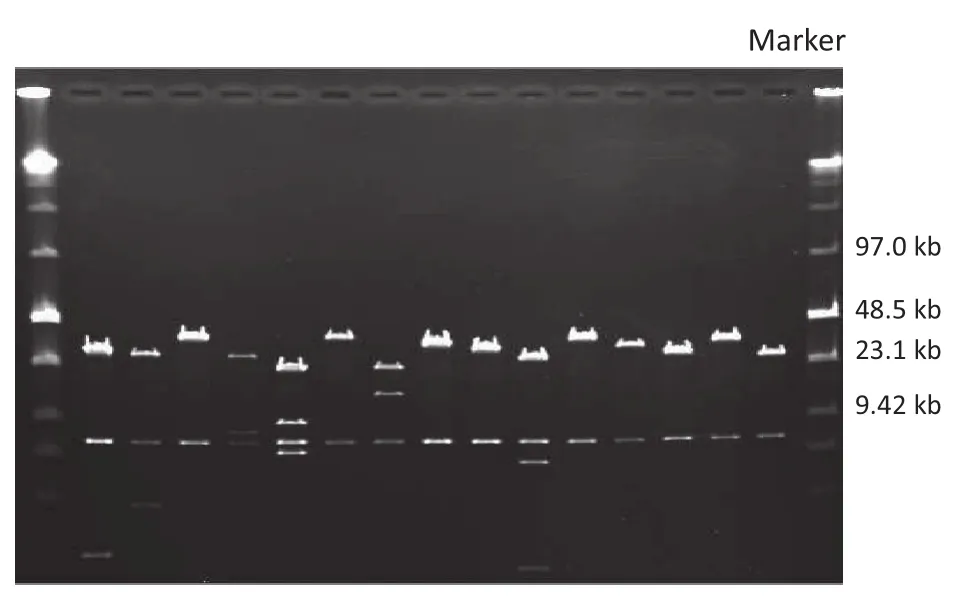
Fig. 3–Fosmid clones digested with restriction enzyme Not I.
3.4. Average insert size of the fosmid library
To estimate the average insert size and insert-size distribution of the Vg1 fosmid library, a random sample of 115 fosmid clones was selected. Fosmid DNA was then completely digested with Not I and the insert size was estimated by PFGE (Fig. 3). These clones contained insert fragments with an average size of 34.6 kb. Thus, the total size of this fosmid library was estimated as 19,860 Mb, covering the maize genome 7.9 times. The majority of fosmid clones contained inserts of 30–45 kb, accounting for 86.96% of the total clones. 10.43% and 2.61% of the remaining harbored inserts<25 kb and>45 kb, respectively (Fig. 4). The library was thus suitable for screening Vg1.

Fig. 4–Distribution of insert size of fosmid clones.

Fig. 5–Digestion patterns of 10 fosmid clones from generations 0 and 100. Lane pairs A to J contain the 10 fosmid clones. M: marker. Numbers 1 and 6 denote fosmid clones at generations 0 and 100, respectively.
3.5. Fosmid stability assays
To evaluate the quality of the Vg1 genomic fosmid library, 10 fosmid clones were randomly picked and serially cultured for 100 generations over a period of 6 days. The Hind III digestion patterns of the generation 0 and generation 100 clones were compared. There were no insertions, deletions, or rearrangements in the 10 fosmid clones (Fig. 5). This finding indicated that the fosmid clones could stably propagate DNA inserts.
3.6. Genomic library screening for Vg1 candidate gene
For target clone screening, a simple method was adopted. Twenty microliters of infected bacteria containing approximately 70 clones was added to 200 μL LB and shaken for 6 h, and then 1 μL aliquot of culture was used for colony PCR to screen for positive clones. Among 17,000 clones, one positive clone was identified (Fig. 6). The PCR products were sequenced and the results showed that the clone contained the target gene (Fig. 7).
4. Discussion
Maize genes of large size or complex structure are usually isolated with a BAC genomic library. Compared with BAC insert sizes, the small (approximately 40 kb) insert sizes offosmid clones allow easier handling of clones, and the use of fosmid clones avoids restriction site bias by use of DNA shearing rather than DNA digestion. The fosmid system thus offers advantages over other genomic libraries and has been widely used in metagenomic research [22,23], genome sequencing [24,25], gap closing [26,27], and other applications.
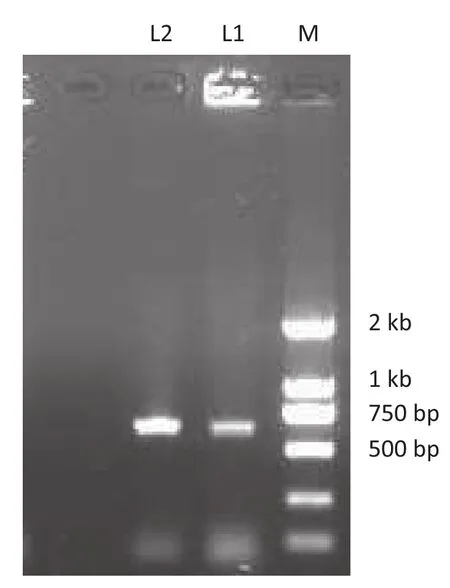
Fig. 6–Vg1 genomic library screening. M, marker; L1, apositive clone identified by bacteria liquid PCR; L2, control.
Here we report Vg1 fosmid library construction and library screening. There are two critical steps in fosmid library construction. First, DNA quality is one of the most important factors, greatly influencing the subsequent ligation efficiency that determines the number of fosmid clones. The extraction of high-quality and high-molecular-weight DNA is thus critical [28]. We adopted a conventional CTAB method for DNA extraction. Vg1 genomic DNA was purified three times after dissolution in TE buffer until most proteins and other impurities were eliminated, giving an A260/A280ratio of 1.79. In contrast, DNA of low quality for Rg1 and Bt fosmid library construction was purified only once and the library size was greatly reduced to 350,000 and 340,000 clones (unpublished data). Second, size selection of end-repaired DNA is another important factor determining the insert size of a fosmid library. According to the protocol, the insert DNA recovered is of size≥25 kb. DNA fragments smaller than 25 kb may result in unwanted chimeric clones. In the Vg1 fosmid library, 10.43% of the fosmid clones had insert sizes of<25 kb. This result could be improved by increasing electrophoresis time for DNA fractionation or re-fractionating DNA fragments after the first recovery of insert DNA.
In summary, we have constructed a high-quality Vg1 fosmid library containing 570,000 clones with average insert size 34.6 kb representing 7.9-fold coverage of the Vg1 genome. Using this fosmid library, we identified a positive clone. PCR product sequencing indicated that this clone contained a Vg1 candidate gene. This result will help to elucidate the function of Vg1 in the maize vegetative and reproductive phases.
Acknowledgments
This study was supported by a Chongqing Postdoctoral Science Foundation funded project (Xm201344), China Postdoctoral Science Foundation funded project (2014M552303),and Fundamental Research Funds for the Central Universities (XDJK2013C023, 2362015xk05).
R E F E R E N C E S
[1] S.S. Woo, J. Jiang, B.S. Gill, A.H. Paterson, R.A. Wing, Construction and characterization of bacterial artificial chromosome library of Sorghum bicolor, Nucleic Acids Res. 22 (1994) 4922–4931.
[2] J.S. Ammiraju, M. Luo, J.L. Goicoechea, W. Wang, D. Kudrna, C. Mueller, J. Talag, H. Kim, N.B. Sisneros, B. Blackmon, The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza, Genome Res. 16 (2006) 140–147.
[3] Y. Hu, Y. Lu, D. Ma, W. Guo, T. Zhang, Construction and characterization of a bacterial artificial chromosome library for the A-genome of cotton (G. arboreum L.), J. Biomed. Biotechnol. 2011 (2010) 1–5.
[4] Y. Yu, J. Tomkins, R. Waugh, D. Frisch, D. Kudrna, A. Kleinhofs, R. Brueggeman, G. Muehlbauer, R. Wise, R. Wing, A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes, Theor. Appl. Genet. 101 (2000) 1093–1099.
[5] C.C. Wu, P. Nimmakayala, F. Santos, R. Springman, C. Scheuring, K. Meksem, D. Lightfoot, H.B. Zhang, Construction and characterization of a soybean bacterial artificial chromosome library and use of multiple complementary libraries for genome physical mapping, Theor. Appl. Genet. 109 (2004) 1041–1050.
[6] X. Li, L. Tan, H. Huang, Z. Zhu, C. Li, S. Hu, C. Sun, Construction of a bacterial artificial chromosome (BAC) library of common wild rice (Oryza rufipogon Griff.) for mapbased cloning of genes selected during the domestication of rice, Biotechnol. Lett. 30 (2008) 555–561.
[7] U.J. Kim, H. Shizuya, J. Sainz, J. Garnes, S.M. Pulst, P. de Jong, M.I. Simon, Construction and utility of a human chromosome 22-specific Fosmid library, Genet. Anal.-Biomol. Eng. 12 (1995) 81–84.
[8] F. Thieme, R. Koebnik, T. Bekel, C. Berger, J. Boch, D. Büttner, C. Caldana, L. Gaigalat, A. Goesmann, S. Kay, Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence, J. Bacteriol. 187 (2005) 7254–7266.
[9] J.S. Ammiraju, Y. Yu, M. Luo, D. Kudrna, H. Kim, J.L. Goicoechea, Y. Katayose, T. Matsumoto, J. Wu, T. Sasaki, Random sheared fosmid library as a new genomic tool to accelerate complete finishing of rice (Oryza sativa spp. Nipponbare) genome sequence: sequencing of gap-specific fosmid clones uncovers new euchromatic portions of the genome, Theor. Appl. Genet. 111 (2005) 1596–1607.
[10] W. Traut, C. Lange, D. Holtgräwe, B. Schulz, B. Weisshaar, H. Himmelbauer, Construction and characterization of a sugar beet (Beta vulgaris) fosmid library, Genome 51 (2008) 948–951.
[11] D. Hao, L. Yang, P. Xiao, The first insight into the Taxus genome via fosmid library construction and end sequencing, Mol. Genet. Genomics 285 (2011) 197–205.
[12] International Human Genome Sequencing Consortium, Finishing the euchromatic sequence of the human genome, Nature 431 (2004) 931–945.
[13] Q. Li, L.Z. Xu, T. Zou, P. Ai, G.H. Huang, P. Li, A.P. Zheng, Complete genome sequence of Bacillus thuringiensis strain HD521, Stand. Genomic Sci. 10 (2015) 62.
[14] D. Bovee, Y. Zhou, E. Haugen, Z. Wu, H.S. Hayden, W. Gillett, E. Tuzun, G.M. Cooper, N. Sampas, K. Phelps, Closing gaps in the human genome with fosmid resources generated from multiple individuals, Nat. Genet. 40 (2008) 96–101.
[15] X. Wang, Q. Zhang, X. Sun, Y. Chen, T. Zhai, W. Zhuang, J. Qi, Z. Wang, Fosmid library construction and initial analysis of end sequences in female half-smooth tongue sole (Cynoglossus semilaevis), Mar. Biotechnol. 11 (2009) 236–242.
[16] V. Magrini, W.C. Warren, J. Wallis, W.E. Goldman, J. Xu, E.R. Mardis, J.D. McPherson, Fosmid-based physical mapping of the Histoplasma capsulatum genome, Genome Res. 14 (2004) 1603–1609.
[17] T.H. Park, B.S. Park, J.A. Kim, J.K. Hong, M. Jin, Y.J. Seol, J.H. Mun, Construction of random sheared fosmid library from Chinese cabbage and its use for Brassica rapa genome sequencing project, J. Genet. Genomics 38 (2011) 47–53.
[18] L.F. Cadavid, A.E. Powell, M.L. Nicotra, M. Moreno, L.W. Buss, An invertebrate histocompatibility complex, Genetics 167 (2004) 357–365.
[19] M.L. Nicotra, A.E. Powell, R.D. Rosengarten, M. Moreno, J. Grimwood, F.G. Lakkis, S.L. Dellaporta, L.W. Buss, A hypervariable invertebrate allodeterminant, Curr. Biol. 19 (2009) 583–589.
[20] G.F. Sprague, Heritable characters in maize 50-vestigial glume, J. Hered. 30 (1939) 143–146.
[21] U.J. Kim, H. Shizuya, P.J. de Jong, B. Birren, M.I. Simon, Stable propagation of cosmid sized human DNA inserts in an F factor based vector, Nucleic Acids Res. 20 (1992) 1083–1085.
[22] J. Lu, L.Q. Du, H. Pang, G. Ma, Y.T. Wei, R.B. Huang, Construction of a metagenomic library from hot spring soil and cloning of pullulanase gene, J. South. Agric. 45 (2014) 725–730.
[23] C.M. Lee, S.H. Seo, S.J. Kim, J.S. Sim, Screening and characterization of a novel cellulase gene from the gut microflora of Hermetia illucens using metagenomic library, J. Microbiol. Biotechnol. 24 (2014) 1196–1206.
[24] S. Li, G. Liu, Z. Chen, Y. Wang, P. Li, J. Hua, Construction and initial analysis of five Fosmid libraries of mitochondrial genomes of cotton (Gossypium), Chin. Sci. Bull. 58 (2013) 4608–4615.
[25] M. Oshiki, K. Shinyako-Hata, H. Satoh, S. Okabe, Draft genome sequence of an anaerobic ammonium-oxidizing bacterium,“Candidatus Brocadia sinica”, Genome Announc. 3 (2015), e00267-15.
[26] S. Weidner, A. Becker, I. Bonilla, S. Jaenicke, J. Lloret, I. Margaret, A. Pühler, J.E. Ruiz-Sainz, S. Schneiker-Bekel, R. Szczepanowski, Genome sequence of the soybean symbiont Sinorhizobium fredii HH103, J. Bacteriol. 194 (2012) 1617–1618.
[27] S. Schneiker-Bekel, D. Wibberg, T. Bekel, J. Blom, B. Linke, H. Neuweger, M. Stiens, F.J. Vorhölter, S. Weidner, A. Goesmann, The complete genome sequence of the dominant Sinorhizobium meliloti field isolate SM11 extends the S. meliloti pan-genome, J. Biotechnol. 155 (2011) 20–33.
[28] L. Li, J. Cai, S. Zhang, L. Wang, F. Hu, Construction of whole genome fosmid library in Africa wild rice (Oryza longistaminata), Rice Genomics Genet. 3 (2012) 66–71.
杂志排行
The Crop Journal的其它文章
- Single nucleotide polymorphisms linked to quantitative trait loci for grain quality traits in wheat
- Characterization of QTL for unique agronomic traits of new-plant-type rice varieties using introgression lines of IR64
- A multivariate partial least squares approach to joint association analysis for multiple correlated traits
- Comparative QTL analysis of maize seed artificial aging between an immortalized F2population and its corresponding RILs
- Analysis of simple sequence repeats in rice bean (Vigna umbellata) using an SSR-enriched library
- Intra-population genetic variance for grain iron and zinc contents and agronomic traits in pearl millet
