Discrimination for minimal hepatic encephalopathy based on Bayesian modeling of default mode network
2015-03-01JiaoYunWangXunhengTangTianyuZhuXiqiTengGaojun
Jiao Yun Wang Xunheng Tang Tianyu Zhu Xiqi Teng Gaojun
(1 Medical School, Southeast University,Nanjing 210009, China)(2 College of Life Information Science and Instrument Engineering, Hangzhou Dianzi University, Hangzhou 310018, China)(3 Radiology Department, The Second Hospital of Nanjing, Nanjing 210003, China)
Discrimination for minimal hepatic encephalopathy based on Bayesian modeling of default mode network
Jiao Yun1Wang Xunheng2Tang Tianyu1Zhu Xiqi3Teng Gaojun1
(1Medical School, Southeast University,Nanjing 210009, China)(2College of Life Information Science and Instrument Engineering, Hangzhou Dianzi University, Hangzhou 310018, China)(3Radiology Department, The Second Hospital of Nanjing, Nanjing 210003, China)
Abstract:In order to classify the minimal hepatic encephalopathy (MHE) patients from healthy controls, the independent component analysis (ICA) is used to generate the default mode network (DMN) from resting-state functional magnetic resonance imaging (fMRI). Then a Bayesian voxel-wised method, graphical-model-based multivariate analysis (GAMMA), is used to explore the associations between abnormal functional integration within DMN and clinical variable. Without any prior knowledge, five machine learning methods, namely, support vector machines (SVMs), classification and regression trees (CART), logistic regression, the Bayesian network, and C4.5, are applied to the classification. The functional integration patterns were alternative within DMN, which have the power to predict MHE with an accuracy of 98%. The GAMMA method generating functional integration patterns within DMN can become a simple, objective, and common imaging biomarker for detecting MHE and can serve as a supplement to the existing diagnostic methods.
Key words:graphical-model-based multivariate analysis; Bayesian modeling; machine learning; functional integration; minimal hepatic encephalopathy; resting-state functional magnetic resonance imaging (fMRI)
Received 2015-07-17.
Biographies:Jiao Yun (1983—), male, doctor, lecturer, yunjiao@seu.edu.cn; Teng Gaojun (corresponding author), male, professor, gjteng@vip.sina.com.
Foundation items:The National Natural Science Foundation of China (No.81230034, 81271739, 81501453), the Special Program of Medical Science of Jiangsu Province (No.BL2013029), the Natural Science Foundation of Jiangsu Province (No.BK20141342).
Citation:Jiao Yun, Wang Xunheng, Tang Tianyu, et al.Discrimination for minimal hepatic encephalopathy based on Bayesian modeling of default mode network[J].Journal of Southeast University (English Edition),2015,31(4):582-587.[doi:10.3969/j.issn.1003-7985.2015.04.026]
Minimal hepatic encephalopathy (MHE) is a common neurocognitive complication of liver cirrhosis, which can result in a wide spectrum of neurocognitive impairments[1]. Resting-state functional magnetic resonance imaging (fMRI) can probe the cerebral intrinsic functional architecture and reflect spontaneous neuronal activity at baseline state[2], which is a useful technique for investigating the neuropathological mechanisms in MHE[3].
Brain activity at rest can be spatially organized into a set of large-scale coherent patterns, namely intrinsic connectivity networks (ICNs), including the default mode network(DMN), attention networks and other networks[4]. The DMN, a resting-state network characterized by consistent task-induced deactivation, is negatively correlated with activity in the attention networks[5]. The spontaneous activities in the DMN were proposed to represent inherent patterns for expected usages[6]and the altered intrinsic connectivity correlated with MHE in the DMN[7-10]. There is a vast body of literature regarding the quantification of voxel-wised intrinsic connectivity levels of certain ICNs for MHE[7-10]. Most of these studies were based on general linear mixed models (GLMMs). One widely used GLMM-based method models intrinsic interactions by computing the t-statistic between groups. Another GLMM-based approach is based on the regression model, in which the connectivity change of an ICN is the dependent variable, and a clinical variable is the independent variable. In this approach, associations among clinical variables and rates of change in brain measurements are estimated by the coefficients of the resulting regression model.
The major limitations of the GLMM-based approach were its focus on functional specialization, and its inability to model multivariate interactions between brain regions[11]. To address these problems, previous studies developed a set of algorithms that model interactions among brain regions and a clinical variable which was called graphical-model-based multivariate analysis (GAMMA)[11-13]. The GAMMA is a Bayesian approach to detecting complex nonlinear multivariate associations between image features and a clinical variable. The GAMMA was employed in many neuroscience related application, such as the brain volume morphometry of sickle cell disease[14], and the fMRI data of Alzheimer’s disease[15].
In this study, we aimed to test the hypothesis that the alterative functional integration patterns within the DMN can characterize MHE from healthy controls by using the GAMMA, and the functional integration patterns are sufficiently powerful to predict MHE. To test this hypothesis, we first generated the DMN from resting-state fMRI for each subject. We then applied the GAMMA method to find the abnormalities of functional integration patterns within the DMN. Finally, machine learning algorithms were employed to find the predictive power of these related integration patterns for MHE.
1Materials and Methods
1.1 Subjects
This study was approved by the Research Ethics Committee of Affiliated Zhongda Hospital, Southeast University. Thirty-two cirrhotic patients with MHE and twenty healthy controls were enrolled after written consent. General information about subjects was summarized in Tab.1. Exclusion criteria included current overt HE or other neuropsychiatric diseases, severe organic diseases (such as cardiac disease, advanced pulmonary disorders, and renal failure), taking psychotropic medications, uncontrolled endocrine or metabolic diseases (such as diabetes mellitus and thyroid dysfunction), and alcohol abuse 6 months prior to the study.

Tab.1 Demographic and clinical characteristics of subjects
MHE was diagnosed by the neuropsychiatric tests, including trail making test A (TMT-A), trail making test B (TMT-B), digit symbol test (DST), and block design test (BDT). DST and BDT are subtests of the Wechsler Adult Intelligence Scale-Revised for China (WAIS-RC). MHE was defined if at least two of four tests were impaired two standard deviations beyond normative performance (i.e., TMT-A>68 s, TMT-B>156 s, the raw DST score<23, and the raw BDT score<16). The normative values for the local population were determined by the results of the neuropsychiatric tests for a sample of 160 healthy controls, who were age- and education-matched to the patients in this study.
1.2 Data acquisition
MRI data was collected using a 1.5 T scanner (Vantage Atlas, TOSHIBA). The participants were instructed to rest with their eyes closed, “not to think of anything in particular”, and keep their heads still during fMRI scanning. Functional images were collected with an echo planar imaging sequence (TR/TE=2 500 ms/40 ms, FOV=24 cm×24 cm, matrix=64×64, FA=90°, slice thickness/gap=5 mm/1 mm, 22 axial slices) to measure 120 brain volumes. The high-resolution, three-dimensional T1-weighted images were also obtained with following parameters: 108 sagittal slices, FOV=256 mm×256 mm, matrix=256×256, slice thickness/gap=1.5 mm/0 mm. The MR images were reviewed by an experienced radiologist for quality. We excluded the subjects with poor MR image quality.
1.3 Preprocessing of resting-state fMRI
Fig.1 and Fig.2 display the data- and image-processing pipelines, respectively.
fMRI data processing was carried out using FSL (FMRIB’s Software Library, www.fmrib.ox.ac.uk/fsl)[16-17]. The preprocessing procedures were applied in the following steps: motion correction using MCFLIRT[18]; slice-timing correction using Fourier-space time-series phase-shifting; spatial smoothing using a Gaussian kernel of FWHM 5 mm; mean-based intensity
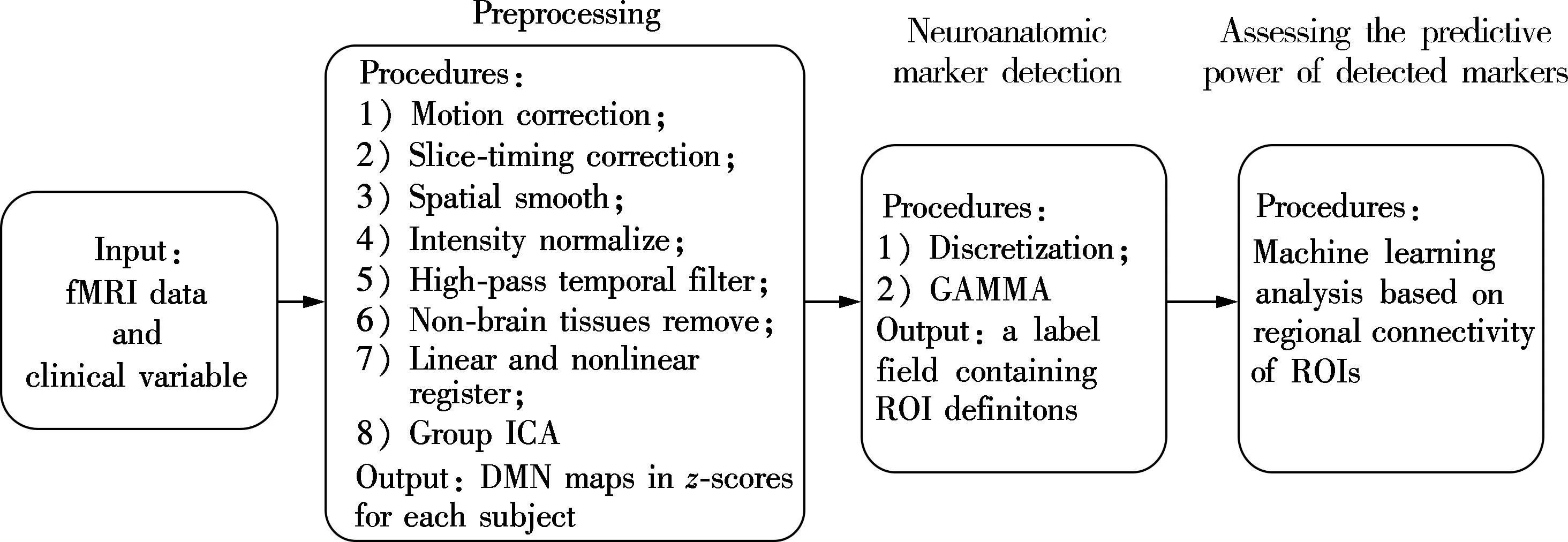
Fig.1 Pipeline for data processing

Fig.2 Pipeline for DMN generation and discretization
normalization of all volumes by the same factor; high-pass temporal filtering (Gaussian-weighted least-squares straight line fitting, with high-pass filter cut-off of 100 s); and removal of non-brain tissue. After preprocessing, the fMRI volumes were registered to the subject’s high-resolution T1-weighted scan using affine registration (FLIRT)[18-19]and subsequently to standard space (MNI152) images using nonlinear registration (FNIRT) with a warp resolution of 10 mm.
1.4 DMN generation
The preprocessed data for all subjects was analyzed with group ICA for the fMRI toolbox (GIFT)[20], which includes double principal component analysis (PCA) reduction, application of ICA, and back-reconstruction for each subject. The reason for using the GIFT was due to the fact that the GIFT has more flexible and adjustable parameters, a more user-friendly interface, and that it can more easily obtain data for the following GAMMA compution.
Our group ICA processing procedures were carried out according to the previous ICNs studies[21-22]. Prior to PCA, the optimal number of components was set to be 54 independent components for all participants, which was estimated using the GIFT dimensionality estimation tool. First, the data from each subject were reduced using PCA according to the selected number of components. Secondly, the data were separated by ICA using the extended infomax algorithm. Finally, independent components and time courses for each subject were back-reconstructed. Following back-reconstruction, the mean spatial maps for all subjects were transformed toz-scores for display. Thez-scores were scaled by the standard deviation of the error term. It can reflect the degree of correlation between the time series of a given voxel and the time series of a specific ICA component[22]. Therefore, thez-scores can be used to measure how much of the standard deviation of the signal was from the background noise; i.e., thez-scores represented the intensity of resting activity at each voxel for each subject in the ICNs studies.
DMN for all participants (32 MHE and 20 controls) were selected by decomposing all the data for each individual into 45 independent components according to the former studies of these three networks[6, 23-24]. The masks were used to analyze brain region changes in the following GAMMA performance.
1.5 Graphical-model-based multivariate analysis
We used a Bayesian voxel-wised method, GAMMA, to explore the associations between brain morphology and the clinical variable. The GAMMA is a between-subject analysis approach that is different from the methods based on general linear models. The GAMMA is nonparametric. It can represent complex nonlinear and linear associations, and it is fully automatic; that is, the GAMMA does not require user-specified parameters such as a significance threshold.
To identify abnormal integration patterns for intrinsic connectivity, we discretized the DMNz-scores maps (normal distribution: mean=0, and standard deviation=1) which were generated from groups ICA. So, for subjecti, if voxelXwas larger than 1, we labeled this voxel as “1” (high connectivity within DMN, active in DMN); otherwise, we labeled it as “0” (lack of connectivity in DMN, non-active in DMN). The result of the discretization step for subjectiwas a high connectivity mapDi. Examples of high connectivity maps were demonstrated in Fig.2. Then, the GAMMA can be applied to automatically detect the representative voxels and generate the ROI for each representative voxel. Also, an embedded model validation step can determine whether the model is a statistical artifact.
We provide a simple description of the GAMMA algorithm in the following. The input of GAMMA was the collection of difference maps and a clinical variableC. The clinical variableCwas a categorical variable representing group membership. In this study,Crepresents either MHE or health control. The region of interests (ROIs), which can predictC, can be identified by the GAMMA. These ROIs can be used as the neuroanatomical markers ofC.
The GAMMA then detected a set of brain regions that were highly predictive ofC. However, because our analysis was at voxel level, brain regions were not predefined. Therefore, the GAMMA also needed to determine group voxels into regions. The two steps, identifying brain regions characterizing group differences and grouping voxels into regions, were referred to as representative voxel detection and brain region delineation, respectively. The GAMMA performed representative voxel detection and brain region delineation in an iterative fashion.
For representative voxel detection, the GAMMA is the forward selection strategy for searching a set of representative voxels. Let RV denote the set of representative voxels. Initially, RV is empty, and the searching space for representative voxels, [VS], contains all voxels. In iterationk, RV containsk-1 representative voxels, and RV={RV1,…,RVi,…,RVk-1}. For voxels in [VS], we identify one voxel that can mostly improve the predictive power of the representative voxel set when adding it to RV. The predictive power of RV is quantified based on the Bayesian Dirichlet equivalent score. Then, we add this voxel RVkto RV. If no such voxel exists, GAMMA then stops.
For brain region delineations, in iterationk, after identifying a representative voxel RVk, we searched [VS] to identify voxels that were probabilistically equivalent to RVk. That was, the probabilistic association between this voxel andCwas similar to that of RVkandC. We used a belief map learning algorithm to solve this problem. RVkand voxels that were probabilistically equivalent to RVkconsist of a single ROI in the label field. An example of such ROI is shown in Fig.3, which shows the spatial distribution abnormalities among MHE and the health controls.
The output of GAMMA was a conditional probability table representing Pr(C,RV1,…,RVi,…,RVn) and a label field to store ROI information. This information can be further analyzed to investigate the predictive power of certain RV.
1.6 Predictive powers evaluation
Machine learning algorithms were used to identify the predictive powers of spatial distribution abnormalities (ROIs generated by GAMMA) for MHE. The inputs of machine learning algorithms were the conditional probability table representing Pr(C,RV1,…,RVi,…,RVn) and a label field mentioned in the above section.
To avoid algorithms bias, we applied several commonly used machine-learning methods, i.e., support vector machines (SVMs), classification and regression trees (CART), logistic regression, Bayesian network, and C4.5, to identify the predictive powers of ROIs.
We evaluated the predictive powers of ROIs based on 10-fold cross-validation. We evaluated the performance of the predictive powers based on the following metrics: sensitivity, specificity and accuracy. Finally, the accuracy predictive model can be obtained according to the three metrics:sensitivity, specificity and accuracy mentioned above. This model was then applied to discriminate MHE patients from normal controls.
2Results
Demographic and clinical characteristics of subjects were displayed in Tab.1. There were no significant differences between groups in age, gender, and education. The neuropsychiatric tests scores were significantly different among MHE and controls.
The Bayesian-based GAMMA methods generated a set of brain regions (altered functional integration patterns) in each of the ICNs to represent group membership (MHE or controls). Then, we tested the predictive powers of these sets of brain regions for DMN. The performances are shown in Tab.2. The predictive model of DMN obtained an accuracy of 98%.

Tab.2 The performances of five machine learning methods for DMN %
We found that C4.5 selected only one set of patterns (one-node tree model) for DMN (see Fig.3). We displayed the corresponding patterns representing root node for DMN (see Fig.4), because the C4.5 algorithm can choose the most predictive variable as its root node.
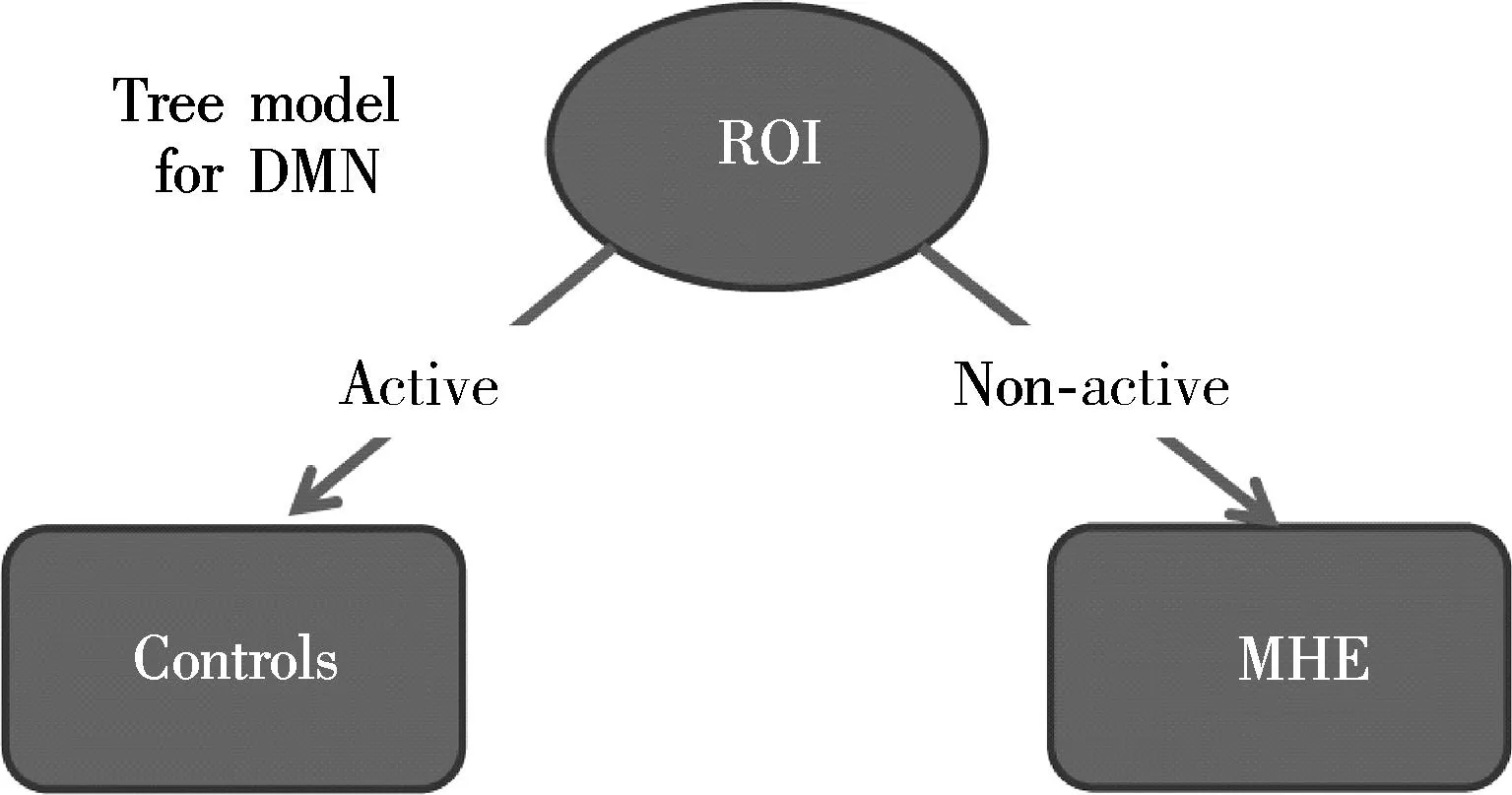
Fig.3 The predictive model tree of all sets of ROIs for DMN generated by C4.5

Fig.4 Subjects in the MHE group with decreased activation in the red ROIs selected by GAMMA
3Discussion
To the best of our knowledge, there has been no previous attempt to focus on the abnormalities functional integration patterns within DMN for MHE. We found that the alterative functional integration patterns in DMN can be potential and complementary markers in early diagnosis of MHE from normal controls.
We used the GAMMA to examine alterative functional integration patterns within DMN for MHE and to identify alterative patterns strongly associated with MHE. Compared with the voxel-wise analysis algorithm based on the general linear model, such as the voxel-wise t-test, the GAMMA is nonparametric and makes no assumption about the probabilistic distribution between DMN functional integration patterns and clinical variables. The GAMMA is an automatic algorithm and it does not require the user to select parameters, such as a significance threshold required for a T-map in the voxel-wise t-test.
In our GAMMA analysis, we detected that MHE patients tended to have lower intrinsic connectivity within DMN. The phenomenon may be associated with metabolic disturbances by ammonia[9], cerebral edema[8], and macroscopical cerebral atrophy[25]. A recent study found that the DMN was impaired in MHE patients through a cross-sectional comparison[9]. Longitudinally, Qi and his colleagues[7]found that progressive abnormalities in resting-state brain activity are associated with MHE development. Thus, it is expected that altered functional connectivity within DMN can be a potential indicator for differential diagnosis of MHE. Abnormal functional integration patterns indicated that the reduction of reaction capabilities of endogenous and exogenous stimuli were reduced in MHE patients. This finding may be an important neuropathophysiological mechanism and may lead to various neurological impairments in MHE (e.g., difficulties in learning ability, working memory, response inhibition, visual perception, and visuoconstructive ability), considering the fundamental role of attention in these cognitive abilities.
More importantly, we used five machine-learning methods to generate classifiers, in order to avoid bias with respect to the functional form of the classifier. We found that the altered functional integration in DMN can be used as a potential biomarker to distinguish MHE patients from patients without MHE or controls. This result is exciting, especially given the fact that there is currently no standardized diagnostic test available for MHE[1, 26]. In addition, it should be noted that resting-state fMRI is a convenient technique that can be easily performed by clinicians.
Furthermore, MHE patients can benefit from early diagnosis and subsequent treatment, and they can have the possibility to reduce negative effects for patients’ cognitive functions. However, MHE patients have few recognizable clinical symptoms. From the research point of view, our findings suggest that in MHE studies of the mechanisms underlying neurocognitive deficits, we need to include a functional integration patterns alterative for DMN as a factor. That is, the diagnostic model for MHE should include not only commonly used neuropsychiatric tests variable, but also functional integration patterns within DMN.
4Conclusion
In this paper, we find that alternative functional integration patterns in DMN can distinguish MHE from healthy controls by using the GAMMA, and the functional integration patterns of DMN have strong power in predicting MHE. The GAMMA method generating functional integration patterns of DMN may become a simple, objective, and common imaging biomarker for detecting MHE and can serve as a supplement to existing diagnostic methods.
References
[1]Bajaj J S, Wade J B, Sanyal A J. Spectrum of neurocognitive impairment in cirrhosis: implications for the assessment of hepatic encephalopathy [J].Hepatology, 2009, 50(6): 2014-2021.
[2]Fox M D, Raichle M E. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging [J].NatRevNeurosci, 2007, 8(9):700-711.
[3]Zhang L J, Qi R, Zhong J, et al. Disrupted functional connectivity of the anterior cingulate cortex in cirrhotic patients without overt hepatic encephalopathy: a resting state fMRI study [J].PLoSOne, 2013, 8(1): e53206.
[4]Zhang D, Raichle M E. Disease and the brain’s dark energy [J].NatRevNeurol, 2010, 6(1): 15-28.
[5]Fox M D, Snyder A Z, Vincent J L, et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks [J].ProcNatlAcadSciUSA, 2005, 102(27): 9673-9678.
[6]Fox M D, Corbetta M, Snyder A Z, et al. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems[J].ProcNatlAcadSciUSA, 2006, 103(26): 10046-10051.
[7]Qi R, Zhang L, Wu S, et al. Altered resting-state brain activity at functional MR imaging during the progression of hepatic encephalopathy [J].Radiology, 2012, 264(1): 187-195.
[8]Lin W C, Hsu T W, Chen C L, et al. Connectivity of default-mode network is associated with cerebral edema in hepatic encephalopathy [J].PLoSOne, 2012, 7(5): e36986.
[9]Qi R, Zhang L J, Xu Q, et al. Selective impairments of resting-state networks in minimal hepatic encephalopathy [J].PLoSOne, 2012, 7(5): e37400.
[10]Chen H J, Jiao Y, Zhu X Q, et al. Brain dysfunction primarily related to previous overt hepatic encephalopathy compared with minimal hepatic encephalopathy: resting-state functional MR imaging demonstration [J].Radiology, 2013, 266(1):261-270.
[11]Chen R, Herskovits E H. Graphical model based multivariate analysis (GAMMA): an open-source, cross-platform neuroimaging data analysis software package [J].Neuroinformatics, 2012, 10(2):119-127.
[12]Chen R, Herskovits E H. Graphical-model-based morphometric analysis [J].IEEETransMedImaging, 2005, 24(10): 1237-1248.
[13]Chen R, Herskovits E H. Graphical-model-based multivariate analysis of functional magnetic-resonance data [J].Neuroimage, 2007, 35(2): 635-647.
[14]Chen R, Pawlak M A, Flynn T B, et al. Brain morphometry and intelligence quotient measurements in children with sickle cell disease [J].JDevBehavPediatr, 2009, 30(6): 509-517.
[15]Chen R, Herskovits E H. Clinical diagnosis based on bayesian classification of functional magnetic-resonance data [J].Neuroinformatics, 2007, 5(3): 178-188.
[16]Smith S M, Jenkinson M, Woolrich M W, et al. Advances in functional and structural MR image analysis and implementation as FSL [J].Neuroimage, 2004, 23(Sup 1): 208-219.
[17]Woolrich M W, Jbabdi S, Patenaude B, et al. Bayesian analysis of neuroimaging data in FSL [J].Neuroimage, 2009, 45(Sup): 173-186.
[18]Jenkinson M, Bannister P, Brady M, et al. Improved optimization for the robust and accurate linear registration and motion correction of brain images [J].Neuroimage, 2002, 17(2): 825-841.
[19]Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images [J].MedImageAnal, 2001, 5(2):143-156.
[20]Calhoun V D, Adali T, Pearlson G D, et al. A method for making group inferences from functional MRI data using independent component analysis [J].HumBrainMapp, 2001, 14(3): 140-151.
[21]Qi Z, Wu X, Wang Z, et al. Impairment and compensation coexist in amnestic MCI default mode network [J].Neuroimage, 2010, 50(1):48-55.
[22]Zhang L, Qi R, Wu S, et al. Brain default-mode network abnormalities in hepatic encephalopathy: a resting-state functional MRI study [J].HumBrainMapp, 2012, 33(6): 1384-1392.
[23]Greicius M D, Srivastava G, Reiss A L, et al. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI [J].ProcNatlAcadSciUSA, 2004, 101(13): 4637-4642.
[24]Mantini D, Perrucci M G, Del Gratta C, et al. Electrophysiological signatures of resting state networks in the human brain [J].ProcNatlAcadSciUSA, 2007, 104(32): 13170-13175.
[25]Chen H J, Zhu X Q, Shu H, et al. Structural and functional cerebral impairments in cirrhotic patients with a history of overt hepatic encephalopathy [J].EurJRadiol, 2012, 81(10): 2463-2469.
[26]Ferenci P, Lockwood A, Mullen K, et al. Hepatic encephalopathy-definition, nomenclature, diagnosis, and quantification: final report of the working party at the 11th World Congresses of Gastroenterology, Vienna, 1998 [J].Hepatology, 2002, 35(3): 716-721.
doi:10.3969/j.issn.1003-7985.2015.04.026
杂志排行
Journal of Southeast University(English Edition)的其它文章
- Mitigation of inter-cell interference in visible light communication
- Modified particle swarm optimization-based antenna tiltangle adjusting scheme for LTE coverage optimization
- Distribution algorithm of entangled particles for wireless quantum communication mesh networks
- Kernel principal component analysis networkfor image classification
- CFD simulation of ammonia-based CO2 absorption in a spray column
- Simulation and performance analysis of organic Rankine cycle combined heat and power system
