Prevalence of shiga toxins (stx1, stx2), eaeA and hly genes of Escherichia coli O157:H7 strains among children with acute gastroenteritis in southern of Iran
2015-12-08MohammadKargarMaryamHomayoon
Mohammad Kargar, Maryam Homayoon
1Department of Microbiology, Islamic Azad University, Jahrom, Iran
2Young Reseasrchers and Elite Club, Marvdasht Branch, Islamic Azad University, Marvdasht, Iran
Prevalence of shiga toxins (stx1, stx2), eaeA and hly genes of Escherichia coli O157:H7 strains among children with acute gastroenteritis in southern of Iran
Mohammad Kargar1*, Maryam Homayoon2
1Department of Microbiology, Islamic Azad University, Jahrom, Iran
2Young Reseasrchers and Elite Club, Marvdasht Branch, Islamic Azad University, Marvdasht, Iran
ARTICLE INFO
Article history:
Received 26 October 2014
Received in revised form 10 November 2014
Accepted 22 December 2014
Available online 20 January 2015
Escherichia coli O157:H7
Acute gastroenteritis
Virulence genes
Multiplex PCR
Objective: To survey the prevalence severe diarrhea arising from these bacteria in children under 5 years old in Marvdasht. Methods: In this study faecal sample from 615 children aged <5 years old who were hospitalized for gastroenteritis in Fars hospitals in Iran were collected and then enriched in Escherichia coli (E. coli) broth and modified tryptone soy broth with novobiocin media. Fermentation of sorbitol, lactose and β-glucoronidase activity of isolated strains was examined by CT-SMAC, VRBA and chromogenic media respectively. Then isolation of E. coli O157:H7 have been confirmed with the use of specific antisera and with multiplex PCR method presence of virulence genes including: stx1, stx2, eaeA, hly has been analyzed. Results: E. coli O157:H7 was detected in 7 (1.14%) stool specimens. A significant difference was seen between detection rate of isolated bacteria from age groups 18-23 months and other age groups (P=0.004). Out of considered virulence genes, only 1 of the isolated strains (0.16%) the stx1and eaeA genes were seen and also all isolated bacteria had resistance to penicillin, ampicillin and erythromycin antibiotics. Conclusions: We found that children < 2 years of age were at highest risk of infection with E. coli O157:H7. Regarding severity of E. coli O157:H7 pathogenesis, low infectious dose and lack of routine assay for detection of these bacteria in clinical laboratory, further and completed studies on diagnosis and genotyping of this E. coli O157:H7 strain has been recommended.
1. Introduction
Shiga toxin-producing Escherichia coli (E. coli) (STEC), also called verocytotoxin-producing E. coli (VTEC), have emerged as pathogens that can cause food-borne infections and severe and potentially fatal illnesses in humans, such as haemorrhagic colitis (HC) and haemolytic uraemic syndrome (HUS) which is the main cause of acute renal failure in children. Most outbreaks of HC and HUS have been attributed to strains of the enterohemorrhagic serotype O157:H7[1,2]. The ability of E. coli O157:H7 to cause severe diseases in humans is related to their capacity to secrete shiga toxins (Stx1and Stx2) or verocytotoxins (VT1 and VT2) and variants of these toxins[1,3]. Another virulenceassociated factor of most STEC isolates associated with severe disease is intimin, a 94-kDa outer membrane protein, which is encoded by the eae gene on a ca. 34 kb chromosomal pathogenicity island termed the locus of enterocyte effacement (LEE). This locus is associated
with the intimate adherence of E. coli to epithelial cells, initiation of host signal transduction pathways, and the formation of attaching-and-effacing intestinal lesions[3]. A factor that may also affect virulence of E. coli O157:H7 is the enterohemolysin (Ehly), also called enterohemorrhagic E. coli haemolysin (EHEC- HlyA), encoded by the hly gene[1].
E. coli O157:H7 infection has been often associated with the consumption of contaminated ground beef, raw milk and other bovine products, thus cattle’s are suspected to be a primary reservoir. But bacteria also have been isolated from domestic and wild animals. Moreover, recent outbreaks of food borne illness associated with eating fresh products have heightened concerns that these foods contaminated with STEC may be an increasing source of illness. In the past decades, outbreaks of diseases caused by STEC have been associated with the consumption of leaf lettuce, potatoes, radish sprouts and raw vegetables. Fruit-related outbreaks have also been caused by the consumption of fresh-pressed apple juice[1,4].
Detection of E. coli O157:H7 in the clinical laboratory depends on distinguishing the pathogenic serotypes from normal faecal flora containing commensal strains of E. coli. Fortunately, E. coli O157:H7 has two unusual biochemical markers; delayed fermentation of D-sorbitol and lack of β-D-glucuronidase activity, which help to phenotypically separate O157:H7 isolates from nonpathogenic E. coli strains. One of these markers (delayed sorbitol fermentation) is able to develop selective media (Sorbitol-MacConkey; SMAC) which aids in the initial recognition of suspicious colonies isolated from bloody stools. The selectivity of SMAC agar has been improved with the addition of cefiximerhamnose (CR-SMAC), cefixime-tellurite (CT-SMAC) and 4-methylumbelliferyl-β-D-glucuronide (MSA-MUG)[4].
Fratamico et al described a multiplex PCR capable of detecting stx1, stx2, eaeA and EHEC hlyA sequences[5]. However, this PCR was not tested with faecal samples; primers for each target gene sequence showed differential sensitivities. Paton and Paton developed a multiplex PCR utilizing four PCR primer pairs for the detection of stx1, stx2, eaeA and EHEC hlyA in human feces and foodstuffs[6,7]. The multiplex PCR system for detecting virulence genes of STEC, reported by Paton et al, Pradel et al, Osek, Blanco et al, and Mohsin et al[8-11]. The aim of this study was to evaluate the prevalence of virulence factors and antibiotic resistance of E. coli O157:H7 in Fars providence, Iran.
2. Materials and methods
2.1. Sample collections
Between September 2008 and September 2009, stool samples were collected from children aged <5 years old were hospitalized with gastroenteric symptoms in Hospitals in Fars Province, Iran. A detailed history of the patients is obtained, including information on the age, sex, source of drinking water, clinical presentation and antibiotic usages.
2.2. Enrichment procedures
For E. coli O157:H7 detection, each faecal sample (1 g) was enriched in 5 mL E. coli (EC) broth (Oxoid) and modified tryptone soy broth (Difco) with 20 mg/L of novobiocin (Sigma) and incubated overnight at 37 ℃.
2.3. Culture methods
The enrichment broth was inoculated into sorbitol-MacConkey agar (Lab.M) supplemented with cefixime (0.05 mg/L) and tellurite (2.5 mg/L) (Oxoid) (CT-SMAC) for isolation (for 24 h at 37 ℃)[12,13]. Sorbitol-negative colonies were confirmed as E. coli by biochemical tests and then colonies transferred on chromogenic E. coli O157 agar and incubated at 37 ℃ for 24 h. Up to 10 colonies were tested for agglutination with E. coli O157:H7 latex tests[13,14].
2.4. Multiplex PCR
Presence of virulence genes were detected by multiplex PCR using specific primers for amplification of stx1, stx2, eaeA and hly genes (Table 1)[15] and isolates were confirmed as E. coli O157:H7 by using specific primers for rfb O157 and flic H7 genes[16].
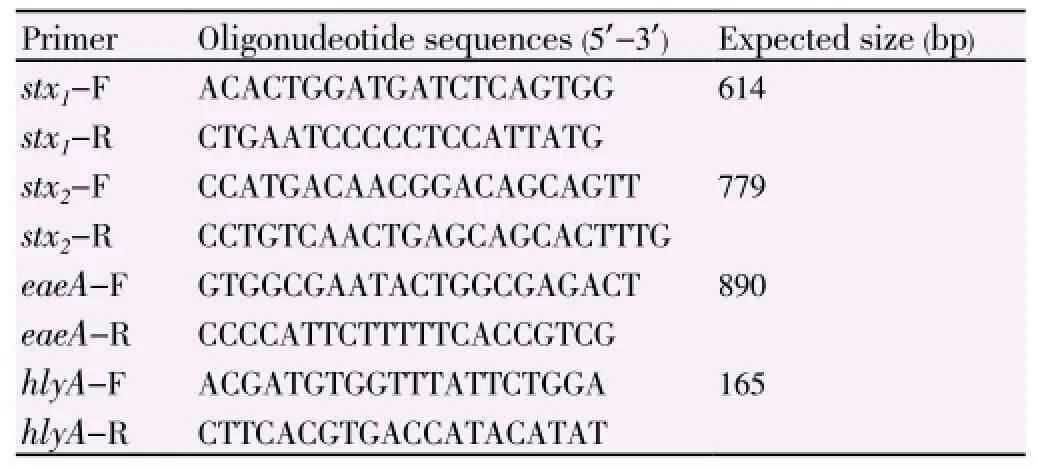
Table 1 Primers used in multiplex PCR.
E. coli O157:H7 strain used as control was 933j. Bacterial DNA was extracted from antisera-positive samples with a DNPTMkit (CinnaGene). PCR assays were carried out in a 50 μL volume containing 4 μL of nucleic acid templates prepared from cultures. And 10 mM Tris-HCl (pH 8.4), 10 mM KCl, 3 mM MgCl2, 20 pmol concentrations of each primer, 0.2 mM dNTPs, and 1 U of Taq DNA polymerase were added to the reaction mixtures. PCR conditions consisted of an initial 95 ℃ denaturation step for 3 min followed by 35 cycles of 95 ℃ for 20 s, 58 ℃ for 40 s and 72 ℃ for 90 s. The final extension cycle was followed by at 72 ℃for 5 min. Amplified DNA fragments were resolved by gel electrophoresis using 1.5% agarose gels. Gels were stained with 0.5 μL of ethidium bromide (EtBr) per mL, visualized and photographed under UV illumination[4].
2.5. Antimicrobial susceptibility test
Antimicrobiol susceptibility testing was based on the disk diffusion method as recommended by the Clinical and Laboratory Standard Institute (CLSI) guidelines, with ampicillin (10 μg), penicillin (10 μg), cephalexin (30 μg), chloramphenicol (30 μg), erythromycin (15 μg), gentamicin (10 μg), kanamycin (30 μg), tetracycline (30 μg) and trimethoprim /sulfamethoxazole (25 μg)[4,17].
2.6. Statistic analysis
Statistical analysis were conducted with the use of SPSS version 14 and the χ2test, Fisher`s exact test. P<0.05 was considered statistically significant.
3. Results
In this study, a total of 615 samples were collected. The results showed that 77 of 89 sorbitol nonfermenting colonies from CT-SMAC were lactose ferment (12.52%) and 7(1.14%) were E. coli O157:H7. Out of 7 E. coli O157:H7 strains 42.86% were isolated from girls and 57.14% from boys. No significant correlation was found between E. coli O157:H7 isolation and sex. The mean age of patients was 11.7 months (range 1-24 months). A significant association was found between E. coli O157:H7 isolation among different age groups (P = 0.004). The highest rates of E. coli O157:H7 isolation (42.86%) was detected in children 18-23 months of age. Of the 7 patients infect with E. coli O157:H7; 4 (57.14%) reported diarrhea, 2 (28.57%) had vomiting, 1 (14.29%) had fever and 1 (14.29%) were hospitalized for 5 day and 3 (42.86%) of patients reported that they received antibiotics.
71.43% of isolates were susceptible to chloramphenicol and 57.14% to tetracycline and trimethoprim/ sulfamethoxazole. All E. coli O157:H7 isolated were resistant to penicillin, ampicillin and erythromycin.
As shown by PCR, 1(14.29%) of the isolates harbored stx1and eaeA genes (Figure 1) and none of the isolates had stx2and hly genes. All of these isolates confirmed to be E. coli O157:H7 by using specific primers for rfb O157 and flic H7 genes (Figure 2).
4. Discussion
Gastroenteritis is one of the most frequent diseases in the world and continues to be one of the main causes of death in developing countries. The high morbidity of E. coli O157:H7 around the world and its presence in five continents has focused a major public health concern. A lot of attention has been given to these pathogens in developed countries and there is a relatively clear picture regarding their prevalence[11]. STEC strains that cause human infections belong to a large number of O:H serotypes. Most outbreaks of HC and HUS have been attributed to strains of the enterohemorrhagic serotype O157:H7. However, as Non-O157 STEC is more prevalent in animals and as contaminant in foods, humans are probably more exposed to these strains[1].
In Bangladesh Shiga toxin genes (stx) were detected by multiplex PCR in nine samples (2.2%) from hospitalized patients and 11 samples (6.9 %) from the community patients. Two isolates were positive for the E. coli attachingand-effacing (eae) gene and four were positive for the enterohaemolysin (hlyEHEC) gene and enterohaemolysin production[12]. In Pakistan results showed that 11 (78.5%), 6 (42.8%), 3 (21.4%) and 11 (78.5%) STEC isolates were positive for stx1, eae, hly and stx2genes respectively[11]. Seropathotypes O157:H7 stx1-eae, are only observed in E. coli O157:H7 that cause human infections in South of Iran. Within the human disease-associated strains, those producing Shiga toxin type 2 (Stx2) appear to be more commonly responsible for serious complications such as HUS than those producing only Shiga toxin type 1 (Stx1)[7]. Kawano et al[18] conclude that stx genotype is one of the important factors of clinical outcome of STEC O157 infection and that pathogenicity for humans was higher in the stx2genotype strains. The eae gene, which has been shown to be necessary for attaching and effacing activity, encodes a protein which is termed intimin. Numerous investigators have underlined the strong association between the carriage of eae gene and the capacity of STEC strains to cause severe human illnesses, especially HUS. This important virulence gene was detected in 14.29% of E. coli O157:H7 in the present study. Nevertheless, production of intimin is not essential for pathogenesis, because a number of sporadic cases of HUS have been caused by eae-negative O157 STEC strains[1].
Antimicrobial resistance patterns were observed most commonly to ampicillin (25.4%), tetracycline (23.8%) and less frequently to cephalothin (11,1%), nalidixic acid (6.4%) in India. The USA study about antibiotic resistance showed that all isolates were resistant to tilmicosin, and most isolates were susceptible to trimethoprim/ sulfamethoxazole and ciprofoloxacin[4]. In Malaysia, resistance was observed mostly towards bacitracin (100%), ampicillin (57%), cephalothin (53%) and carbenicillin (30%). The antibiotic resistant patterns to ampicillin, fosfomycin, kanamycin and vancomycin were observed in Japan[4]. From these data, E. coli O157:H7 was mainly resistant to ampicillin and tetracycline. Resistance patterns of Iran isolates were approximately similar to those of the USA and Malaysia. However, antibiotics are a risk factor for HUS and their use is therefore contraindicated in patients with STEC infection[17].
In this study it was found that E. coli O157:H7 strains could be isolated from diarrheal as well as asymptomatic children. Epidemiologic data were not collected regarding contaminated water as a possible source of E. coli O157:H7 infection in the patients. Stool cultures of all patients with acute bloody diarrhea should be tested for E. coli O157:H7 to identify those at risk of HUS[19]. However, serotyping, cytotoxicity assays or genotyping for E. coli O157:H7 are not routinely performed in Iran. Further studies are needed to identify the pathogenic mechanisms of this E. coli O157:H7 strains and to determine the faecal carriage rate in healthy children.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
The authors are grateful to the Islamic Azad University, Jahrom branch, for their executive support of this project.
[1] Mora A, Blanco M, Blanco JE, Dahbi G, lópez C, Justel P, et al. Serotypes, virulence genes and intimin types of shiga toxin (verocytotoxin)- producing Escherichia coli isolates from minced beef in Lugo (Spain) from 1995 through 2003. BMC Microbiol 2007; 7: 1-9.
[2] Prendergast DM, Lendrum L, Pearce R, Ball C, McLernon J, O’Grady D, et al. Verocytotoxigenic Escherichia coli O157 in beef
and sheep abattoirs in Ireland and characterisation of isolates by Pulsed-Field Gel Electrophoresis and Multi-Locus Variable Number of Tandem Repeat Analysis. Int J Food Microbiol 2011; 144: 519-527.
[3] Leotta GA, Miliwebsky ES, Chinen I, Espinosa EM, Azzopardi K, Tennant SM, et al. Characterisation of Shiga toxin-producing Escherichia coli O157 strains isolated from humans in Argentina, Australia and New Zealand. BMC Microbiol 2008; 8: 1-8.
[4] Kim JY, Kim S, Kwon N, Bae WK, Lim JY, Koo HC, et al. Isolation and identification of Escherichia coli O157:H7 using different detection methods and molecular determination by multiplex PCR and RAPD. J Vet Sci 2005; 6: 7-19.
[5] Fratamico PM, Sackitey SK, Wiedmann M, Deng MY. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol 1995; 33: 2188-2191.
[6] Fagan PK, Hornitzky MA, Bettelheim KA, Djordjevic SP. Detection of shiga-like toxin (stx1and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl Environ Microbiol 1999; 65: 868-872.
[7] Paton AW, Paton JC. Detection and characterization of shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol 1998; 36: 598-602.
[8] Paton AW, Voss E, Manning PA, Paton JC. Shiga toxin producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect Immun 1997; 65: 3799-3805.
[9] Osek J. Development of a multiplex PCR approach for the identification of shiga-toxin producing Escherichia coli strains and their major virulence factor genes. J Appl Microbiol 2003; 95: 1217-1225.
[10] Blanco M, Blanco JE, Mora A, Rey J, Alonso JM, Hermoso M, et al. Serotypes, virulance genes , and intimin types of shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J Clin Microbiol 2003; 41: 1351-1356.
[11] Mohsin M, Hussain A, Butt MA, Bashir S, Tariq A, Babar S, et al. Prevalence of shiga toxin-producing Escherichia coli in diarrhoeal patients in Faisalabad region of Pakistan as determined by multiplex PCR. J Infect Developing 2007; 1: 164-169.
[12] Islam MA, Heuvelink AE, de Boer E, Sturm PD, Beumer RR, Zwietering MH, et al. Shiga toxin-producing Escherichia coli isolated from patients with diarrhoea in Bangladesh. J Med Microbiol 2007; 56: 380-385.
[13] Pao S, Patel D, Kalantari A, Tritschler JP, Wildeus S, Sayre BL. Detection of Salmonella strains and E. coli O157:H7 in faeces of small ruminants and their isolation with various media. Appl Environ Microbiol 2005; 71: 2158-2161.
[14] Phan Q, Mshar P, Rabatsky-Ehr T, Welles C, Howard R, Hadler J. Laboratory-confirmed non-O157 shiga toxin -producing Escherichia coli-connecticut, 2000-2005. MMWR 2007; 56: 29-31.
[15] Santaniello A, Gargiulo A, Borrelli L, Dipineto L, Cuomo A, Sensale M, et al. Survey of shiga-toxin producing Escherichia coli in urban pigeons (Columba Livia) in the City of Napoli, Italy. Ital J Anim Sci 2007; 6: 313-316.
[16] Badouei MA, Zahraei Salehi T, Rabbani Khorasgani M, Tadjbakhsh H, Nikbakht Brujeni G. Occurrence and characterisation of enterohaemorrhagic Escherichia coli isolates from diarrhoeic calves. Comp Clin Pathol 2010; 19: 295-300.
[17] Bidet P, Mariani-Kurkdjian P, Grimont F, Brahimi N, Courroux C, Grimont P, et al. Characterization of Escherichia coli O157:H7 isolates causing haemolytic uraemic syndrome in France. J Med Microbiol 2005; 54: 71-75.
[18] Kawano K, Okada M, Haga T, Maeda K, Goto Y. Relationship between pathogenicity for humans and stx genotype in shiga toxinproducing Escherichia coli serotype O157. Eur J Clin Microbiol Infect Dis 2008; 27: 227-232.
[19] Mattar S, Vasquez E. Escherichia coli O157:H7 infection in Colombia. Emerg Infect Dis 1998; 4: 126-127.
ment heading
10.1016/S1995-7645(14)60182-6
*Corresponding author: Mohammad Kargar, Ph.D, Associate Professor, Department of Microbiology, Islamic Azad University, Jahrom Branch, Iran.
Tel: +98 9173149203
E-mail: mkargar@jia.ac.ir
杂志排行
Asian Pacific Journal of Tropical Medicine的其它文章
- Inhibition of advanced glycation endproducts formation by Korean thistle, Cirsium maackii
- Anti-hypercholesterolemic effect of kenaf (Hibiscus cannabinus L.) seed on high-fat diet Sprague dawley rats
- Susceptibility to temephos, permethrin and deltamethrin of Aedes aegypti (Diptera: Culicidae) from Muang district, Phitsanulok Province, Thailand
- Cloning, expression, purification and bioinformatic analysis of 2-methylcitrate synthase from Mycobacterium tuberculosis
- Larvicidal, ovicidal and repellent activities of marine sponge Cliona celata (Grant) extracts against Anopheles stephensi Liston (Diptera: Culicidae)
- Characterization of mycobacterium isolates from pulmomary tuberculosis suspected cases visiting Tuberculosis Reference Laboratory at Ethiopian Health and Nutrition Research Institute, Addis Ababa Ethiopia: a cross sectional study
