Characterization of mycobacterium isolates from pulmomary tuberculosis suspected cases visiting Tuberculosis Reference Laboratory at Ethiopian Health and Nutrition Research Institute, Addis Ababa Ethiopia: a cross sectional study
2015-12-08BiniamMathewosNigatuKebedeTesfuKassaAdaneMihretMuluworkGetahun
Biniam Mathewos, Nigatu Kebede, Tesfu Kassa, Adane Mihret, Muluwork Getahun
1Department of Immunology and Molecular biology, School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia
2Aklilu Lemma Institute of Pathobiology, College of Health Sciences, Addis Ababa University
3Armaour Hanson Research Institute, Adiss Ababa
4Ethiopian Health and Nutrition Research Institute, Addis Ababa
Characterization of mycobacterium isolates from pulmomary tuberculosis suspected cases visiting Tuberculosis Reference Laboratory at Ethiopian Health and Nutrition Research Institute, Addis Ababa Ethiopia: a cross sectional study
Biniam Mathewos1*, Nigatu Kebede2, Tesfu Kassa2, Adane Mihret3, Muluwork Getahun4
1Department of Immunology and Molecular biology, School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia
2Aklilu Lemma Institute of Pathobiology, College of Health Sciences, Addis Ababa University
3Armaour Hanson Research Institute, Adiss Ababa
4Ethiopian Health and Nutrition Research Institute, Addis Ababa
ARTICLE INFO
Article history:
Received 26 October 2014
Received in revised form 10 November 2014
Accepted 22 December 2014
Available online 20 January 2015
NTM
Mycobacterium tuberculosis complex
Mycobacteria growth indicator tube
Lowenstein Jensen media
Objective: To characterize mycobacterium isolates from pulmomary tuberculosis suspected cases visiting National Tuberculosis Reference Laboratory at Ethiopian Health and Nutrition Research Institute, for diagnosis of pulmonary tuberculosis from January 4 to February 22, 2010 with total samples of 263. Methods: Sputum specimens were collected and processed; the deposits were cultured. For culturing Lowenstein Jensen medium (LJ) and Mycobacteria Growth Indicator Tube (BACTEC MGIT 960) were used. Capilia Neo was used for detecting NTM isolates from isolates of BACTEC MGIT 960. In Armauer Hansen Research Institute, Addis Ababa Ethiopia, Deletion typing PCR method for species identification (from confirmed Mycobacterium tuberculosis complex (MTBC) isolates by Capilia Neo) was done. Results: Out of 263 enrolled in the study, 124 and 117 of them were positive for mycobacterium growth by BACTEC MGIT 960 and LJ culture method, respectively. From BACTEC MGIT 960 positive media of 124 isolates, 117 were randomly taken to perform Capilia TB Neo test. From these 7 (6%) of them were found to be NTM and 110 (94%) were MTBC. From these 110 MTBC isolates, 81 of them were randomly taken and run by the deletion typing RD9 PCR method of molecular technique. Out of these 78 (96.3%) were found to be species of Mycobacterium tuberculosis and 3 (3.7%) were found to be not in the MTBC. Regarding the types of methods of culture media, Mycobacteria Growth Indicator Tube (BACTEC MGIT 960) method was found to have excellent agreement (with kappa value of 0.78) with the routine method of LJ. Conclusions: Pulmonary tuberculosis suspected cases visiting the National Tuberculosis Reference Laboratory at EHNRI that were confirmed to be pulmonary tuberculosis are caused by the species of Mycobacterium tuberculosis, hence treatment regimen including pyrazinamide can be applied to the patients as the first choice in the study area in Addis Ababa, Ethiopia. There is indication of the presence of NTM in patients visiting the tuberculosis reference laboratory and this is important because NTM is known to cause pulmonary disease similar with sign and symptom of pulmonary tuberculosis but different in treatment. BACTEC MGIT 960 has excellent agreement with LJ media but it has high tendency of having high contamination rate unless a better decontamination method is designed.
1. Introduction
Mycobacteria are aerobic and nonmotile bacteria (except for the species Mycobacterium marinum, which has been shown to be motile within macrophages) that are characteristically acid-alcohol fast. There are different classes of isolates of mycobacterium. These are mycobacterium complex, Mycobacterium leprae and non tuberculosis mycobacterium (NTM); Mycobacterium tuberculosis (M. tuberculosis) complex (MTBC) members are causative agents of human and animal tuberculosis. Species
in this complex include M. tuberculosis, the major cause of human tuberculosis, Mycobacterium bovis (M. bovis), M. bovis BCG, Mycobacterium africanum (M. africanum), Mycobacterium canetti, Mycobacterium microti and Mycobacterium pinnipedii[1].
NTM are widely distributed in the environment, particularly in wet soil, marshland, streams, rivers and estuaries; different species of NTM prefer different types of environment. Human disease is believed to be acquired from environmental exposures, and unlike tuberculosis and leprosy, there has been no evidence of animal-tohuman or human-to-human transmission of NTM, hence the alternative label environmental bacteria is used and NTM are frequently isolated from Oregon residents; more than one-half of all isolates likely represent true disease. Pulmonary NTM is most common among elderly women, and Mycobacterium avium causes most disease[2,3].
Most NTM disease cases involve the species Mycobacterium avium complex, MAC, Mycobacterium abscessus, Mycobacterium fortuitum and Mycobacterium kansasii. Mycobacterium abscessus is being seen with increasing frequency and is particularly difficult to treat; rapidly growing NTMs are implicated in catheter infections, postlasic, skin and soft tissue (especially post-cosmetic surgery) and pulmonary infections[3]. But there is no much scientific information about the existence of NTM in Ethiopia so far.
As to M. tuberculosis complex isolates is concerned, there is an urgent need to evolve and apply techniques that not only rapidly identify but also characterize tubercle bacilli to facilitate epidemiological studies. Investigations on the epidemiology of tuberculosis need strain or species specific markers, which can be used to differentiate M. tuberculosis isolates. DNA based technology is now available for molecular characterization of M. tuberculosis[4].
It is generally accepted that different strains or species of M. tubetculosis complex have distinctive epidemiological and clinical characterstics such as virulence and clinical presentation, and that beahaviour in animal models appear to be strain or species dependent for example some M. tuberculosis strain are noted for their dissimination and acquisition of drug resistance while others tend to predominate limited locals. Therefore molecular typing of M. tuberculosis isolates is useful in elucidating the natural history of the tuberculosis epidemic and evaluating tuberculosis control efforts[5]. If we review different literature about the proportion of different species of M. tuberculosis complexes, 10%-15% of human tuberculosis infection in developing countries is caused by M. bovis. However, the contribution of M. bovis to the current tuberculosis epidemic is unknown in developing countries. In addition, little is known about the species/strains of mycobacterium that circulate in many developing countries, including Ethiopia[6].
Based on review of the literatures, there is no current study on characteriaton of mycobacterium isolates in Addis Ababa, Ethiopia. Therefore the purpose of this study was to provide preliminary information on the existence and or extent of NTM and type of species of M. tuberculosis complex isolates circulating cases in Addis Ababa, Ethiopia, from pulmonary tuberculosis suspected cases, in particular from cases visiting the National Tuberculosis Reference Laboratory at Ethiopian Health and Nutrition Research Institute (EHNRI).
2. Materials and methods
2.1. Study area
The study was conducted at Addis Ababa, Ethiopia. Addis Ababa is the capital city of Ethiopia and it was established in 1889 and is now a city of 2.7 million people. Addis Ababa is a grassland biome, located at 9°1′48″N 38°44′24E″. The city lies at the foot of Mount Entoto. From its lowest point, around Bole International Airport, at 2 326 meters (7 631 ft) above sea level in the southern periphery, the city rises to over 3 000 metres (9 800 ft) in the Entoto Mountains to the north. Based on the 2007 Census conducted by the Central Statistical Agency of Ethiopia (CSA), Addis Ababa has a total population of 2 739 551, of whom 1 305 387 are men and 1 434 164 women; all of the populations are urban inhabitants. All Ethiopian ethnic groups are represented in Addis Ababa due to its position as capital of the country[7].
2.2. Study design and target populations
A cross sectional study was conducted from January 4 to February 22 and target populations were pulmomary tuberculosis suspected cases visiting for diagnosis of pulmonary tuberculosis at National Tuberculosis Reference Laboratory of EHNRI.
2.2.1. Sample size and sampling
All pulmomary tuberculosis suspected cases who requested for sputum examination during the study period were included in the study and a total of 263 specimens were collected.
2.2.2. Specimen collection and processing
After getting signed informed consent from each study participant, sputum was collected. The amount of the sputum used was 2-5 mL and then these amounts of the sputum were transferred into centrifuge tube and mixed with equal volume of N-acetyl-L-cysteine-sodium hydroxide (NALC-NAOH) solution. Then vortexing, not more than 20
sec, was done and it was kept for 15 min at 20℃-25℃, for giving time for decontamination process. Then Phosphate Buffer was filled to the top of the tube (50 mL marks on the tube) and vortexing were done. After it was centrifuged at 3 000 g for 20 min and the supernatant were poured off (discarded) and some portion of the deposits were used for culture and the other portion was used for Ziehl Neelsen (ZN) staining method[8].
2.2.3. Culture
The deposits (100 μL) were inoculated on to the slopes of Lowenstein Jenson (LJ) medium labeled with the ID number of the study participants. On Mycobacterium Growth Indicator Tube (BACTEC MGIT 960), 0.8 mL of the resultant enrichment was added to each Mycobacterium Growth Indicator Tube (BACTEC MGIT 960 tube) prior to inoculation and 0.5 mL of the processed/concentrated specimen (the deposits) were inoculated to the tubes (BACTEC MGIT 960 tubes).
After inoculation, LJ medium were incubated at 37 ℃ in incubator and the BACTEC MGIT 960 tube were loaded into slots of the BACTEC MGIT 960 machine. LJ media were incubated until typical growth were seen (rough, buff, creamy colonies) and in case of BACTEC MGIT 960, the tubes were incubated in the instrument until instrument flags positive or negative by its indicator lights. For control purpose distilled water were inoculated before the sample inoculation (start control) and the distilled water were inoculated after sample inoculation (end control).
BACTEC MGIT 960 positive tubes were further sub cultured to blood agar plate (BAP) and slide were made from the culture of the BACTEC MGIT 960 tubes for ZN staining. This was intended to distinguish whether the positivity of the BACTEC MGIT 960 is because of mycobacterium or contaminants. This also helps for calculating the contamination rate of BACTEC MGIT 960 machine.
Those isolates which were grown on BAP after overnight incubation but no acid fast bacilli (AFB) seen on Ziehl Neelson staining were considered as contaminants. And also those which shown no growth on BAP after overnight incubation and no AFB seen on ZN staining but other bacteria (not AFB) seen in the ZN staining were considered as contaminants but those which shown no growth on BAP but AFB were seen on Ziehl Neelsen staining were considered mycobacterium isolates. When there were growths on BAP after overnight incubation and also AFB seen on Ziehl Neelsen staining from the BACTEC MGIT 960 culture, it was considered as presence of both contaminants and mycobacterium isolates. To calculate the contamination rate of BACTEC MGIT 960 machine, the calculation used were: (Contaminated BACTEC MGIT 960 tubes÷total inoculated BACTEC MGIT 960 tubes) ×100[8].
2.2.4. Capilia TB-Neo test (Immunochromatographic method)
Those samples that showed growth in BACTEC MGIT 960 tubes were taken for Capilia TB Neo test to differentiate whether the isolate (the mycobacterium) is Mycobacterium complex or NTM. The test was performed by taking isolates from BACTEC MGIT 960 (liquid culture) and 80-100 μL of the specimen were droped on the specimen placing area of the test place of Capilia TB-Neo test device and after waiting 15 min, the reading area of the test plate were observed. Test results were interpreted within 60 min. a positive result indicate that the mycobacterium is MTBC but a negative result indicate the isolate is NTM[9].
2.3. Molecular techniques
2.3.1. Polymerase chain reaction (Deletion typing method)
From the isolates of MTBC that were isolates which were positive by Capilia TB-Neo Test, a molecular techniques were applied in order to differentiate which of these isolates are M. tuberculosis, M. bovis or M. africanum by a molecular technique. The Capilia Neo positive isolates (MTBC) which were grown in BACTEC MGIT 960 liquid media were heat killed by 800for one hour. The heat killed isolates were packed by biosefty plastics and sent to Armauer Hansen Research Institute (AHRI) for molecular characterization of the isolates. Polymerase chain reaction (PCR) was performed to characterize mycobacterium isolates. This is made by PCR amplification of species-specific DNA fragments. The method is based on specific “Region of Difference”. These are different like RD4, RD9 and RD10. In this study, RD9 was used. A strain with RD9 present is M. tuberculosis. This deletion typing is designed as a multiplex PCR using three different primers which are RD9-flankFW (5’-AAC ACG GTC ACG TTG TCG TG-3’), RD9-internalRev (5’-TTG CTT CCC CGG TTC GTC TG-3’) and RD9-FlankRev (5’-CAA ACC AGC AGC TGT CGT TG-3’). In this method mycobacterium control strain H37Rv (ATCC 25618) for M. tuberculosis and AF2122/97(ATCC BAA-935) for M. bovis were used[10].
Ultra violet light was applied in the safety cabinet for at least 15 min before starting the work. With “DNA away” (DNA degrading agent) the safety cabinet and the surrounding areas were cleaned. The HotStarMaster mix (which includes DNA polymerase, buffer, MgCl2and dNTPs.) was prepared on the day of use then Water (7.1 μL), the HotStarMaster Mix (10 μL), and the three primers mentioned above each 0.3 μL, were mixed in a sterile epppendorf tube by using filter tips. Working in a cabinet, 18 μL (7.1 μL+10 μL+0.9 μL) of the total mix were liquated into PCR tubes and each tube was labeled. Eventually these tubes were moved to the designated area in the laboratory for addition of the DNA templates. Therefore, 2 μL of DNA templates
(samples) were added to the respective tubes. These tubes were placed in the PCR Thermocycler and the corresponding PCR programme was started[10].
2.3.2. Agarose gel electrophoresis
Agarose gel electrophoresis was performed to verify the amplification of the desired gene of interest. The PCR products were analyzed by electrophoresis through 1.5% agarose gels containing 0.3 μg/mL ethidium bromide (Bio-Rad Laboratories, Germany) at 100 volts and 50 ampere for 30 to 45 minutes in 50× Tris acetate EDTA (Sigma, Chem. Corp. USA) buffer with pH 8.0. The isolate was considered be M. tuberculosis when a 396 base pair (bp) (RD9-internalRev+RD9-FlankFW) fragment was detected, and when 575 bp fragment was detected, it was considered as M. bovis or M. africanum. This was judged by comparing with the DNA ladder and the positive control[11].
2.4. Statistical analysis
Demographic and laboratory data were collected using Case Record Form. The data were double entered using SPSS 20 software by two data entry clerks on two different computers. And for analyzing the degree of agreement of LJ and BACTEC MGIT 960 methods, Kappa analysis was done. A P-value less than 0.05 were considered statistically significant.
2.5. Ethical considerations
This research project obtained ethical clearance from Institutional Review Board of Aklilu Lemma Institute of Pathobiology before commencement of the actual activities. Informed consent was also obtained from each study participant and guardians before collection of sputum. Informed consent signatures were requested and obtained from each adult study participants and from parents or guardians for adolescents and children.
3. Results
3.1. Age and sex distribution of pulmonary tuberculosis suspected cases
A total of 263 pulmonary tuberculosis suspected cases with the mean age of 32.84 who have come to National Tuberculosis Reference Laboratory of EHNRI, from January 4 to February 22, 2010 were included in the study. From all the study participants, 132 (50.2%) of them were males and 131 (49.8%) of them were females. The age group of 16-30 was the highest with 48.7% percentage of the total study population (Table 1).
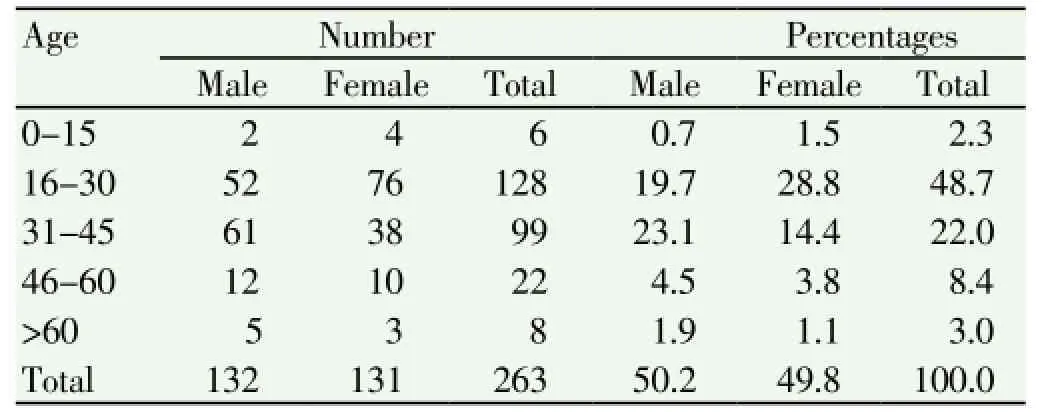
Table 1 Distribution of the visiting pulmonary tuberculosis suspected cases by sex and age.
3.2. Evaluation of BACTEC MGIT 960, diagnostic method for isolation of Mycobacterium isolates
Out of 263 pulmonary tuberculosis suspected cases, 117(44.4%) of them were LJ (solid culture) positive and 124 (47.1%) of them were positive by BACTEC MGIT 960 technique. 129 (49%) of the results were negative by both methods and 106 (40.3%) were positive by both methods but 17(6.4%) of the results were positive by MGIT and negative by LJ media whereas 11(4.1%) were positive by LJ and negative by MGIT (Table 2).
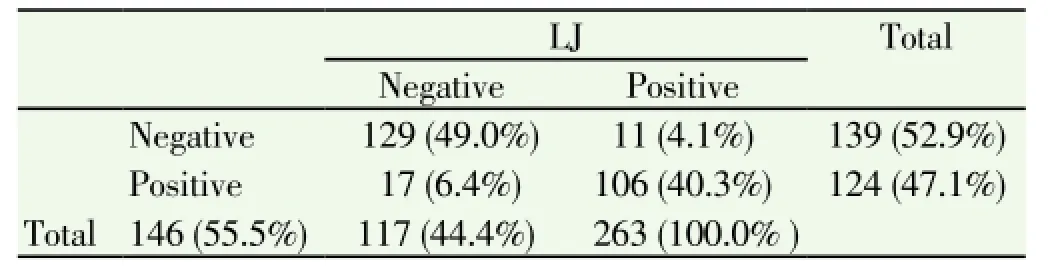
Table 2 Agreement between LJ media (solid culture) and MGIT (liquid culture).
3.3. Kappa analysis for measuring the agreement of MGIT 960 with the routine LJ method
The level of agreement between BACTEC MGIT 960 tubes and LJ method were determined by statistical method of data analysis using kappa. The kappa value was 0.78 which indicated the level of agreement between the two methods was excellent.
3.4. AFBContamination rate of BACTEC MGIT 960
From 263 total samples inoculated into MGIT 960, 58 of them were grown on BAP overnight and were not grown on AFB staining indicating the MGIT 960 tubes were contaminated and this showed a contamination rate of BACTEC MGIT 960 were 22%.
3.5. Determination of extent of NTM by using Capilia TB Neo test
Out of 124 positive samples obtained by MGIT 960, 117 isolates was taken randomly and tested for Capilia Neo and from this 110 (94%) of them were positive showing that they
were under MTBC and 7 (6%) of them were negative in which it indicated that the isolates were NTM.
3.6. Identification of the species of Mycobacterium isolates
From 110 samples that were confirmed to be MTBC (confirmed by Capilia Neo method), 81 of them were randomly taken (because of shortage of primers available) and were run by the deletion typing RD9 PCR which is a method for characterization of members of the MTBC. From these 81 PCR samples, 3 (3.7%) samples were not in the MTBC and all other 78 (96.3%) isolates were confirmed to be species of M. tuberculosis therefore it indicates no other species were identified which are members of MTBC (M. bovis and M. africanum were not identified) (Figure 1).
4. Discussion
In the present study, the isolates identified from patients visiting the National Tuberculosis Reference Laboratory in EHNRI were 7 NTM (6%) and 110 (94%) MTBC from 117 BACTEC MGIT 960 positive isolates. From these MTBC (110 isolates), 81 of them were randomly selected and typed by deletion typing-RD9 characterization method, 78 (96.3%) were found to be species of M. tuberculosis and 3 (3.7%) were not in the MTBC and this result confirmed the low or no level of involvement of M. bovis in human tuberculosis in the study area.
The percentage of NTM found in this study which is 6% were with little differences with a study done in 2005 at Saint Peter tuberculosis specialized hospital laboratory in Addis Ababa in which a 7% of NTM were obtained. This indicates the extent of isolation of NTM in the study area is relatively stable in different study periods[12].
A similar study on characterization of mycobacterium isolates in Egypt showed that from 45 mycobacterium isolates, 44 were M. tuberculosis and only one of them was M. bovis[13]. This difference from our findings may be because of general geographical and/ or habits and cultural difference between the two different populations.
Study conducted in Ghana Mbarara University Teaching Hospital Tuberculosis ward from September 2004 to January 2005 showed similar result with our study findings in that out of 70 similar samples with our study, 69 samples were M. tuberculosis and 1 was not mycobacterium species[14].
The same with the above mentioned studies a study was conducted in Bangladesh to identify M. tuberculosis clinical isolates by a species distinguishable multiplex PCR. The species of mycobacterium isolates identified were similar with our study; some 350 isolates were used for identification and all species of the isolates were founded to be M. tuberculosis[15].
Regarding the assessment of BACTEC MGIT 960 machine, it was evaluated by the present study in the context of the extent of its contamination rate. It was assessed since it was the method that was used to culture the isolates and the isolates grown on it was used in the deletion typing method for molecular characterization. With the Kappa value of 0.78, there was excellent agreement with LJ media which indicated that the method was appropriate to get the isolates. On the other hand, the BACTEC MGIT 960 method showed a 22% contamination rate which shows a high rate of contamination. These indicate there is high contamination rate when we compared it with other similar studies.
Because of the above mentioned facts regarding evaluation BACTEC MGIT 960 which indicated that the method favors almost all types of bacterial growth, there should be further study on effective decontamination process so that having effective decontaminator specifically kills all bacteria except mycobacterium isolates.
In addition to this, in the present study, high number of positivity was obtained that means, 17 out of 263 results (6.4%) which were negative by LJ it was positive by BACTEC MGIT 960. Out of these positive isolates 8 (47%), of them were randomly selected and Capilia Neo test were done and all were positive showing the high tendency of true positivity of BACTEC MGIT 960 machine.
Our finding on comparing the degree of detection of mycobacterium isolates of the specimens demonstrates that 47% and 44% for BACTEC MGIT 960 and Lowenstein Jenson medium (LJ) method respectively. This indicates the former is better in this criterion. Additionally, this finding was similar with the study done in the Gambia with 147 samples which demonstrated that BACTEC MGIT 960 degree of detection to be 57.1% and LJ medium degree to be 43.5%. In the rate of contamination also, 12% was obtained in the study done of Gambia but in our study higher contamination rate that was (22%) obtained. Kappa value between the methods of our study (BACTEC MGIT 960 and LJ media) is 0.78 which is different from the study conducted in the Gambia which
demonstrates a 0.69 kappa value (interpreted as fair to good agreement)[16].
On the other hand, similar study done in Taiwan with the sample size of 121, to evaluate BACTEC MGIT 960, the contamination rate was found to be 13% which is lower than our study of 22% of contamination rate[17].
Our study had also some limitations, like in the case of detecting NTM isolates; the test was done only once for each study participant even though for confirmation repetition of the test is important. Besides, eventhough we were be able to distinguish whether the mycobacterium isolates were MTBC or NTM isolates, we could not show what were the species under the NTM and this is because of the lack of opportunity in obtaining enough set up for species differentiation of the NTM isolates.
Because pulmonary tuberculosis suspected cases visiting the National Tuberculosis Reference Laboratory of EHNRI that were confirmed to be pulmonary tuberculosis are caused by the species of M. tuberculosis, treatment regimen including pyrazinamide can be applied to the patients as the first choice in study area in Addis Ababa.
There is indication of the presence of NTM in pulmomary tuberculosis suspected cases patients visiting the reference laboratory and this is important because NTM is known to cause pulmonary disease similar with sign and symptom of pulmonary tuberculosis.
Regarding the methods of culture for the growth of the isolates, BACTEC MGIT 960 has been founded to have excellent agreement with LJ media even though it had high contamination rate. Hence, a better decontamination method should be designed.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgements
We would like to appreciate and give our heartfelt thanks to EHNRI for helping me in providing some literatures and the laboratory work. Our thanks also go to Aklilu Lemma Institute of Pathobiology for giving me this chance of conducting the research project.
[1] Ryan KJ, Ray CG. Sherris medical microbiology. 4th edition. McGraw Hill; 2004.
[2] Maureen Cassidy P, Ashlen S, Erin M, Winthrop KL. Nontuberculous mycobacterial disease prevalence and risk factors: A changing epidemiology. Clin Infect Dis 2009; 49: 12.
[3] American Lung Association. [Online] Available at: http:// www.lung.org/lung-disease/nontuberculosis-mycobacterium/ understanding-nontuberculous.html.
[4] Chauhan A, Chauhan DS, Parashar D, Gupta P, Sharma VD, Sachan AS, et al. DNA fingerprinting of Mycobacterium tuberculosis isolates from agra region by IS6100 probe. Ind J Medical Microbiol 2004; 22(4): 238-240.
[5] Gencer B, Shinnick T. Molecular genotyping of Mycobacterium tuberculosis isolates from Turkey. American J Infectious Dis 2005; 1(1): 5 -11.
[6] NUFU project: Studies of molecular epidemiology, clinical epidemiology and immunology of tuberculosis in pastoral communities and their livestock in Ethiopia. Part Ⅱ. Cambridge University. [Online] Available at: http:// www.med.uio.no/iasam/ inthel/english/research/molecularepidemiology.
[7] Census. Addis Ababa, Central Statistical Agency; 2007.
[8] MGIT procedure, manual. Foundation for Innovative New Diagnostics. 2006. [Online] Available at: http://www. finddiagnostics.org/export/sites/default/resourcecentre/ presentations/iuatld.
[9] Paramasivan CN. Foundation for innovative new diagnostics (FIND), Status of Capilia evaluation and demonstration projects. 2007. [Online] Available at: http://www.finddiagnostics.org/export/ sites/default/resourcecentre/presentations/iuatld_38th_union_ conf_capetown_2007/status_of_capilia_studies.pdf.
[10] Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Nati Acad Sci 2002; 99: 3684-3689.
[11] Smith NH, Gordon SV, Rua-Domenech RD, Clifton-Hadley RS, Hewinson RG. Molecular evolution of Mycobacterium bovis. Nat Rev Microbial 2006; 4: 670-681.
[12] Desta K, Asrat D, Lemma E, Gebeyehu M, Feleke B. Prevalence of smear negative pulmonary tuberculosis among patients visiting St. Peter’s Tuberculosis Specialized Hospital, Addis Ababa, Ethiopia. Ethiop Med J 2009; 47(1): 17-24.
[13] Abbadi S, Hadidy G, Gomaa N, Cooksey R. Strain differentiation of Mycobacterium tuberculosis complex isolated from sputum of pulmonary tuberculosis patients. Int J Infect Dis 2009; 3(2): 236-242.
[14] Byarugaba F, Etter MC, Godreuil S, Grimaud P. Pulmonary tuberculosis and Mycobacterium bovis, Uganda. EID Journalhome 2009; 15(1): 124-125.
[15] Nakajima C, Rahim Z, Fukushima Y, Sugawara I, Zanden A, Tamaru A, et al. Identification of Mycobacterium tuberculosis clinical isolates in Bangladesh by a species distinguishable multiplex PCR. BMC Infectious Dis 2010; 10: 118.
[16] Otu J, Antonio M, Cheung Y.B, S Donkor S, De Jong BC, Corrah T, et al. Comparative evaluation of BACTEC MGIT 960 with BACTEC 9000 MB and LJ for isolation of mycobacteria in The Gambia. J Infect Dev Countries 2008; 2(3): 200-205.
[17] Huang TS, Chen CS, Susan Lee SJ, Huang WK, Liu YC. Comparison of the BACTEC MGIT 960 and BACTEC 460TB systems for detection of mycobacteria in clinical specimens. Ann Clin & Laboratory Sci 2001; 31: 279-283.
ment heading
10.1016/S1995-7645(14)60184-X
*Corresponding author: Biniam Mathewos, Department of Immunology and Molecular biology, School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia.
Tel: 251-911-855020
E-mail: fikirbinny@gmail.com
杂志排行
Asian Pacific Journal of Tropical Medicine的其它文章
- Inhibition of advanced glycation endproducts formation by Korean thistle, Cirsium maackii
- Anti-hypercholesterolemic effect of kenaf (Hibiscus cannabinus L.) seed on high-fat diet Sprague dawley rats
- Susceptibility to temephos, permethrin and deltamethrin of Aedes aegypti (Diptera: Culicidae) from Muang district, Phitsanulok Province, Thailand
- Cloning, expression, purification and bioinformatic analysis of 2-methylcitrate synthase from Mycobacterium tuberculosis
- Prevalence of shiga toxins (stx1, stx2), eaeA and hly genes of Escherichia coli O157:H7 strains among children with acute gastroenteritis in southern of Iran
- Larvicidal, ovicidal and repellent activities of marine sponge Cliona celata (Grant) extracts against Anopheles stephensi Liston (Diptera: Culicidae)
