Role of IL-17 in nucleus pulposus cell proliferation and metabolism cultured in vitro
2015-12-08XiLinQiLinJunJianYe
Xi Lin, Qi Lin, Jun-Jian Ye
1Emergency Department, Affiliated First Hospital of Fujian Medical University, Fuzhou 350005, China
2Pharmacy Department, Affiliated Union Hospital of Fujian Medical University, Fuzhou 350005, China
3Orthopedics Department, Affiliated First Hospital of Fujian Medical University, Fuzhou 350005, China
Role of IL-17 in nucleus pulposus cell proliferation and metabolism cultured in vitro
Xi Lin1, Qi Lin2, Jun-Jian Ye3*
1Emergency Department, Affiliated First Hospital of Fujian Medical University, Fuzhou 350005, China
2Pharmacy Department, Affiliated Union Hospital of Fujian Medical University, Fuzhou 350005, China
3Orthopedics Department, Affiliated First Hospital of Fujian Medical University, Fuzhou 350005, China
ARTICLE INFO
Article history:
Received 20 October 2014
Received in revised form 15 November 2014
Accepted 20 December 2014
Available online 15 Jaunary 2015
Interleukin-17
Cytokine
Inflammation
Degenerative disc disease
Objective: To explore the role of cytokine, interleukin-17 (IL-17) in human degenerative disc disease. Methods: Through magnetic resonance imaging, human degenerative disc tissues were confirmed from the isolated nucleus pulposus cells, which were then cultured in vitro. The ells were cultured with and without different concentrations of IL-17. 2 ng/mL, 5 ng/mL, 10 ng/ mL, 15 ng/mL and 20 ng/mL IL-17 concentrations were used for stimulation. After 72 hours, the nhibition rate of proliferation was measured by MTS method. For 48 and 96 hours, the nucleus pulposus cells were cultured with and without the appropriate IL-17 concentrations. The mRNA nd protein expression levels of the matrix macromolecules and degrading tissue genes were measured by Real-time PCR and Western blot analysis. Results: It was noted that nucleus pulposus cell proliferation was inhibited after culturing in vitro with IL-17 stimulation, and t was further observed that the inhibition effect was significantly stronger with 15 ng/mL IL-7 concentration. With the dosage of 15 ng/mL, IL-17 stimulation induced multiple cellular esponses, such as the significant increase in mRNA expression for both aggrecan (ACAN) and ype Ⅰ collagen (COL1A1) genes (P<0.05), and the significant decrease in mRNA expression of both degrading tissue genes, MMP3 and TIMP3 (P<0.05). Western blot results also showed that the protein level of COL1A1 was significantly decreased (t=3.199, P=0.006), while the protein level f one peptidases (ADAMTS5) significantly increased (t=2.667, P=0.021). Conclusions: These indings suggest that IL-17 can inhibit proliferation and affect the metabolism of the cultured nucleus pulposus cells in vitro, and these findings could possibly contribute to the degenerative changes that occur in DDD through extracellular matrix synthesis inhibition, promoting nucleus pulposus extracellular matrix degradation and disrupting the metabolic balance.
1. Introduction
Degenerative Disc Disease (DDD) is a common health problem in invertebrates, including humans[1]. It is a chronic disease that involves multiple-factors that progressively destroys the composition, structure and function of the disc[2,3]. The main pathological features that characterize DDD include collagen type conversion, progressive loss of extracellular matrix proteoglycans, disc dehydration, increased expression and activity of matrix-degrading enzymes and inflammatory factors accumulation[1,4,5]. Immune cell accumulation and increased inflammatory cytokine expressions have been identified in degenerative disc tissues[6-10], indicating that inflammatory cytokines may play an important role in the disc degeneration process[11,12].
Interleukin-17 (IL-17), a powerful inflammatory cytokine, plays critical roles in the pathogenesis of various inflammatory disorders[13]. Studies on arthritis found that IL-17 can, by itself or combined with other inflammatory cytokines, induce inflammatory cytokine
aggregation, inhibit aggrecan synthesis, and up-regulate the aggrecan enzyme and degrading collagen molecules; thus, contributing to joint degeneration[14,15]. Chondrocytelike cells, similar to articular chondrocytes, is the main cell type found in the nucleus pulposus (NP) of the human disc, suggesting that IL-17 may be involved in disc degeneration similar to joint degeneration. Scuderi et al revealed that the epidural space lavage in patients with lumbar intervertebral disk herniation and radiculopathy contained an increased expression of IL-17[16], suggesting that IL-17 may be involved in intervertebral disc tissue degeneration which may also be associated with the development of pain. With further analysis, Shamji et al[17] identified that nucleus pulposus cells cultured in vitro not only responded with a catabolic phenotype to IL-17 stimulation, but also expressed surface ligands with a consequential potential to recruit additional lymphocytes and immune cells into the DDD microenvironment. Perhaps IL-17 may be an important inflammation regulator in DDD pathologies. So far, studies have shown that IL-17 may be involved in the process of disc degeneration; however, the contribution of inflammatory IL-17 cytokines in disc degeneration through inhibiting cell proliferation, the extracellular matrix synthesis of intervertebral disc cells, as well as promoting the degradation of aggrecan enzyme expressions and collagen molecules, still needs to be investigated.
Based on this background, we found the importance of examining the response of isolated nucleus pulposus cells from IL-17 stimulated degenerative disc tissues; and hypothesized that IL-17 can inhibit the proliferation and destruction of extracellular matrix tissue homeostasis in nucleus pulposus cells cultured in vitro.
2. Materials and methods
2.1. Clinical cases
Out of the 36 patients that visited our hospital for lumbar disc herniation surgery, between December 2013 and March 2014, 20 patients (12 males, 8 females) were enrolled as subjects for this study, after obtaining informed consents. The patients had average age of (68.4±7.7) years old. The extent of disc degeneration of each subject was from levelsⅢ to Ⅳ, which was graded using Thompson’s classification by magnetic resonance imaging[18]. The subjects had no prior history of surgery, serious infection, cancer, hypotension, and diabetes, which was confirmed by questionnaire and pathological examination after operation.
2.2. Cell separation and culturing
During surgery, NP tissues was dissected, stripped from the annulus and cartilage tissues, and immersed in saline for subsequent analysis, within 30 minutes. Using scissors, the collected human NP tissues were cut into small pieces. The NP cells were enzymatically released from the NP tissues by digestion, using a combined solution of 0.25% trypsin and 0.025% collagenase for half an hour at 37 ℃. After 10 minutes of centrifugation at 1 000 rpm, the supernatant was removed. Subsequently, cell clumps were resuspended with Ham’s F12 media (Gibco), combined with 10% fetal bovine serum, 100 μg/mL of penicillin and 100 μg/mL of streptomycin (culture medium), for cell counting; and it was inoculated on a gelatin-coated tissue culture plastic with 5% CO2at 37 ℃. All cells were seeded at an approximate density of 25 000 cells per cm2, the medium was changed every other day. Cell morphology and density were observed under an inverted microscope DIAVERT. Cell passage was established when approximately 80% cell fusion was achieved[19]. Cells after three passages were used in all experiments.
2.3. IL-17 stimulation
For cell proliferation analysis: NP cells were plated at a density of 50 000 cells per well in 48-well plates (n=6 replicates) with an overlaid culture medium of 150 μL. In order to determine the optimal IL-17 dosage for all experiments, the inhibition effects of IL-17 on NP cells were first evaluated over a broad range of doses (0-20 ng/ mL). After 72 hours, the cultured cells were collected and the proliferation rate were measured by MTS analysis.
For cell metabolism analysis: NP cells were plated at a density of 50 000 cells per well in 48-well plates (n=6 replicates), overlaid with 300 μL of the following culture mediums: fresh culture medium without IL-17 (unstimulated group), fresh culture medium with concentrations identified during theIL-17 cell proliferation analysis (stimulated group). After culturing for 48 and 96 hours, the mRNA expression and protein levels were measured by real-time PCR and Western blot analysis.
2.4. Cell proliferation and metabolism measurement
2.4.1. Cell proliferation measurement
Cell proliferation was determined by MTS assay; the CellTiter 96® AQueous One Solution Cell Proliferation Assay kit was used, based on manufacturer’s instructions. Briefly, 3 hours before each of the desired time points, 20 μL of
MTS reagent was added into each well, and the cells were incubated at 37 ℃ for 3 hours. Then, a 550 BioRad platereader (Bio-Rad, Hertfordshire, UK) was used, and an optical density (OD) value of OD490was detected. All experiments were repeated three times[20]. Cell suppression ratio was calculated, according to the following formula: (stimulated group OD - unstimulated group OD)/(unstimulated group OD - background OD).
2.4.2. Reagents and instruments
DMEM/F12 Medium (Gibco, USA); fetal bovine serum (Gibco, USA):0.25% trypsin (Invitrogen, USA); CollagenaseⅡ (Invitrogen , USA):DMSO(Invitrogen, USA); Penicillin and Streptomycin(Invitrogen, USA); SuperScript® Ⅲ First Strand kits (Takara, China): CellTiter 96® AQueous One Solution Cell Proliferation Assay kit (Promega, USA).
Cell incubator (Heraeus, Germany); Inverted microscope (Olympus, Japan); Clean bench (Shenggong Tecnology Company, Shanghai): Digital thermostat water bath (Shenggong Tecnology Company, Shanghai): Spectrophotometer.
2.4.3. Total RNA extraction and RT-PCR analysis
Total RNA was isolated from the two groups of cells using TRIzol Reagent (Gibco) and the cells were immediately reverse-transcribed using oligo (dT); then, cDNA was generated from 5 μg of total RNA in 20 μL reaction mixes using SuperScript® Ⅲ First Strand kits (Takara), and stored at -20 ℃ for later use. All procedures were conducted according to manufacturer’s instructions. Primers for the following genes were designed by using Oligo 6.0 (National Biosciences Inc., Plymouth, MN) and synthesized by Sangon Biotech (Shanghai, China): glyceraldehyde-3-phosphate dehydrogenase (GAPDH), aggrecan (ACAN), collagen, typeⅠ, alpha 1 (COL1A1), collagen, type Ⅱ, alpha 1 (COL2A1), matrix metalloprotease-3 (MMP-3), stromolysin, matrix metalloprotease-1 (MMP-1), collagenase, a disintegrin and metalloproteinase with thrombospondin motifs 4 (ADAMTS4), a disintegrin and metalloproteinase with thrombospondin motifs 5 (ADAMTS5), tissue inhibitor of matrix metalloprotease-1 (TIMP-1), tissue inhibitor of matrix metalloprotease-13 (TIMP-13). The detailed sequences of the primer pairs are shown in Table 1.
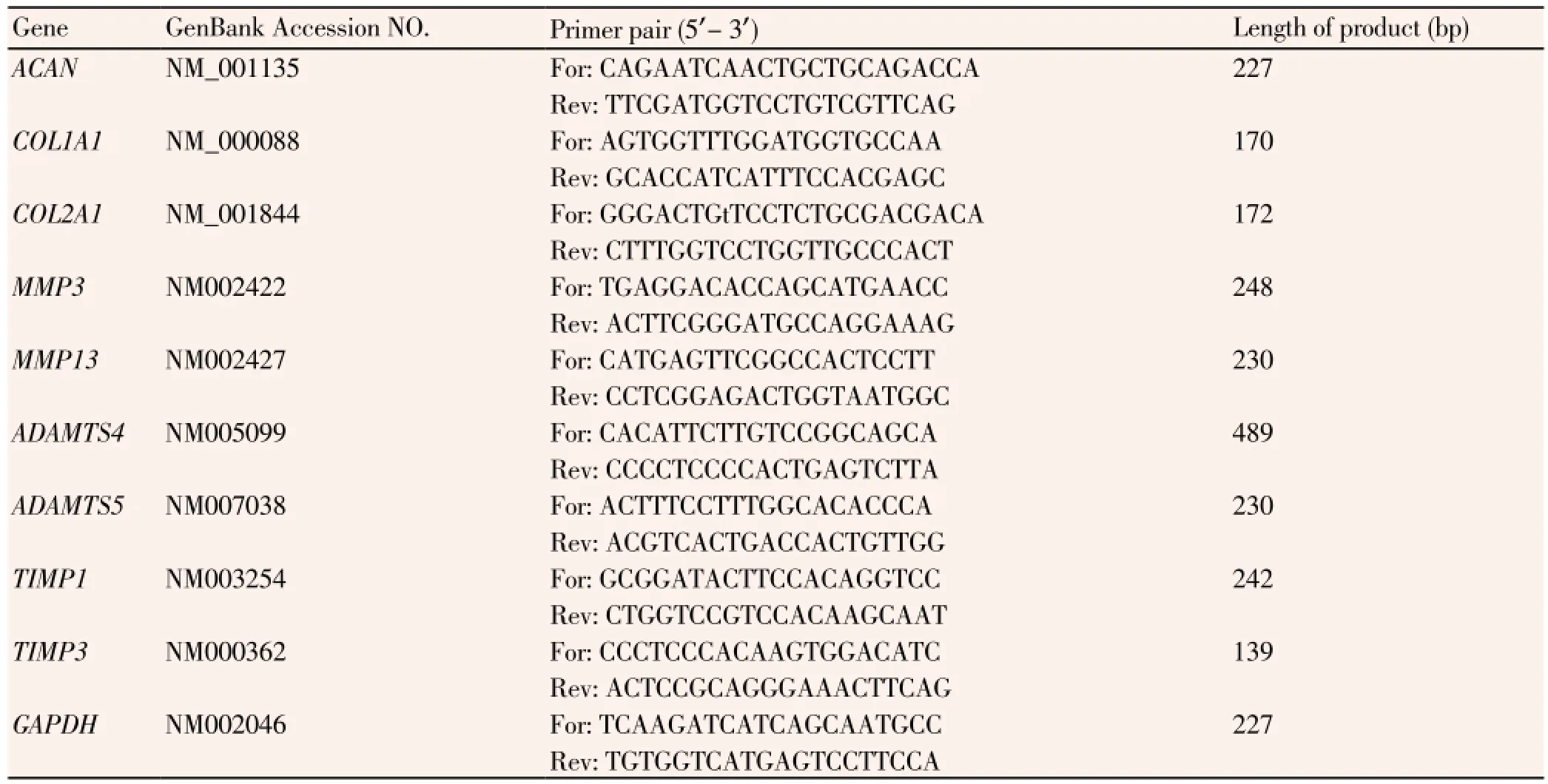
Table 1 Gene sequences and primer information.
The mRNA expression levels were quantified by real-time PCR using the Mas tercycler®ep Rea lplex Thermal Cycler (Eppendorf, Hamburg, Germany) and Thunderbird SYBR® qPCR Mix (Toyobo, Osaka, Japan) to detect PCR products. The 25 μL PCR reaction mixture contained 5 μL of cDNA, 12.5 μL of Thunderbird SYBR® qPCR Mix, 0.2 mM of dNTP, and 0.5 μM of each primer (Toyobo). The reaction mixture was denatured at 95 ℃ for 2 minutes, followed by 40 cycles of PCR reactions in the following settings: 95 ℃, 15 seconds; 60 ℃, 15 seconds; and 72 ℃, 45 seconds. The expression level of each target gene was normalized to GAPDH expression. Normalized mRNA expression (in ΔCtunits) was calculated by subtracting the cycle threshold (Ct) of the target gene from the Ctvalue of the house-keeping genes: ΔCt(target gene)=Ct(target gene) - Ct(GAPDH).
2.4.4. Western blot analysis
After the IL-17 stimulation of NP cells cultured in vitro, a Western blot analysis was conducted to measure the protein levels of ACAN, COL1A1, COL2A1, ADAMTS4, and ADAMTS5 genes with polyclonal goat anti-ACAN antibody, polyclonal goat anti-COL1A1 antibody , polyclonal goat anti-COL2A1 antibody, polyclonal goat anti-ADAMTS4 antibody, and polyclonal goat anti-ADAMTS4 antibody (sc-16492, sc-8784, sc-7764, sc-16534, Santa Cruz Biotechnology Company; LSC124924-100, LifeSpanBioSciences Inc.).
Cells were harvested and washed twice with ice-cold PBS. Lysates were obtained with a TEN-T buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.4, 5 mM EDTA, pH 8.0, 1% Triton X-100, 1 mM PMSF, 2 μg/mL aprotinin) and was subjected to 10 000×g centrifugation at 4 ℃ for 20 minutes. The BCA Protein Assay Kit (Beyotime) was used to measure the protein concentration of the supernatants. Per lane, 50μg of protein was electrophoresed in 12% SDS polyacrylamide gels and transferred onto nitrocellulose membranes (Sigma). After blocking with TTBS (50 mM Tris-HCl, pH 7.4, 0.5 M NaCl, 0.05% Tween-20) containing 5% (w/v) skimmed milk, the membranes were incubated with first antibodies overnight at 4 ℃, followed by horseradish peroxidase-conjugated second antibody incubation (Santa Cruz Biotechnology Company). Immunoreactive bands were detected by Super ECL Plus Detection Reagent (Amersham).
2.5. Statistical analysis
All tests were repeated three times. Statistical analysis was carried out using Student’s t-test with SPSS software. Results are presented as mean±SD. P<0.05 was considered statistically significant.
3. Results
3.1. IL-17 stimulation effects on cell proliferation
In 72 hours, IL-17 stimulation induced a dose-dependent decrease in proliferation rate. With the IL-17 dosage range (0-20 ng/mL), the suppression ratios were 3.89%±0.65%, 6.87%±1.05%, 8.35%±0.77%, 12.89%±0.64%, and 12.80%± 1.56%, respectively. The 15 ng/mL concentration showed significantly stronger inhibition effect on nucleus pulposus cell proliferation than 2 ng/mL (t=17.089, P=0.000), 5 ng/ mL (t=8.479, P=0.001), and 10 ng/mL (t=7.854, P=0.001); however, the difference was not significant compared with 20 ng/mL (t=0.015, P=0.99). These results suggest that IL-17 stimulation induced the proliferation rate to decrease in cultured NP cells, and the inhibition effect on 15 ng/mL of IL-17 was significantly stronger. A 15 ng/mL concentration of IL-17 was used to stimulate nucleus pulposus cells cultured in vitro for metabolic analysis.
3.2. IL-17 stimulation effects on cell metabolism
3.2.1. IL-17 stimulation effects on gene mRNA expression
After 48 and 96 hours of treatment with 15 ng/mL IL-17, a significant decrease on ACAN expression levels were observed; MMP3 expression levels were significantly elevated; a significant decrease of COL1A1 mRNA was noted after 48 hours (P<0.05), and the significance disappeared after 96 hours; a significant increase of TIMP3 mRNA was noted after 48 hours (P<0.05), and the significance also disappeared after 96 hours. The expression levels of COL2A1, MMP13, ADAMTS4, ADAMTS5 and TIMP1 also responded to IL-17 stimulation, but no significant changes were noted (Table 2).

Table 2 Normalized mRNA expressions (ΔCtunit) in annulus fibrosus cells exposed to interleukin-17 for (A) 48 hrs and (B) 96 hrs compared with unstimulated cells.
3.2.2. IL-17 stimulation effects on protein expression
The effects of IL-17 (15 ng/mL) on the protein levels of ACAN, COL1A1, COL2A1, ADAMTS4 and ADAMTS5 were measured by Western blot analysis (Figure 3). NP cells demonstrated a greater decrease in COL1A1 protein expression after 48 hours (t=3.199, P=0.006) and a significant increase in ADAMTS5 protein expression (t=2.667, P=0.021), compared with the unstimulated group. However, both
significant changes disappeared after 96 hours (t=0.359, P=0.724; t=0.538, P=0.568). At this time point, COL2A1 protein levels greatly increased, compared with the unstimulated group (t=4.411, P=0.000).
4. Discussion
IL-17 is a cytokine factor that has recently shown to be elevated in degenerated human intervertebral disc tissues[6], suggesting that an in-depth investigation of its involvement in disc degeneration may help us better understand the DDD process. In this study, through establishing cultured nucleus pulposus cells in vitro, the inhibiting effects of different IL-17 concentrations on nucleus pulposus cell proliferation was analyzed. After IL-17 exposure, nucleus pulposus cell metabolic changes were explored in order to identify the possible role of IL-17 in the pathogenesis of disc degeneration from an extracellular matrix balance aspect.
Within 72 hours, IL-17 stimulation in cultured NP cells induced a dose-dependent proliferation inhibition rate. The suppression ratio increased as the concentrations were increased, and the suppression ratio of IL-17 at a concentration of 15 ng/mL had the highest results. Based on the analysis of Gabr et al, they found that NP cells increased its nitric oxide (NO) production over the 72 hour culture period in the presence of IL-17, with a maximal effect noted for IL-17 doses higher than 10 ng/mL[17]. This result was basically consistent with the results of our experiments. The suppression ratio of IL-17 at 20 ng/mL concentration also showed no significant difference, perhaps due to the complete saturation of the IL-17 receptors in NP cells[21]. Until now, the mechanism of the proliferation inhibiting effects of IL-17 on NP cells is still unknown. However, it is known that NO, another inflammatory factor, is involved in DDD development, through promoting apoptosis[22], and IL-17 can significantly induce the expression of nitric oxide synthase, thereby, contributing to NO molecule generation[23]. IL-17 may also play a role in inhibiting cell proliferation by indirectly inducing apoptosis in the NP cells.
A IL-17 at 15 ng/mL concentration was used to stimulate the NP cells cultured in vitro for metabolic analysis. We found that ACAN and COL1A1 mRNA levels were significantly down regulated by IL-17 after 48-h culture. After 96 hours, the ACAN mRNA levels in stimulated group were still significantly down regulated. Western blot results showed that COL1A1 protein levels in stimulated group were significantly higher than the unstimulated group. Collagen type Ⅱ mRNA expression was down regulated at two time points, but no significant differences were noted, compared with unstimulated group. Based on the experiments of Seguin et al, NP cells formed in vitro were treated with TNF-α, a proinflammatory cytokine, over 48 hours[24], and the tissues were assessed for proteoglycan and collagen synthesis. They found that TNF-α induced the decreased expressions of both aggrecan and type Ⅱ collagen genes, suggesting that decreasing the synthesis of matrix macromolecules may be one of the main functions of inflammatory cytokines in disc degeneration. Besides the decrease in matrix macromolecules synthesis[25,26], a gradual transition from type Ⅰ collagen to type Ⅱ collagen was observed[27]. In our analysis, the down regulation of type Ⅰ collagen was not observed after IL-17 stimulation, but the COL2A1 gene protein level was significantly lower, compared with the unstimulated group (time point: 96 hours). One possible explanation is that within 48 hours, NP cells responded to IL-17 stimulation with a decreased type Ⅰ collagen protein synthesis; and also, the collagen type transition enabled the observation of increased type Ⅰ collagen protein level and decreased type Ⅱ protein level. The results suggest that IL-17 may be involved in tissue matrix homeostasis destruction through inhibiting matrix macromolecules synthesis and collagen type transition.
Nucleus pulposus cells not only synthesize extracellular matrix macromolecules, but also synthesize a variety of enzymes for matrix degradation[28,29]. A variety of enzyme changes in IL-17 stimulated NP cells were noted in our
analysis. Compared with the unstimulated group, the mRNA levels of the MMP3 gene significantly increased after 48 and 96 hours of stimulation; and the significant increase in TIMP3 mRNA levels were noted at the 48-hour time point; an additional evidence that DDD develops, as specific gene/ protein expression increases, such as the MMP3 and TIMP3 genes identified by Liang et al[30,31], which indicated that IL-17 may directly or indirectly contribute to disc degeneration through other mechanisms. ADAMTS4, ADAMTS5, and TIMP1 mRNA levels also increased after IL-17 stimulation, but compared with the unstimulated group, no significant differences were noted. A possible reason, for the absence of significant response to IL-17 stimulation, is that the expression of these genes has already been up-regulated in vivo since the time the NP cells were isolated from degenerative human discs. After 48 hours of stimulation, ADAM-TS5 gene protein levels were significantly upregulated, probably due to the response of ADAMTS5 mRNA translation from IL-17 stimulation. All these results have shown that IL-17 stimulation increased the synthesis and activity of degrading enzymes in NP cells, which therefore enhanced disc extracellular matrix degradation and further promoted the degradation process.
IL-17 stimulation effects on the proliferation and metabolism of nucleus pulposus cells cultured in vitro were analyzed. Our findings suggest that IL-17 can inhibit proliferation and possibly contribute to the degenerative changes that occur in disc disease, through inhibiting extracellular matrix synthesis, promoting nucleus pulposus extracellular matrix degradation and disrupting metabolic balance.
Despite the effectiveness of using a model where nucleus pulposus cells were cultured in vitro to study the process of disc degeneration, previous studies have also demonstrated gene expression changes in culturing intervertebral disc cells[32-35]. In order to minimize any potential phenotypic changes, we used third passage cells for all experiments. Clearly, the in vitro results presented in this study cannot be directly used to interpret the function of IL-17 in vivo; however, we hope that the identified responses of IL-17 stimulation would be further considered for DDD. Future studies are needed to identify the specific changes of normal and degenerative cells that respond to IL-17 only or in combining IL-17 with other inflammatory cytokinesboth in vitro and in vivo; and how these results may be further exploited to facilitate its beneficial effects on tissue matrix homeostasis.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
The study was supported by Natural Science Youth Foundation of China (NO.81201395).
[1] Boos N, Weissbach S, Rohrbach H, Weiler C, Spratt KF, Nerlich AG. Classification of age-related changes in lumbar intervertebral discs: 2002 Volvo Award in basic science. Spine 2002; 27(23): 2631-2644.
[2] Gazzeri R, Galarza M, Alfieri A. Controversies about interspinous process devices in the treatment of degenerative lumbar spine diseases: Past, present, and future. Biomed Res Int 2014; 2014: 975052.
[3] Cunningham C, Srivastava A, Collin E, Grad S, Alini M, Pandit A, et al. Isolation and characterisation of a recombinant antibody fragment that binds NCAM1-expressing intervertebral disc cells. PloS One 2013; 8(12): e83678.
[4] Le Maitre CL, Pockert A, Buttle DJ, Freemont AJ, Hoyland JA. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soctrans 2007; 35(Pt 4): 652-655.
[5] Kang JD, Stefanovic-Racic M, McIntyre LA, Georgescu HI, Evans CH. Toward a biochemical understanding of human intervertebral disc degeneration and herniation. Spine 1997; 22(10): 1065-1073.
[6] Shamji MF, Setton LA, Jarvis W, So S, Chen J, Jing L, et al. Proinflammatory cytokine expression profile in degenerated and herniated human intervertebral disc tissues. Arthritis Rheum 2010; 62(7): 1974-1982.
[7] Le Maitre CL, Hoyland JA, Freemont AJ. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1beta and TNFalpha expression profile. Arthritis Res Ther 2007; 9(4): R77.
[8] Weiler C, Nerlich AG, Bachmeier BE, Boos N. Expression and distribution of tumor necrosis factor alpha in human lumbar intervertebral discs: a study in surgical specimen and autopsy controls. Spine 2005; 30(1): 44-53.
[9] Kang JD, Georgescu HI, McIntyre-Larkin L, Stefanovic-Racic M, Donaldson WF, Evans CH. Herniated lumbar intervertebral discs spontaneously produce matrix metalloproteinases, nitric oxide, interleukin-6, and prostaglandin E2. Spine 1996; 21(3): 271-277.
[10] Igarashi T, Kikuchi S, Shubayev V, Myers RR. Exogenous tumor necrosis factor-alpha mimics nucleus pulposus-induced neuropathology. Molecular, histologic, and behavioral comparisons in rats. Spine 2000; 25(23): 2975-2980.
[11] Sun Z, Zhang M, Zhao XH, Liu ZH, Gao Y, Samartzis D, et al. Immune cascades in human intervertebral disc: the pros and cons. Int J Clin Exp Pathol 2013; 6(6): 1009-1014.
[12] Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol 2014; 10(1): 44-56.
[13] Kirkham BW, Lassere MN, Edmonds JP, Juhasz KM, Bird PA, Lee CS, et al. Synovial membrane cytokine expression is predictive of joint damage progression in rheumatoid arthritis: a two-year prospective study (the DAMAGE study cohort). Arthritis Rheum 2006; 54(4): 1122-1131.
[140] Dudler J, Renggli-Zulliger N, Busso N, Lotz M, So A. Effect of interleukin 17 on proteoglycan degradation in murine knee joints. Ann Rheum Dis 2000; 59(7): 529-532.
[15] Liu DF, Yan J, Guo MY, Wang C, Hu YH, Yang M, et al. Correlation study between interleukin-17 and ESR and CRP in serum and the synovial fluid of rheumatoid arthritis patients of accumulated dampness-heat obstruction in joints syndrome. Zhongguo Zhong Xi Yijie He Zazhi 2014; 34(3): 272-275.
[16] Scuderi GJ, Brusovanik GV, Anderson DG, Dunham CJ, Vaccaro AR, Demeo RF, et al. Cytokine assay of the epidural space lavage in patients with lumbar intervertebral disk herniation and radiculopathy. J Spinal Disord Tech 2006; 19(4): 266-269.
[17] Gabr MA, Jing L, Helbling AR, Sinclair SM, Allen KD, Shamji MF, et al. Interleukin-17 synergizes with IFN gamma or TNF alpha to promote inflammatory mediator release and intercellular adhesion molecule-1 (ICAM-1) expression in human intervertebral disc cells. J Orthop Res 2011; 29(1): 1-7.
[18] Thompson JP, Pearce RH, Schechter MT, Adams ME, Tsang IK, Bishop PB. Preliminary evaluation of a scheme for grading the gross morphology of the human intervertebral disc. Spine 1990; 15(5): 411-415.
[19] Zhang C, Cui G, Shao C, Zhou X, Xiao Y, Zhou J. Effects of recombinant adenovirus vector carrying human insulin-like growth factor 1 gene on the apoptosis of nucleus pulposus cells in vitro. Zhongguo Xiufu Chongjian Waike Zazhi 2013; 27(11): 1375-1379.
[20] Liu W, Xu C, Wan H, Liu C, Wen C, Lu H, et al. MicroRNA-206 overexpression promotes apoptosis, induces cell cycle arrest and inhibits the migration of human hepatocellular carcinoma HepG2 cells. Int J Mol Med 2014; 47(11): 13-37.
[21] Kuestner RE, Taft DW, Haran A, Brandt CS, Brender T, Lum K, et al. Identification of the IL-17 receptor related molecule IL-17RC as the receptor for IL-17F. J Immunol 2007; 179(8): 5462-5473.
[22] Kohyama K, Saura R, Doita M, Mizuno K. Intervertebral disc cell apoptosis by nitric oxide: biological understanding of intervertebral disc degeneration. Kobe J Med Sci 2000; 46(6): 283-295.
[23] Miljkovic D, Trajkovic V. Inducible nitric oxide synthase activation by interleukin-17. Cytokine Growth Factor Rev 2004; 15(1): 21-32.
[24] Seguin CA, Pilliar RM, Roughley PJ, Kandel RA. Tumor necrosis factor-alpha modulates matrix production and catabolism in nucleus pulposus tissue. Spine 2005; 30(17): 1940-1948.
[25] Little CB, Flannery CR, Hughes CE, Mort JS, Roughley PJ, Dent C, et al. Aggrecanase versus matrix metalloproteinases in the catabolism of the interglobular domain of aggrecan in vitro. Biochem J 1999; 344 Pt 1: 61-68.
[26] Lotz JC, Colliou OK, Chin JR, Duncan NA, Liebenberg E. Compression-induced degeneration of the intervertebral disc: an in vivo mouse model and finite-element study. Spine 1998; 23(23): 2493-2506.
[27] Guiot BH, Fessler RG. Molecular biology of degenerative disc disease. Neurosurgery 2000; 47(5): 1034-1040.
[28] Kozaci LD, Guner A, Oktay G, Guner G. Alterations in biochemical components of extracellular matrix in intervertebral disc herniation: role of MMP-2 and TIMP-2 in type Ⅱ collagen loss. Cell Biochem Funct 2006; 24(5): 431-436.
[29] Bachmeier BE, Nerlich A, Mittermaier N, Weiler C, Lumenta C, Wuertz K, et al. Matrix metalloproteinase expression levels suggest distinct enzyme roles during lumbar disc herniation and degeneration. Eur Spine J 2009; 18(11): 1573-1586.
[30] Liang QQ, Ding DF, Xi ZJ, Chen Y, Li CG, Liu SF, et al. Protective effect of ligustrazine on lumbar intervertebral disc degeneration of rats induced by prolonged upright posture. Evid Based Complement Alternat Med 2014; 15(22): 508461.
[31] Tsuji T, Chiba K, Imabayashi H, Fujita Y, Hosogane N, Okada Y, et al. Age-related changes in expression of tissue inhibitor of metalloproteinases-3 associated with transition from the notochordal nucleus pulposus to the fibrocartilaginous nucleus pulposus in rabbit intervertebral disc. Spine 2007; 32(8): 849-856.
[32] Wang JY, Baer AE, Kraus VB, Setton LA. Intervertebral disc cells exhibit differences in gene expression in alginate and monolayer culture. Spine 2001; 26(16): 1747-1751.
[33] Kluba T, Niemeyer T, Gaissmaier C, Grunder T. Human anulus fibrosis and nucleus pulposus cells of the intervertebral disc: effect of degeneration and culture system on cell phenotype. Spine 2005; 30(24): 2743-2748.
[34] Wuertz K, Urban JP, Klasen J, Ignatius A, Wilke HJ, Claes L, et al. Influence of extracellular osmolarity and mechanical stimulation on gene expression of intervertebral disc cells. J Orthop Res 2007; 25(11): 1513-1522.
[35] Markova DZ, Kepler CK, Addya S, Murray HB, Vaccaro AR, Shapiro IM, et al. An organ culture system to model early degenerative changes of the intervertebral disc Ⅱ: profiling global gene expression changes. Arthritis Res Ther 2013; 15(5): R121.
ment heading
10.1016/S1995-7645(14)60185-1
*Corresponding author: Jun-Jian Ye, M.D., Chief Physician, Emergency Department, Affiliated First Hospital of Fujian Medical University, Fuzhou 350005, China.
Tel: 059187982105
E-mail: yejunjian@medmail.com
Foundation project: This study was supported by Natural Foundation of Fujian Province (NO. 2012J01126).
杂志排行
Asian Pacific Journal of Tropical Medicine的其它文章
- Inhibition of advanced glycation endproducts formation by Korean thistle, Cirsium maackii
- Anti-hypercholesterolemic effect of kenaf (Hibiscus cannabinus L.) seed on high-fat diet Sprague dawley rats
- Susceptibility to temephos, permethrin and deltamethrin of Aedes aegypti (Diptera: Culicidae) from Muang district, Phitsanulok Province, Thailand
- Cloning, expression, purification and bioinformatic analysis of 2-methylcitrate synthase from Mycobacterium tuberculosis
- Prevalence of shiga toxins (stx1, stx2), eaeA and hly genes of Escherichia coli O157:H7 strains among children with acute gastroenteritis in southern of Iran
- Larvicidal, ovicidal and repellent activities of marine sponge Cliona celata (Grant) extracts against Anopheles stephensi Liston (Diptera: Culicidae)
