Characterization of QTL for unique agronomic traits of new-plant-type rice varieties using introgression lines of IR64
2016-04-05AnlizTgleDisukeFujitLeodegrioEronMryJenieTelencoYnoriKzuhiroSskiTsutomuIshimruYoshimichiFukutNouyKoyshi
Anliz G. Tgle, Disuke Fujit, Leodegrio A. Eron, Mry Jenie Telenco-Ynori, Kzuhiro Sski,,3, Tsutomu Ishimru,, Yoshimichi Fukut, Nouy Koyshi,,*
aInternational Rice Research Institute, DAPO Box 7777, Metro Manila, PhilippinesbJapan International Research Center for Agricultural Sciences, 1-1 Ohwashi, Tsukuba, Ibaraki 305-8686, Japan
Characterization of QTL for unique agronomic traits of new-plant-type rice varieties using introgression lines of IR64
Analiza G. Taglea,1, Daisuke Fujitaa,2, Leodegario A. Ebrona, Mary Jeanie Telebanco-Yanoriaa, Kazuhiro Sasakia,b,3, Tsutomu Ishimarua,b, Yoshimichi Fukutab, Nobuya Kobayashia,b,*,4
aInternational Rice Research Institute, DAPO Box 7777, Metro Manila, Philippines
bJapan International Research Center for Agricultural Sciences, 1-1 Ohwashi, Tsukuba, Ibaraki 305-8686, Japan
A R T I C L E I N F O
Article history:
Received 29 May 2015
Received in revised form
5 October 2015
Accepted 27 November 2015
Available online 3 December 2015
Keywords:
Agronomic traits
Quantitative trait loci
Near-isogenic lines
New-plant-type rice variety Pleiotropic effect
A B S T R A C T
To enhance the yield potential of an elite indica rice cultivar, an introgression (BC3-derived) line of IR64, YTH288, was developed using a new-plant-type cultivar, IR66215-44-2-3, as a donor parent. YTH288 has agronomically valuable characteristics such as large panicles, few unproductive tillers, and large leaves inherited from NPT. To identify the genetic basis of these traits, we used 167 F2plants derived from a cross between IR64 and YTH288 to conduct QTL analysis for five agronomic traits: days to heading (DTH), culm length (CL), flag leaf length (FLL), flag leaf width (FLW), and filled spikelet number per panicle (FSN). Six putative QTL were detected: four on chromosome 4 (for CL, FLL, FLW, and FSN) and two on chromosome 2 (for DTH and FLL). All QTL with the IR66215-44-2-3 allele, except that for FLL on chromosome 2, had positive effects on each trait. To confirm the effects of these putative QTL, we developed NILs with the IR64 genetic background by marker-assisted selection. We observed significant differences in several agronomic traits between IR64 and NILs that carried these QTL on chromosomes 2 and 4. Additionally, four IR64-NILs carrying chromosomal segments derived from different NPT varieties on the long arm of chromosome 4 exhibited similar pleiotropic effects for unique agronomic traits. These NILs can be used as research materials for studying each trait and as breeding materials for yield improvement of indica rice cultivars.
©2015 Crop Science Society of China and Institute of Crop Science, CAAS. Production and hosting by Elsevier B.V. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abbreviations: NPT, new plant type; QTL, quantitative trait loci (locus); GF, grain fertility; DTH, days to heading; CL, culm length; PL, panicle length; LL, leaf length; FLL, flag leaf length; LW, leaf width; FLW, flag leaf width; FSN, filled spikelet number per panicle; TSN, total spikelet number per panicle; PN, panicle number per plant; IRRI, International Rice Research Institute; NIL, near-isogenic line; IL, introgression line; SSR, simple sequence repeat; PCR, polymerase chain reaction
* Corresponding author at: Japan International Research Center for Agricultural Sciences, 1-1 Ohwashi, Tsukuba, Ibaraki 305-8686, Japan.
Tel.: +81 298388809.
E-mail address: nobuya@affrc.go.jp (N. Kobayashi).
Peer review under responsibility of Crop Science Society of China and Institute of Crop Science, CAAS.
1Present address: Graduate School of Agricultural Science, Kobe University, 1-1 Rokkodai, Nada, Kobe 657-8501, Japan.
2Present address: Faculty of Agriculture, Saga University, 1 Honjo-Machi, Saga, 840-8502, Japan.
3Present address: The University of Tokyo, Graduate School of Agricultural and Life Sciences, 1-1-1 Midoricho, Nishitokyo, Tokyo 188-0002, Japan.
4Present address: NARO Institute of Crop Science, 2-1-18, Kannondai, Tsukuba, Ibaraki 305-8518, Japan.
http://dx.doi.org/10.1016/j.cj.2015.10.001
2214-5141/©2015 Crop Science Society of China and Institute of Crop Science, CAAS. Production and hosting by Elsevier B.V. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
1. Introduction
According to recent estimates, rice (Oryza sativa L.) production must be increased by 40% before 2030 to satisfy the demands of the world's growing population [1]. To increase grain production, enhancement of rice yield potential is one of the most important objectives for rice breeding. Since the 1960s, the IRRI has released many high-yielding varieties that have been distributed worldwide [2]. In the late 1980s, a breeding program to develop NPT rice was launched at IRRI to further increase the yield potential of the high-yielding varieties through improvement of plant type. NPT lines had several unique agronomic traits such as large panicles, few unproductive tillers, and large leaves inherited from tropical japonica varieties [3,4]. However, despite the improved plant architecture, the NPT lines yielded less than expected, mainly because of low GF [2,4].
Identification and analyses of QTL have improved our understanding of the basis of natural allelic diversity, which is crucialtosupportingcropimprovement.Inthepasttwodecades, many QTLthatcontroltraitsofagronomicimportancehavebeen detected using molecular markers (http://www.gramene.org/). Some of these QTL have been identified by map-based cloning. For example, QTL for TSN have been identified using various segregating populations, including F2populations, RILs, and double-haploid lines. Several QTL for grain and spikelet number (QSpp8,qSSP1,qSSP2,qSSP3,qSSP7,andgpa7)havebeenidentified andmapped[5–8],andothers(Gn1a,Ghd7,and WFP)havealready been cloned [9–11]. However, many of these QTL cannot be used directlyinrice-breedingprogramsbecausetheirexpressionsvary considerably in different genetic backgroundsandenvironments [12]. Recently, Zhang et al. [13] demonstrated the successful genetic dissection of yield-related traits using ILs with the genetic background of elite rice. ILs carrying small introgressed segments from the donor parents are useful materials for detailed characterization of agronomic traits associated with these specific introgressions.
An indica rice variety, IR64 has been accepted as a high-quality and high-yielding variety in many countries [14]. To further improve yield potential and to analyze the genetic basis of yield-related traits, a total of 334 ILs derived from crosses between IR64 and 10 donor varieties (mostly NPT lines) have been developed in our previous study under the IRRI–Japan Collaborative Research Project [15]. Among the 334 ILs, several lines showed agronomically unique characteristics inherited from the NPT lines, such as later heading, longer culm, wider and longer flag leaf, and higher spikelet number than IR64 [16]. By use of the ILs, chromosomal regions associated with agronomic traits such as days to DTH and TSN have been identified. Detailed locations of QTL controlling TSN were identified on chromosome 4 by use of F2plants derived from crosses between ILs and IR64 [17]. These studies showed that the ILs are useful materials for the genetic analysis of unique agronomic characteristics in high-yielding varieties. Objective of this study was to detect QTL for unique agronomic traits inherited from NPT varieties and valuable in breeding programs. Using marker-assisted selection, NILs carrying these QTL with elite genetic backgrounds were developed and characterized to confirm the effects of the newly detected QTL.
2. Materials and methods
2.1. Plant materials

Fig. 1–Breeding scheme of development of an introgression line with the genetic background of IR64.
In our previous study, F1plants were generated by crosses between IR64 and 10 donor varieties to develop ILs. These F1plants were recurrently backcrossed with IR64 three times (Fig. 1). A single BC3F1plant was self-pollinated to generate BC3F2plants. Similar practice was repeated until the BC3F7generation. Agronomic traits in the 334 ILs were almost fixed genetically in BC3F8and a genome-wide DNA marker survey revealed that most of the chromosomal regions in the ILs were replaced with the genotype of the recurrent parent, IR64 [15]. In the present study, we selectedan IL, IR84639-7-28-5-5-2-2-3-2-2-14-B (renamed YTH288), which showed unique agronomic traits such as large panicles and broadleavesinheritedfroman NPTvarietyandhadhigheryield potential than IR64 (Table 1). A NPT variety, IR66215-44-2-3 (renamed YP8), which is a donor parent of YTH288 was derived from a cross between Gaok (a tropical japonica variety) and IR66072-11-8-5-4 (IRIS; http://www.iris.irri.org/). One hundred sixty-seven F2plants were generated from a cross between IR64 and YTH288 and used for QTL mapping for agronomic traits.

Table 1–Grain yield of IR64 and YTH288 in the experimental field at IRRI (Los Baños, Philippines). Line Year Grain yield (g m−2) YTH288 2009 654.3 2011 558.5 IR64 2009 390.0 2011 422.5 Single plants were planted at 20 cm between rows and 20 cm between hills and grown during the dry season (January–May). Grain was harvested from a 1×5 m plot with 2 replications in 2009 and from a 1×1 m plot with 3 replications in 2011.
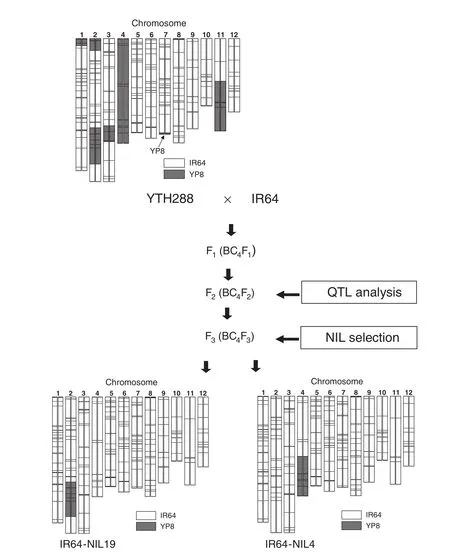
Fig. 2–Graphical genotypes of YTH288 and of two NILs containing QTL alleles for agronomically important traits: IR64-NIL19 and IR64-NIL4. The 12 pairs of vertical bars represent the chromosomes, and are numbered at the top.
From the F2mapping population, we selected two F2plants that carried the newly detected QTL on chromosome 2 or 4and as few other introgressed segments as possible. F3lines generated by self-pollination of the selected plants were considered to be NILs. The two NILs were designated IR64-NIL4 (QTL on chromosome 4) and IR64-NIL19 (QTL on chromosome 2) (Fig. 2), and were used for further characterization in the experimental field at IRRI (Los Baños, Laguna, The Philippines; 121°E, 14°N).
In our previous study [17], four NILs of IR64 for QTL for high TSN on chromosome 4 derived from different parents: IR64-NIL2 from IR65564-2-2-3 (renamed YP4), IR64-NIL3 from IR69093-41-2-3-2 (YP5), IR64-NIL5 from IR68522-10-2-2 (YP9), and IR64-NIL6 from IR66750-6-2-1 (YP11) had been developed (Fig. 3). These NILs were used to confirm the effect of QTL on chromosome 4.
2.2. Phenotypic evaluation
The F2plants, IR64, YTH288, and YP8 were grown in an experimental field at IRRI during the dry season (January to May). The approximate day length was 12.5 h. At 21 days after sowing, individual plants were transplanted into the field at a spacing of 20 cm between hills and 30 cm between rows. The F2plants were individually evaluated for DTH, CL, FLL, FLW, and FSN. DTH was evaluated as the number of days from sowing to flowering. CL was measured from the soil surface to the neck of the tiller that was highest above the ground. FLL and FLW were measured on the flag leaf of the tallest tiller. FSN was evaluated as the number of filled spikelets in a panicle of the tallest tiller. IR64, YTH288, and YP8 were grown together with the F2population and likewise evaluated using 10 plants per variety. TSN was also evaluated for IR64, YTH288, and YP8 as the sum of the number of filled and unfilled spikelets in the tallest tiller.
To characterize two developed NILs, IR64-NIL4 and IR64-NIL19, CL, FLL, FLW, FSN, and TSN were measured during the 2012 dry season following the same method as used for the F2plants. IR64-NIL19 was evaluated for DTH in the 2011 dry season.
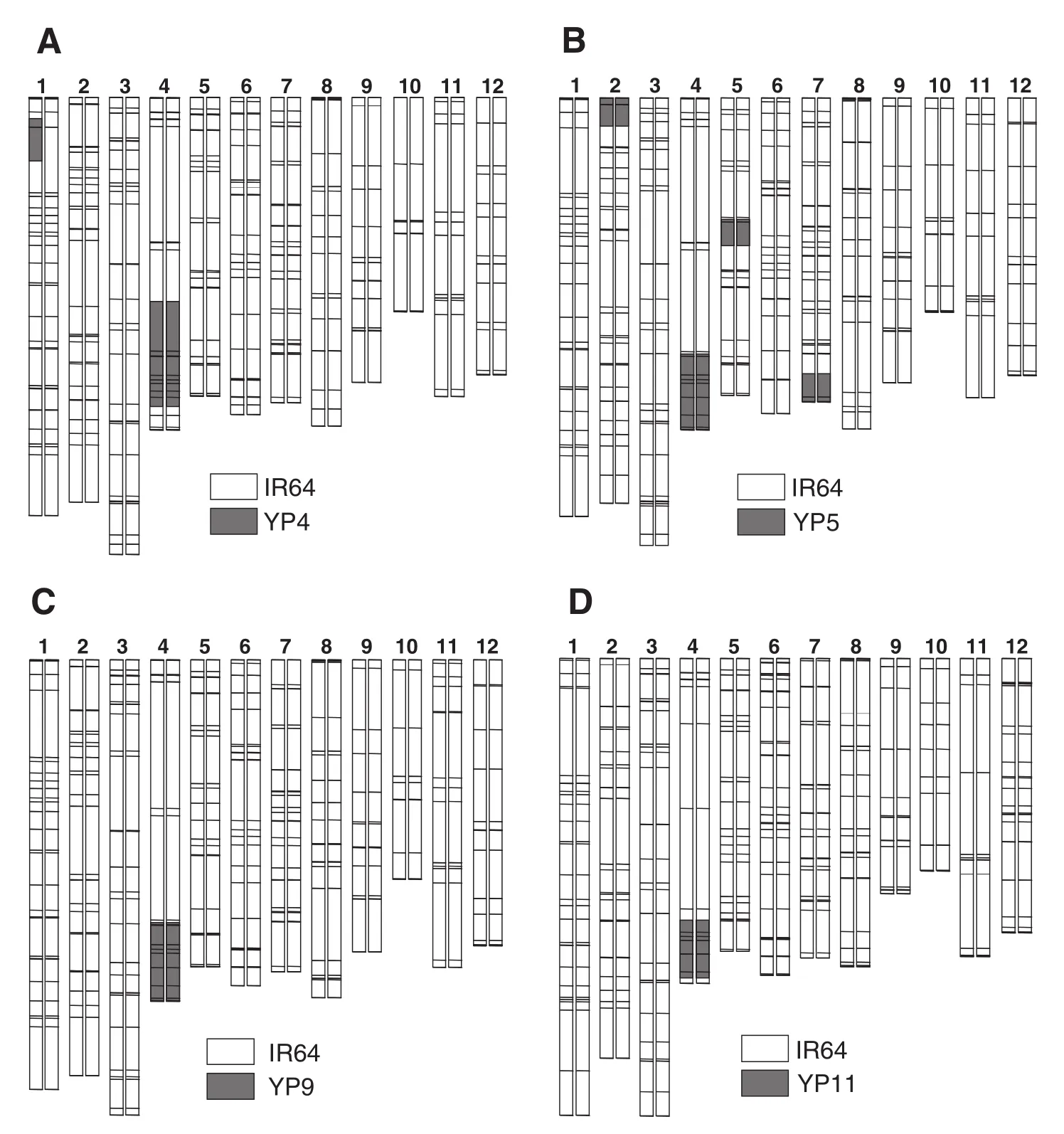
Fig. 3–Graphical genotypes of four NILs with QTL on chromosome 4 derived from NPT varieties. (A) IR64-NIL2; (B) IR64-NIL3; (C) IR64-NIL5; (D) IR64-NIL6. The 12 pairs of vertical bars represent chromosomes, numbered at the top. Horizontal lines across the bars show positions of SSR marker loci. White indicates chromosomal segments from IR64; gray indicates chromosomal segments from the donor parents.
To confirm the effects of QTL on the long arm of chromosome 4, five NILs and IR64 were evaluated for CL, PL,FLL, FLW, and PN in the 2009 dry season. In addition to these traits, LL and LW of the first leaf below the flag leaves were measured for these lines.
2.3. Genotyping using SSR markers
Genomic DNA of YTH288, IR64, YP8, and the F2individuals was extractedfromleaftissueusingthe CTABmethod[18].Atotal of 457 SSRmarkerswith known positionsdistributedacrossthe 12 rice chromosomes were used to survey the polymorphism between IR64 and YP8 [19]. We used 224 of these markers that showed polymorphism between the parents for our genotyping analysis of YTH288. Introgressed segments in YTH288 that derived from YP8 were detected in chromosomes 1, 2, 3, 4, 7, and 11 (see Fig. 2). We used 22 SSR markers that showed the YP8 donor genotype in YTH288 to analyze the F2individuals: RM3252 and RM3148 on chromosome 1; RM5631, RM5472, and RM240 on chromosome 2; RM2334, RM6329, and RM168 on chromosome 3; RM518, RM1155, RM3843, RM3534, RM17470, RM5503, RM6480, RM6089, RM6365, RM17483, RM17486, and RM6909 on chromosome 4; and RM5349 and RM1341 on chromosome 11. Primer information such as forward and reverse sequences was obtained from http://www.gramene.org/.
We used the PCR to determine SSR marker genotypes. The reactions were performed in a 10-μL PCR mixture containing 1.4 μL of sterile H2O, 1 μL each of 10×PCR buffer containing 20 mmol L−1Mg2+(SBS Genetech, Beijing, China), PCR dye (SBS Genetech), dNTPs (1 mmol L−1), 0.1 μL Taq polymerase at 5 U μL−1(SBS Genetech), 0.25 μL of each forward and reverse SSR primer (5 μmol L−1), and 5 μL of DNA (5 ng μL−1) template. PCR amplification was performed in a DNA Engine Dyad thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA) with the following profile: an initial denaturation step at 95°C for 5 min; 35 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 45 s; and a final extension step at 72°C for 5 min and 30 s. PCR products were separated in 4% agarose gels for 1 h at 250 V. Gels were stained with ethidium bromide and visualized under UV light.
2.4. QTL analysis
To construct a linkage map around the introgressed segments, we determined the genetic distances between SSR markers for the genotypes in the F2population. The linkage map was constructed with genetic distances (cM) calculated using the Kosambi map function [20]. The chromosomal locations associated with the QTL for DTH, CL, FLL, FLW, and FSN were determined on the linkage map for the F2population. Associations between phenotypes and marker genotypes were investigated by simple interval mapping using Windows QTL Cartographer v. 2.5 software [21]. The proportion of the observed phenotypic variation attributable to a given chromosomal region was estimated by using the coefficient of determination (R2). Critical threshold values for the LOD scores for QTL detection were calculated with 1000 permutation tests (with significance at P<0.01).
3. Results
3.1. QTL analysis for five agronomic traits Among 334 ILs that had been developed by backcross breeding, YTH288 had several unique agronomic characteristics such as large panicles and broad leaves that were inherited from the NPT rice variety, while the other traits were similar to those of IR64. Compared with IR64, YTH288 had longer DTH and CL, wider FLW, and more FSN (Table 2). YTH288 showed higher FSN and GF than YP8, while TSN of these lines was not significantly different. Continuous distributions were observed for four agronomic traits; CL, FLL, FLW, and FSN in the F2population derived from the cross between IR64 and YTH288, whereas DTH showed a bimodal distribution (Fig. 4).
QTLanalysisusing SSRmarkersontheintrogressedsegments detected six putative QTL for agronomic traits in YTH288 (Fig. 5, Table 3). Four QTL (for CL, FLL, FLW, and FSN) were detected in the same region on the long arm of chromosome 4, between SSR markers RM6480 and RM3843. These QTL are designated by the letter“q”followed by the trait and then the chromosome number: qCL4.1, qFLL4.1, qFLW4.1, and qFSN4.1. The peak LODs for each QTL fell between RM17470 and RM3534 for qCL4.1, between RM6909 and RM3843 for qFLL4.1, between RM3534 and RM17483 for qFLW4.1, and between RM6480 and RM5503 for qFSN4.1. The other two QTL, designated as qDTH2.1 and qFLL2.1, were both located between RM5472 and RM240 on the long arm of chromosome 2. Each of the QTL explained 9% to 28% of the phenotypic variance. All QTL with the YP8 allele, except for qFLL2.1, increased the phenotypic value of the trait they controlled.

Table 2–Characterization of agronomic traits of IR64 and YTH288 in the dry season in 2009. Line Agronomic trait (mean±SD)aDTHb CL (cm) FLL (cm) FLW (cm) FSN TSN GF (%) IR64 85.0±1.0 70.1±3.2 A 38.2±4.3 A 1.3±0.1 A 101.3±14.0 A 138.0±21.8 A 74.0±6.3 B YP8 – 83.6±4.2 B 41.5±2.4 A 1.8±0.1 C 112.7±20.4 A 197.7±34.1 B 57.7±11.1 A YTH288 93.1±2.8 81.4±2.9 B 38.1±4.0 A 1.5±0.1 B 164.1±23.8 B 201.6±22.8 B 81.7±6.7 B Different letters indicate significant differences (P<0.01, Tukey–Kramer test).aDTH, days to heading; CL, culm length (cm); FLL, flag leaf length (cm); FLW, flag leaf width (cm); FSN, filled spikelet number per panicle; TSN, total spikelet number per panicle; and GF, grain fertility.bCalculated from data obtained in three years.
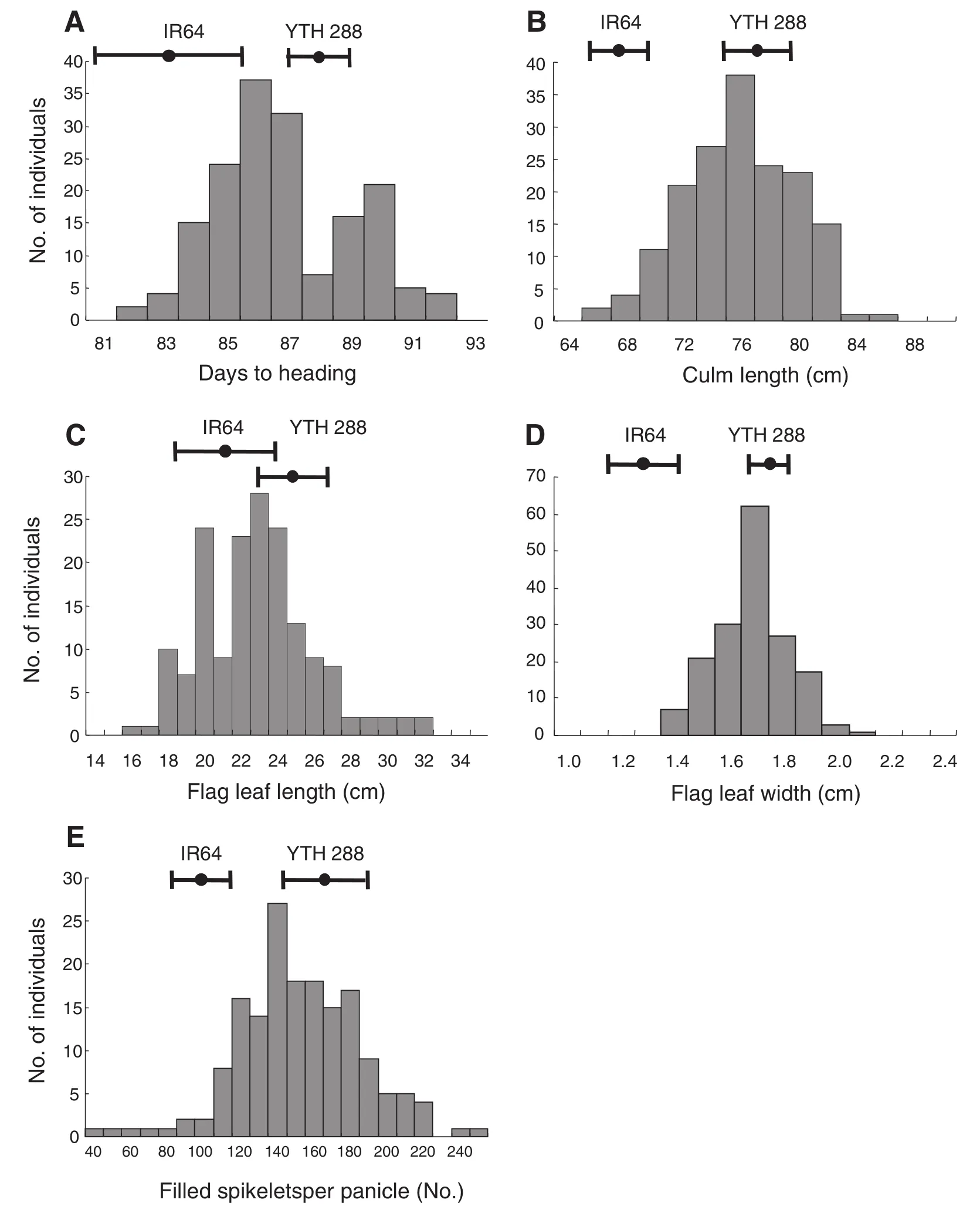
Fig. 4–Frequency distributions of five agronomic traits in the F2population derived from a cross between IR64 and YTH288 (N = 167). (A) days to heading; (B) culm length; (C) flag leaf length; (D) flag leaf width; and (E) filled spikelet number per panicle. The dots and error bars at the top of each graph represent the mean and standard deviation of the agronomic trait of each parent.
3.2. Characterization of NILs for the detected QTL
To confirm the effects of QTL for these agronomic traits, we developed two NILs with the IR64 genetic background that carried the detected QTL on either chromosome 2 or chromosome 4 (Fig. 2). These two NILs, designated as IR64-NIL4 and IR64-NIL19, were evaluated for DTH, CL, FLL, FLW, FSN, and TSN. IR64-NIL19, which carries qDTH2.1 and qFLL2.1 on chromosome 2, flowered two days later than IR64 but showed no significant difference for other traits (Table 4). IR64-NIL4, which carries qCL4.1, qFLL4.1, qFLW4.1, and qFSN4.1 on chromosome 4, showed significantly higher CL, FLL, FLW, and TSN than IR64.
3.3. Characterization of NILs carrying QTL on chromosome 4 derived from different parents
To evaluate effect of the QTL on agronomic traits, five NILs carrying introgressed segments on chromosome 4 derived from different donor parents were compared with IR64. TheseNILs showed significant differences from IR64 for several agronomic traits. All five NILs showed significantly higher CL, FLL, LW, and FLW than IR64 (Table 5). These NILs also showed greater PL than IR64, whereas the difference between IR64 and IR64-NIL2 was not significant. LL of these NILs was also greater than that of IR64, and significantly so in IR64-NIL2, IR64-NIL5, and IR64-NIL6. PN values of IR64-NIL2, IR64-NIL3 and IR64-NIL5 were significantly lower than that of IR64, whereas those of IR64-NIL5 and IR64-NIL6 were not significantly different from that of IR64. These five NILs showed significantly greater FSN than IR64 in our previous study (see Table 3 in [17]).
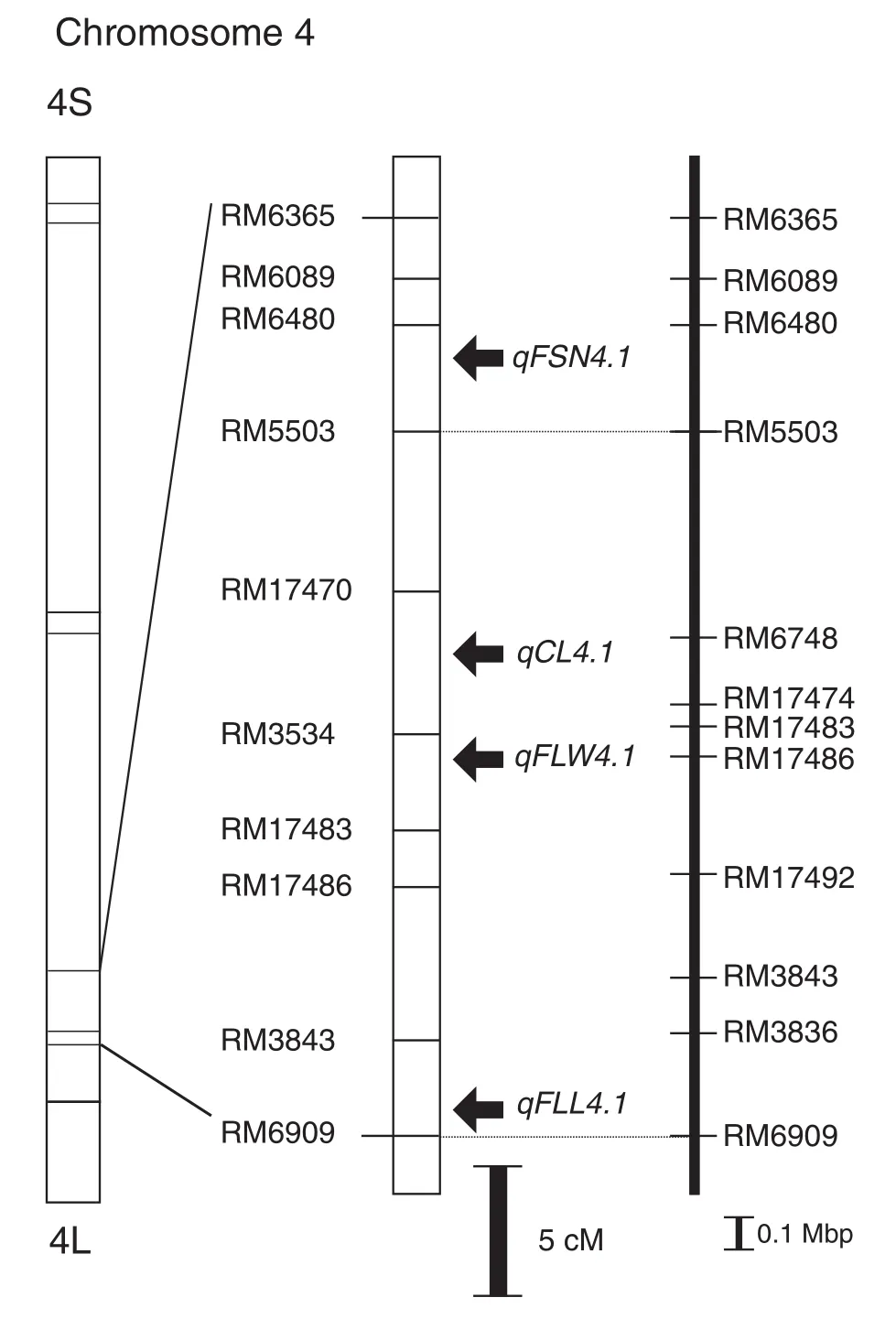
Fig. 5–Mapping of the QTL for agronomic traits onchromosome 4. Positions of the simple sequence repeatmarkers on chromosome 4 were detected. The arrowsindicate the location of four putative QTL: FSN (filled spikelet number per panicle), CL (culm length), FLW (flag leaf width), and FLL (flag leaf length).
4. Discussion
4.1. Newly detected QTL for agronomic traits
QTL for agronomic traits in rice have been identified in all chromosomal regions. Yonemaru et al. [22] reported co-localization of QTL and distribution of QTL clusters associated with multiple morphological traits on chromosomes 1, 3, 4, 6, and 9. Among the QTL clusters, many QTL for morphological traits such as leaf size, spikelet number, PN, and root volume were located on the long arm of chromosome 4 (http://qtaro.abr.affrc.go.jp/). In the present study, four QTL for CL, FLL, FLW, and FSN were colocalized on the long arm of chromosome 4 (Fig. 5). In previous studies, QTL for various agronomic traits were identified in the same region of chromosome 4: GW and plant height [23]; FLL and FLW [24]; FLL, FLW, FLA, PL, PN, and grain yield [25,26]; FLW and PL [27]; and FLW and root volume per tiller [28]. This colocalization of QTL for varioustraitsmayrepresentageneclusterorapleiotropiceffect of a gene underlying the QTL. Precise analysis will be needed to identify the detailed locations of the QTL derived from NPT varieties on the long arm of chromosome 4.

Table 3–QTL associated with five agronomic traits in the F2population derived from a cross between IR64 and YTH288 using simple interval mapping. Traita QTL Marker interval Chromosome LOD score R2b Additive effectc Dominant effectcDTH qDTH2.1 RM5472–RM240 2 4.6 0.12 −1.1 0.2 CL qCL4.1 RM17470–RM3534 4 4.6 0.13 −0.8 2.3 FLL qFLL2.1 RM5472–RM240 2 3.7 0.10 1.3 0.5 FLL qFLL4.1 RM6909–RM3843 4 6.0 0.15 −1.6 −0.7 FLW qFLW4.1 RM3534–RM17483 4 12.2 0.28 −0.1 0.0 FSN qFSN4.1 RM6480–RM5503 4 3.1 0.09 −14.6 −0.1aDTH, days to heading; CL, culm length; FLL, flag leaf length; FLW, flag leaf width; and FSN, filled spikelet number per panicle.bProportion of the phenotypic variation explained by each QTL.cThe values indicate the relative effect of the IR64 alleles.
We detected QTL for unique agronomic traits such as large panicles and broad leaves that may contribute to higher yield of YTH288 than IR64. Among them were QTL associated with leaf size, qFLL4.1 and qFLW4.1, on the long arm of chromosome 4 where Yue et al. [26] detected QFll4, QFlw4, and QFla4. We identified qFLL4.1 between RM3843 (32.2 Mb) and RM6909 (32.6 Mb) and qFLW4.1 between RM3534 (31.7 Mb) and RM17483 (31.7 Mb). In contrast, QFll4, QFlw4, and QFla4 were detected in the interval between RM255 (31.5 Mb) and RM349 (33.2 Mb). Besides leaf size, we detected a QTL associated with panicle structure, qFSN4.1 on the long arm of chromosome 4 in the interval between RM6480 (30.4 Mb) and RM5503 (30.9 Mb), whereas qNOS4-3 was identified around RG214 (32.3 Mb) and qSN-4a was detected in the interval between RM317 (29.7 Mb) and RM255 (31.5 Mb) [25,29]. Furthermore, aQTL for FLW, spikelet number and root volume has been mapped within 38 Kb on the long arm of chromosome 4 [28]. Recently, a gene responsible for increase in TSN on this chromosomal region was identified by Fujita et al. [30]. To determine whether the detected QTL are the same as those previously reported, more detailed analysis is needed.
We also detected QTL for agronomic traits qDTH2.1 and qFLL2.1 on the long arm of chromosome 2 where QTL for similar traits were previously identified. The location of qDTH2.1 is similar to that where Hd7 was reported [31]. C560, the nearest RFLP marker for Hd7, was identified on the long arm of chromosome 2 and Hd7 was located between R3393 (29.2 Mb) and R2511 (35.9 Mb), while qDTH2.1 was detected around RM5472 (31.5 Mb) to RM240 (32.4 Mb). The location of Hd7 overlapped with that of qDTH2.1.

Table 4–Agronomic characteristics of IR64 and developed NILs. Line Introgressed segment on Agronomic traita(mean±SD) DTH CL (cm) FLL (cm) FLW (cm) FSN TSN IR64 – 86±1.8 65.2±1.9 26.6±3.9 1.3±0.2 96.0±18.8 105.7±19.6 IR64-NIL19 Chr. 2 88±1.6* 68.8±3.8 23.3±2.4 1.3±0.1 104.3±13.3 118.3±11.3 IR64-NIL4 Chr. 4 NA 75.6±5.2* 32.4±4.1* 1.9±0.1* 116.6±30.9 197.8±27.5*aDTH, days to heading; CL, culm length; FLL, flag leaf length; FLW, flag leaf width; FSN, filled spikelet number per panicle; and TSN, total spikelet number per panicle. The agronomic traits were calculated from data obtained in the 2012 dry season except for DTH, obtained in the 2011 dry season. Values represent means and standard deviations with significance levels (by t-test). *P<0.05.
4.2. Effects of detected QTL on chromosomes 4 from NPT rice varieties
In 1990s, NPT rice varieties had been developed using tropical japonica varieties to enhance the yield potential of rice. However, the grain yield of NPT variety was not higher than expected, mainly because of low GF [4]. To eliminate genetic factors associated with low GF, the NPT variety YP8 was recurrently backcrossed to IR64 and ILs with unique agronomic traits inherited from NPT varieties were developed [15]. One of the ILs, YTH288, showed unique agronomic traits such as heavy panicles and broad leaves inherited from a NPT variety, YP8, but GF of YTH288 was higher than that of YP8. In the F2population of a cross between YTH288 and IR64, QTL for several agronomic traits were detected on the same location on the long arm of chromosome 4. Effects on multiple traits were commonly observed among NILs for chromosome 4 derived from different NPT parents: YP4, YP5, YP8, YP9, and YP11, in addition to increased TSN as observed previously by Fujita et al. [17]. These results suggested that a QTL cluster or pleiotropic QTL is a major genetic factor influencing the unique agronomic traits of NPT varieties.
IR64-NIL4, which carries an introgressed segment containing qCL4.1, qFLL4.1, qFLW4.1, and qFSN4.1 on the long arm of chromosome 4, showed greater CL, FLL, FLW, and FSN than IR64 (Table 3). Although the difference in FSN between IR64 and IR64-NIL4 was not significant, the TSN of the NIL was significantly greater than that of IR64. These NILs for qTSN4 are promising lines for enhancing yield potential with fewer undesirable traits related to grain yield in rice.
5. Conclusion
Using IL of IR64, we successfully detected QTL for unique agronomic traits in a NPT variety. Effects of the QTL were confirmed by development of two NILs that each carried an introgressed segment from YP8 on chromosome 2 or 4 in the IR64 genetic background. By using developed NILs, only valuable agronomic traits can be isolated from NPT varieties for rice breeding programs. These NILs could thus be used as breeding materials to improve yield potential in rice and as research materials for studying agronomic traits.
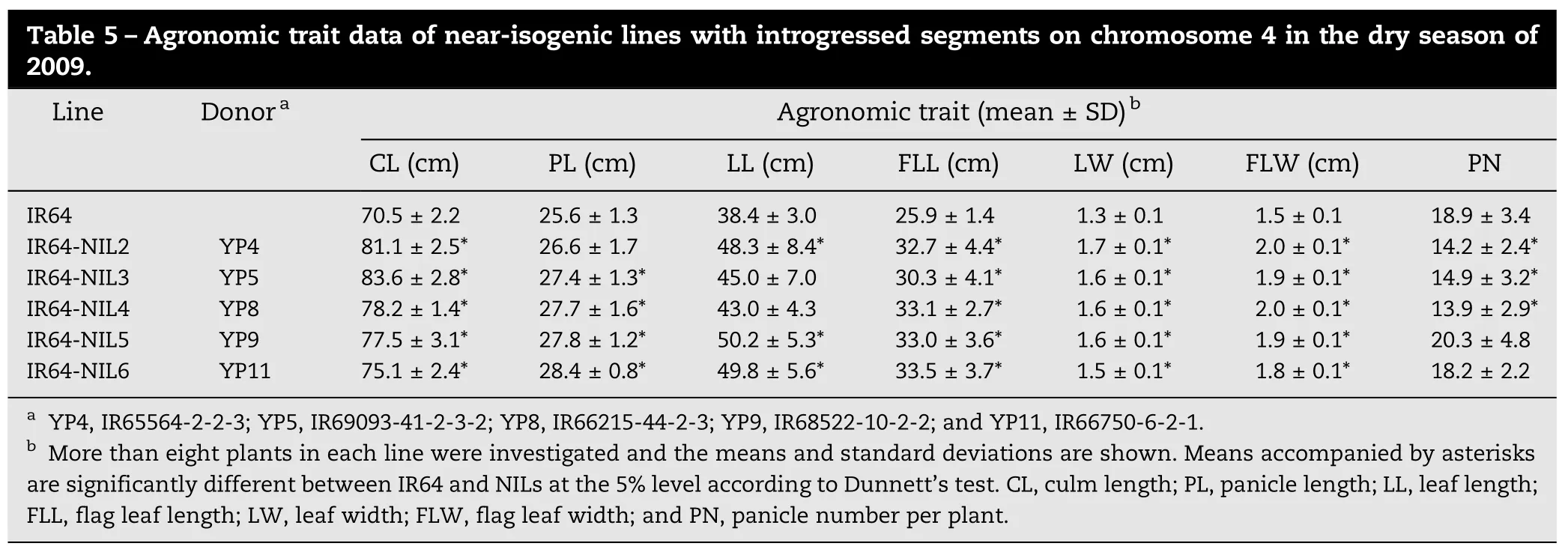
Table 5–Agronomic trait data of near-isogenic lines with introgressed segments on chromosome 4 in the dry season of 2009. Line DonoraAgronomic trait (mean±SD)bCL (cm) PL (cm) LL (cm) FLL (cm) LW (cm) FLW (cm) PN IR64 70.5±2.2 25.6±1.3 38.4±3.0 25.9±1.4 1.3±0.1 1.5±0.1 18.9±3.4 IR64-NIL2 YP4 81.1±2.5* 26.6±1.7 48.3±8.4* 32.7±4.4* 1.7±0.1* 2.0±0.1* 14.2±2.4* IR64-NIL3 YP5 83.6±2.8* 27.4±1.3* 45.0±7.0 30.3±4.1* 1.6±0.1* 1.9±0.1* 14.9±3.2* IR64-NIL4 YP8 78.2±1.4* 27.7±1.6* 43.0±4.3 33.1±2.7* 1.6±0.1* 2.0±0.1* 13.9±2.9* IR64-NIL5 YP9 77.5±3.1* 27.8±1.2* 50.2±5.3* 33.0±3.6* 1.6±0.1* 1.9±0.1* 20.3±4.8 IR64-NIL6 YP11 75.1±2.4* 28.4±0.8* 49.8±5.6* 33.5±3.7* 1.5±0.1* 1.8±0.1* 18.2±2.2aYP4, IR65564-2-2-3; YP5, IR69093-41-2-3-2; YP8, IR66215-44-2-3; YP9, IR68522-10-2-2; and YP11, IR66750-6-2-1.bMore than eight plants in each line were investigated and the means and standard deviations are shown. Means accompanied by asterisks are significantly different between IR64 and NILs at the 5% level according to Dunnett's test. CL, culm length; PL, panicle length; LL, leaf length; FLL, flag leaf length; LW, leaf width; FLW, flag leaf width; and PN, panicle number per plant.
Acknowledgments
This paper reports the results obtained from phases III, IV, V, and VI of the IRRI–Japan Collaborative Research Project, which was supported by the Ministry of Foreign Affairs and the Ministry of Agriculture, Forestry and Fisheries of Japan.
R E F E R E N C E S
[1] G.S. Khush, What will it take to feed 5.0 billion rice consumers in 2030, Plant Mol. Biol. 59 (2005) 1–6.
[2] S. Peng, G.S. Khush, Four decades of breeding for varietal improvement of irrigated lowland rice in the International Rice Research Institute, Plant Prod. Sci. 6 (2003) 157–164.
[3] G.S. Khush, Breaking the yield frontier of rice, GeoJournal 35 (1995) 329–332.
[4] S. Peng, K.G. Cassman, S.S. Virmani, J. Sheehy, G.S. Khush, Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential, Crop Sci. 39 (1999) 1552–1559.
[5] F. Tian, Z. Zhu, B. Zhang, L. Tan, T. Fu, X. Wang, C.Q. Sun, Fine mapping of a quantitative trait locus for grain number per panicle from wild rice (Oryza rufipogon Griff.), Theor. Appl. Genet. 113 (2006) 619–629.
[6] Y. Zhang, L. Luo, C. Xu, Q. Zhang, Y. Xing, Quantitative trait loci for panicle size, heading date and plant height co-segregating in trait-performance derived near-isogenic lines of rice (Oryza sativa), Theor. Appl. Genet. 113 (2006) 361–368.
[7] Y. Zhang, L. Luo, T. Liu, C. Xu, Y. Xing, Four rice QTL controlling number of spikelets per panicle expressed the characteristics of single Mendelian gene in near isogenic backgrounds, Theor. Appl. Genet. 118 (2009) 1035–1044.
[8] Y.Z. Xing, W.J. Tang, W.Y. Xue, C.G. Xu, Q. Zhang, Fine mapping of a major quantitative trait loci, qSSP7, controlling the number of spikelets per panicle as a single Mendelian factor in rice, Theor. Appl. Genet. 116 (2008) 789–796.
[9] M. Ashikari, H. Sakakibara, S. Lin, T. Yamamoto, T. Takashi, A. Nishimura, E.R. Angeles, Q. Qian, H. Kitano, M. Matsuoka, Cytokinin oxidase regulates rice grain production, Science 309 (2005) 741–745.
[10] W. Xue, Y. Xing, X. Weng, Y. Zhao, W. Tang, L. Wang, H. Zhou, S. Yu, C. Xu, X. Li, Q. Zhang, Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice, Nat. Genet. 40 (2008) 761–767.
[11] K. Miura, M. Ikeda, A. Matsubara, X.J. Song, M. Ito, K. Asano, M. Matsuoka, H. Kitano, M. Ashikari, OsSPL14 promotes panicle branching and higher grain productivity in rice, Nat. Genet. 42 (2010) 545–549.
[12] Z.K. Li, S.B. Yu, H.R. Lafitte, N. Huang, B. Courtois, S. Hittalmani, C.H.M. Vijayakumar, G.F. Liu, G.C. Wang, H.E. Shashidhar, J.Y. Zhuang, K.L. Zheng, V.P. Singh, J.S. Sidhu, S. Srivantaneeyakul, G.S. Khush, QTL×environment interactions in rice. I. Heading date and plant height, Theor. Appl. Genet. 108 (2003) 141–153.
[13] H.H. Zhang, Y. Qian Wang, J. Xia, Z. Li, Y. Shi, L. Zhu, J. Ali, Y. Gao, Z. Li, Simultaneous improvement and genetic dissection of grain yield and its related traits in a backbone parent of hybrid rice (Oryza sativa L.) using selective introgression, Mol. Breed. 31 (2013) 181–194.
[14] G.S. Khush, Rice breeding: past, present and future, J. Genet. 66 (1987) 195–216.
[15] D.Fujita,R.E.Santos,L.A.Ebron,M.J.Telebanco-Yanoria,H.Kato, S. Kobayashi, Y. Uga, E. Araki, T. Takai, H. Tsunematsu, T. Imbe, G.S. Khush, D.S. Brar, Y. Fukuta, N. Kobayashi, Development of introgressionlinesofan Indica-typericevariety,IR64,forunique agronomic traits and detection of the responsible chromosomal regions, Field Crop Res. 114 (2009) 244–254.
[16] D. Fujita, R.E. Santos, L.A. Ebron, M.J. Telebanco-Yanoria, H. Kato, S. Kobayashi, Y. Uga, E. Araki, T. Takai, H. Tsunematsu, T. Imbe, G.S. Khush, D.S. Brar, Y. Fukuta, N. Kobayashi, Characterization of introgression lines for yield-related traits with Indica-type rice variety IR64 genetic background, Jpn. Agric. Res. Q. 44 (2010) 277–290.
[17] D. Fujita, A.G. Tagle, L.A. Ebron, Y. Fukuta, N. Kobayashi, Characterization of near-isogenic lines carrying QTL for high spikelet number with the genetic background of an indica rice variety IR64 (Oryza sativa L.), Breed. Sci. 62 (2012) 18–26.
[18] M.G. Murray, W.F. Thompson, Rapid isolation of high-molecular-weight plant DNA, Nucleic Acids Res. 8 (1980) 4321–4325.
[19] S.R. McCouch, L. Teytelman, Y. Xu, K.B. Lobos, K. Clare, M. Walton, B. Fu, R. Maghirang, Z. Li, Y. Xing, Q. Zhang, I. Kono, M. Yano, R. Fjellstrom, G. DeClerck, D. Schneider, S. Cartinhour, D. Ware, L. Stein, Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.), DNA Res. 9 (2002) 199–207.
[20] D.D. Kosambi, The estimation of map distance from recombination values, Ann. Hum. Genet. 12 (1943) 172–175.
[21] S. Wang, C.J. Basten, Z.B. Zeng, Windows QTL Cartographer 2.5, Department of Statistics, North Carolina State University, Raleigh, NC, 2010.
[22] J. Yonemaru, T. Yamamoto, S. Fukuoka, Y. Uga, K. Hori, M. Yano, Q-TARO: QTL annotation rice online database, Rice 3 (2010) 194–203.
[23] T. Yagi, K. Nagata, Y. Fukuta, K. Tamura, I. Ashikawa, T. Terao, QTL mapping of spikelet number in rice (Oryza sativa L.), Breed. Sci. 51 (2001) 53–56.
[24] H.W. Mei, Z.K. Li, Q.Y. Shu, L.B. Guo, Y.P. Wang, X.Q. Yu, C.S. Ying, L.J. Luo, Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two backcross populations, Theor. Appl. Genet. 110 (2005) 649–659.
[25] G.H. Zou, H.W. Mei, H.Y. Liu, G.L. Liu, S.P. Hu, X.Q. Yu, M.S. Li, J.H. Wu, L.J. Luo, Grain yield responses to moisture regimes in a rice population: association among traits and genetic markers, Theor. Appl. Genet. 112 (2005) 106–113.
[26] B. Yue, W.Y. Xue, L.J. Lou, Y.Z. Xing, QTL analysis for flag leaf characteristics and their relationships with yield and yield traits in rice, Acta Genet. Sin. 33 (2006) 824–832.
[27] S. Kobayashi, E. Araki, M. Osaki, G.S. Khush, Y. Fukuta, Localization, validation and characterization of plant-type QTLs on chromosomes 4 and 6 in rice (Oryza sativa L.), Field Crop Res. 96 (2006) 106–112.
[28] X.P. Ding, Z.K. Li, L.Z. Xiong, Evaluation of near-isogenic lines for drought resistance QTL and fine mapping of a locus affecting flag leaf width, spikelet number, and root volume in rice, Theor. Appl. Genet. 123 (2011) 815–826.
[29] S. Hittalmani, N. Huang, B. Courtois, R. Venuprasad, H.E. Shashidhar, J.Y. Zhuang, K.L. Zheng, G.F. Liu, G.C. Wang, J.S. Sidhu, S. Srivantaneeyakul, V.P. Singh, P.G. Bagali, H.C. Prasanna, G. McLaren, G.S. Khush, Identification of QTL for growth- and grain yield-related traits in rice across nine locations of Asia, Theor. Appl. Genet. 107 (2003) 679–690.
[30] D. Fujita, K.R. Trijatmiko, A.G. Tagle, M.V. Sapasap, Y. Koide, K. Sasaki, N. Tsakirpaloglou, R.B. Gannaban, T. Nishimura, S. Yanagihara, Y. Fukuta, T. Koshiba, I.H. Slamet-Loedin, T. Ishimaru, N. Kobayashi, NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars, Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 20431–20436.
[31] T. Yamamoto, H. Lin, T. Sasaki, M. Yano, Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny, Genetics 154 (2000) 885–891.
杂志排行
The Crop Journal的其它文章
- Single nucleotide polymorphisms linked to quantitative trait loci for grain quality traits in wheat
- A multivariate partial least squares approach to joint association analysis for multiple correlated traits
- Comparative QTL analysis of maize seed artificial aging between an immortalized F2population and its corresponding RILs
- Analysis of simple sequence repeats in rice bean (Vigna umbellata) using an SSR-enriched library
- Intra-population genetic variance for grain iron and zinc contents and agronomic traits in pearl millet
- Fosmid library construction and screening for the maize mutant gene Vestigial glume 1
