4-(1H-1,2,4-三唑-1-亚甲基)苯甲酸过渡金属配合物的合成、结构、抑菌活性及DNA 裂解活性
2014-09-21熊萍萍步怀宇陈三平
李 婕 熊萍萍 步怀宇 陈三平
(1西北大学西部资源生物与现代生物技术教育部重点实验室,陕西省生物技术重点实验室,西安710069;2西北大学化学与材料科学学院,合成与天然功能分子化学教育部重点实验室,西安710069)
1 Introduction
Over the past decades,many studies on the rational design of polymeric metal-organic frameworks(MOFs)and their potential applications in catalysis,separation,gas storage,and even biological activity caused much interest in the field of inorganic chemistry.1-4DNAcleavage studies would count for much for the evolution of the new therapeutic reagents and DNAprobes,5-8and drug researches suggest that many anticancer agents,antiviral agents,and antiseptic agents take action through binding to DNA.9-14In addition,transition metal compounds can interact covalently or non-covalently with DNA in the mode of intercalation,groove binding,or external electrostatic binding.15-18This inspires growing interest in the study of the biochemical behavior of these compounds including their interactions with DNA and antifungal activity.19-25
As well-known,triazole derivatives,specifically their corresponding transition metal coordination compounds,have been concerned as a highly effective antifungal fungicide.26In our earlier research on the antifungal activity of copper(II)compounds,novel copper(II)compounds with the ligand(4-(1H-1,2,4-triazol-1-ylmethyl)benzoic acid)showed a higher antifungal effect than those of ligand and CuCl2especially compound 1(the antifungal percentage is 72.5%on Fusarium graminearum).Based on our earlier research,27we are interested in exploring the relationships between DNAcleavage and antifungal activity of the transition metal compounds derived from a triazole ligand.
Hence,we synthesized the ligand(4-(1H-1,2,4-triazol-1-ylmethyl)benzoic acid(HL))and the three transition metal compounds(Cu-L,Ni-L,Co-L).The thermal analyses and luminescent properties of the compounds were investigated.Furthermore,comparative study of the interactions of the compounds with plasmid DNA(pUC 18)as well as the related antifungal activities against five agriculture related fungi was experimentally explored.The remarkable DNA cleavage and antifungal activity suggested that the compounds above would have potential utilization for developing new drugs for agricultures.
2 Experimental
2.1 Materials and methods
All of the reagents were purchased and used without further purification.CuCl2·2H2O(99.9%),NiCl2·6H2O(99.9%),CoCl2·6H2O(99.9%),NaN3(>99.5%),KOH(>86%),methanol anhydrous(>99.5%),N,N-dimethylformamide(>99.5%),4-methylbenzoic acid(>99.5%),succinbromimide(>99.5%),benzoyl peroxide(>99.5%),tetrachloromethane(>99.5%),dichloromethane(>99.5%),and 1H-1,2,4-triazole(>99.5%)were purchased fromXi′anChemblossom Pharmaceutical Technology Co.,Ltd.NaOH(97%)was purchased from Sigma-Aldrich.Elemental analyses(C,H,N)were performed on an Elementar Vario EL III analyzer(USA).Infrared(IR)spectra were recorded on a Tensor 27 spectrometer(Bruker Optics,Ettlingen,Germany)as KBr pellets in the range of 400-4000 cm-1.Powder X-ray diffraction(XRD)patterns were measured on a Bruker D8 Advance X-ray powder diffractometer with Cu Kαradiation(λ=0.15405 nm).Ultraviolet(UV)absorption studies were carried out with a Shimadzu UV-2450 spectrophotometer.Fluorescent spectra were measured at room temperature with an Edinburgh FLSP920 fluorescence spectrometer.Thermogravimetric(TG)measurements were performed with a Netrzsch STA 449C apparatus(Germany)under asimulatednitrogenatmospherewithaheatingrateof 10°C·min-1from room temperature to 1000°C.
2.2 Syntheses
2.2.1 Preparation of 4-(1H-1,2,4-triazol-1-ylmethyl)benzoic acid
The ligand was synthesized according to the literature procedure.28,29A mixture of 4-methylbenzoic acid(5.44 g,40.0 mmol),succinbromimide(7.12 g,40.0 mmol),benzoyl peroxide(0.10 g,412.0 mmol),and tetrachloromethane(60 mL)were refluxed for 5 h.Cooling to room temperature and washing with tetrachloromethane and distilled water.White solid was obtained by recrystallization from dichloromethane.Subsequently,a mixture of KOH(0.30 g,0.50 mmol),the above products(0.22 g,0.10 mmol),and 1H-1,2,4-triazole(0.07 g,0.10 mmol)was dissolved in distilled water(6 mL)and sealed in a 10 mL Teflon-lined stainless steel autoclave after stirring them for 30 min.The mixture was heated at 90 °C for 72 h and cooled to room temperature at a rate of 5 °C·h-1.Colorless crystals(HL)were formed and washed with distilled water.Yield:91.4%.IR(KBr pellet,cm-1)for HL:3453(b),3119(b),2952(w),2363(w),1914(w),1694(b),1515(s),1433(m),1275(s),1141(s),1011(m),919(m),731(s),677(m)(Fig.S1a(see Supporting Information)).m.p.215.1-215.5°C.
2.2.2 Preparation of[Cu0.5L]n(1)
Amixture of HL(20.3 mg,0.10 mmol)and CuCl2·2H2O(17.1 mg,0.10 mmol)was dissolved in distilled H2O(3 mL),Teflonlined stainless reactor at 140 °C for 72 h,cooled to 100 °C at a rate of 5 °C·h-1,and held at this temperature for 10 h.Then,it was cooled to room temperature at the same rate.Purple lump crystals were isolated and washed with distilled water.Yield:45%(based on HL).IR(KBr pellet,cm-1):3455(b),1608(s),1562(m),1371(s),1288(m),1119(m),736(m),674(m)(Fig.S1b).Elemental analyses(%)calculated for C20H16CuN6O4:C 51.29,H 3.41,N 17.95;found:C 51.11,H 3.74,N 17.80.
2.2.3 Preparation of{[Ni(L)2·(H2O)2]·(H2O)2}n(2)
A mixture containing HL(20.3 mg,0.10 mmol),NiCl2·6H2O(23.8 mg,0.10 mmol),NaOH(4.0 mg,0.10 mmol),NaN3(12.6 mg,0.20 mmol),were dissolved in the solution of CH3OH/H2O(6 mL)(1:2,V/V),Teflon-lined stainless reactor at 160°C for 10 h,cooled to 100 °C at a rate of 5 °C·h-1,and held at this temperature for 72 h.Then,it was cooled to room temperature at the same rate.The resulting solution was filtered and transfered in a vial for two weeks.Light blue fusiform crystals were formed and washed with EtOH and dried in air.Yield:90%(based on HL).IR(KBr pellet,cm-1):3455(w),1600(s),1555(s),1398(s),1292(m),1138(m),730(m),678(m)(Fig.S1c).Elemental analysis(%)calculated for C20H24NiN6O8:C 44.85,H 4.48,N 15.70;found:C 44.93,H 4.16,N 15.94.
2.2.4 Preparation of{[Co(L)2·(H2O)2]·(H2O)2}n(3)
Compound 3 was synthesized by the identical pathway with compound 2 except that NiCl2·6H2O was replaced by CoCl2·6H2O(23.8 mg,0.10 mmol)and pink fusiform crystals were obtained in the mother liquor after it standing for seven days.Yield:90%(based on HL).IR(KBr pellet,cm-1):3455(w),1600(s),1555(s),1398(s),1292(m),1138(m),730(m),678(m)(Fig.S1d).Elemental analysis(%)calculated for C20H24CoN6O8:C 44.83,H 4.48,N 15.69;found:C 44.97,H 4.08,N 15.95.
2.3 Single-crystal structure determination
All single crystal X-ray experiments were collected on a Bruker Smart Apex II CCD diffractometer(Germany)equipped with graphite monochromated Mo Kαradiation(λ=0.071073 nm)using ω and φ scan mode at 296(2)K.The single-crystal structures of compounds were both solved by direct methods and refined with full-matrixleast-squares refinements based on F2using SHELXS-97 and SHELXL-97.30,31All non-hydrogen atoms were refined anisotropically.Crystallographic data,data collection parameters,and refinement for 1-3 are listed in Table 1,selected bond distances(nm)and angles(°)are given in Table S1(see Supporting Information).CCDC numbers:970263 for 1,969965,969966 for 2 and 3,respectively.
2.4 Antifungal activity tests
The tested microorganisms in the present study are Fusarium graminearum,Vasa mali,Macrophoma kawatsukai,Colletotrichum,gloeosporioides,and Alternaria alternate.Due to their close relationship to crop,such as wheat,apple,and tobacco,we selected these fungi to carry out the research and valued the compounds in agriculture.The method of determining antifungal activity was according to the radial growth method.32Sterilized hot PDA(potato dextrose agar)nutrient medium(composition:potato(200.0 g),dextrose(15.0 g),agar(18.0 g),and distilled water(1000 mL))and 4 mm diameter hole punch were used in the method.4.64 mg Cu-L(1),5.35 mg Ni-L(2),and 5.35 mg Co-L(3)were used to prepare the 2.5 mmol·L-1mother liquid with distilled water(the compound solutions were prepared by formerly dis-solving in a small quantity of DMSO and then diluting using distilled water).Five final concentration solutions in PDA,0,40,80,120,and 160 µmol·L-1were prepared for compounds 1-3 to against five kinds of fungi respectively.The 0 µmol·L-1treatment was regarded as control.
The titled compounds were mixed with 40 mL PDA in 50 mL centrifuge tubes at each concentration,divided into three Petri dishes,and allowed to solidify.Then dishes were incubated with 4 mm diameter of the fungi culture which was obtained by the hole punch.Then these Petri dishes were cultured at 28°C for four days.The diameter of fungi growth was for measuring the antifungal activities.All procedures of the above were repeated three times.
2.5 DNA cleavage tests
Plasmid DNA(pUC 18)was acquired from Takara Biotechnology(Da Lian)Co.Ltd.Aliquots of plasmid DNA(0.06 μg·μL-1)was mixed with a series of concentrations of the three compounds from 0 to 0.8 µmol·L-1.The mixtures were mixed in the same sequence,with plasmids being added first,followed by the titled three compounds and H2O which was used to replenish the volume in the control.Then,those mixtures were incubated in a DK-8D Thermostatic water bath for 150 min at 37°C.
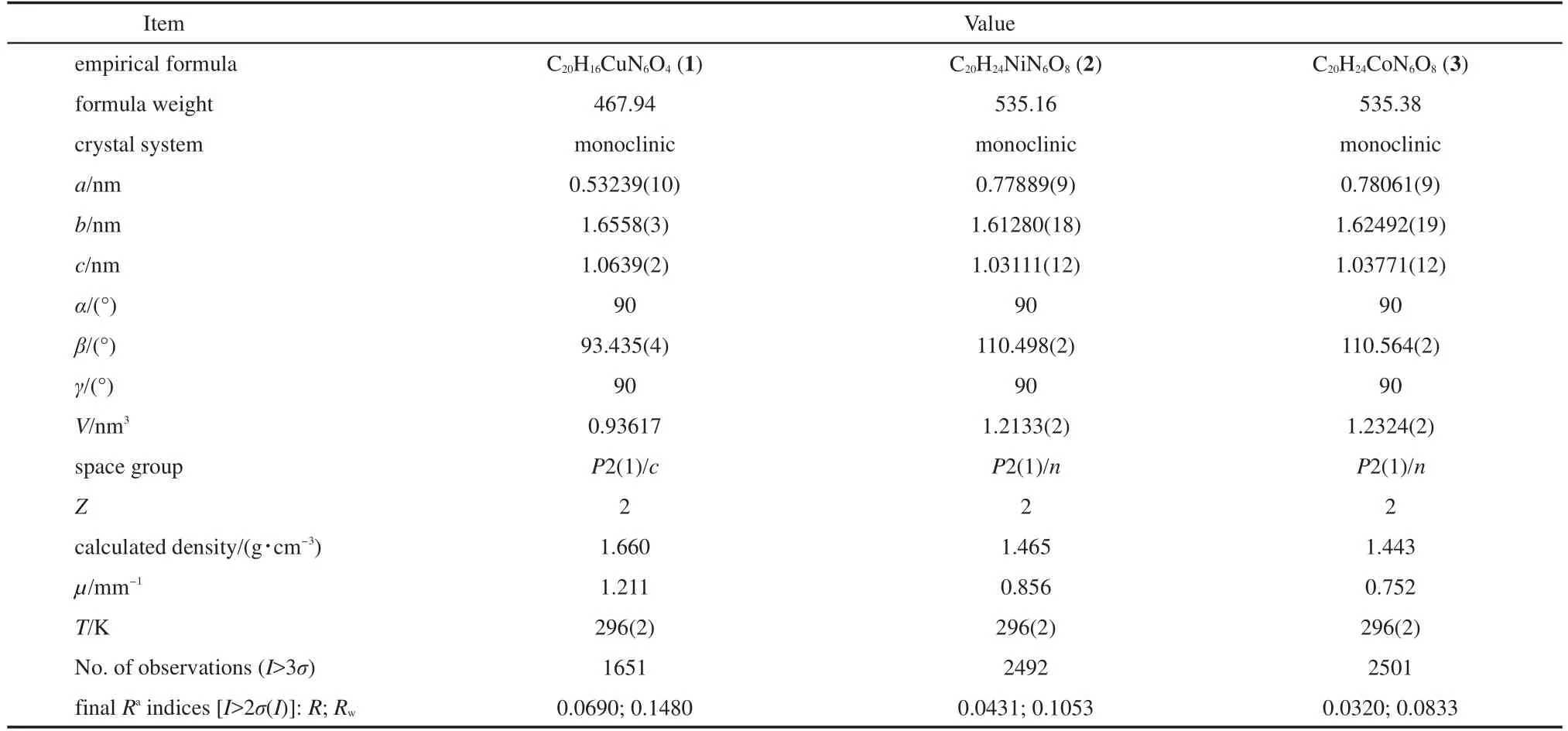
Table 1 Crystallographic data for compounds 1-3
The cleavage reaction could be monitored by agarose gel electrophoresis(AGE).Agarose was purchased from Promega Co.(Germany).Gel electrophoresis experiments were worked out with plasmid DNA,in 0.8%agarose solution,at 100 V 40 min using TAE(tris-acetate-EDTA)buffer(45 mmol·L-1Tris(tris(hydroxymethyl)aminomethane),1 mmol·L-1EDTA(ethylene diamine tetraacetic acid),pH 7.44).The cleavage reactions were terminated by the addition of EDTA and bromphenol blue.The plasmid DNA was stained with 1 µL·mL-1gold view.The cleavage products were analyzed by Gel Doc XR gel documentation and analysis system(Bio-rad).
2.6 Absorption peak of DNA displacement tests
This part was performed by absorption spectral titration,33keeping the concentration of DNA(Salmon sperm)constant while varying the title compound concentration.The displacement was measured at 260 nm HALO DB-30 UV-Vis spectrophotometer(Dynamica).The solution of the compound was dropwise added into 3 mL salmon sperm DNA(50 µg·mL-1),and the mixture solution was monitored by UV-Vis spectrum.
3 Results and discussion
3.1 Crystal structure analysis
Over the past few decades,the metal-triazole-based materials have attracted tremendous attention.Undoubtedly,hydrothermal synthesis provides a convenient method for preparation of such composite materials,allowing more routine structural characterization by single crystal X-ray diffraction.34,35The structures of 1-3 were determined using single crystal X-ray diffraction.1-3 crystallize in space groups P2(1)/c,P2(1)/n,P2(1)/n,respectively.
In compound 1,the crystallographically unique copper(II)atom adopts a distorted octahedral coordination sphere,with four carboxylate O atoms from the different carboxylate groups,bidentate coordination in the equatorial plane,and two N atoms from triazoles in the axial positions(Fig.1(a)).As shown in Fig.1(b),each copper(II)atom coordinates with four ligands,and each ligand coordinates to two Cu ions.As a bridging ligand,triazole N atoms and carboxylate O atoms are involved in coordination with copper(II)to form an infinite one-dimensional(1D)Z-shaped double-chain.
Compounds 2 and 3 have the same structure(Fig.2(a,b)).Hence,only the structure of 2 is described in detail.Single-crystal structure analysis shows a three-dimensional(3D)extended high density framework based on ligand and metal salts building blocks.Each nickel(II)center adopts a distorted octahedral coordination sphere,which is occupied by two O atoms from the coordinated water molecules and two carboxylate O atoms from the different carboxylate groups in the equatorial plane.The coordination sphere is completed by two N atoms from triazoles occupying the axial positions(Fig.2(a)).Anotable feature for the difference between compounds 1 and 2 is that carboxylate group of 2 is monodentate coordination.As shown in Fig.2(c),each nickel(II)atom coordinates with four ligands,and each ligand coordinates to two nickel(II)ions.Like compound 1,as a bridging ligand,the ligand takes end to end coordination modes with the metal ions and forming an infinite 1D Z-shaped double-chain.
There are two guest water molecules in each coordination unit(Fig.2(a)),and each guest water molecule connects to three adjacent chains through hydrogen bonds.As shown in Fig.2(d),the hydrogen bond systems make the whole framework into a network structure.The O…H―O distance is 0.1904 nm and 0.1928 nm,N…H―O distance is 0.2052 nm.In this case,these 1D chains are linked together by such interchain hydrogen bond systems into a 3D framework(Fig.2(e)).
3.2 X-ray powder diffraction and thermal analyses
As shown in the Fig.3,X-ray powder diffraction patterns of the samples of 1-3 are quite similar to the simulated data of the crystal structure.
Compounds 1-3 are air stable,the typical TG curves for the crystal samples of 1-3 were performed between room temperature up to 1000 °C at a heating rate of 10 °C·min-1under nitrogen atmosphere.The polymers are thermally stable up to 285.2,57.8,57.6°C for 1-3,respectively.In the TG curve of 1 shown in Fig.4,with increasing temperature,the whole framework of the compound collapses with a huge mass loss of 42.21%.Thermogravimetric analyses reveal that compounds 2 and 3 have a similar thermal decomposition behavior due to that they possess the same polymeric motif.2 remains stable up to 57.8°C and then undergoes one-step mass loss of 14.2%from 57.8 to 103.4°C,attributing to the loss of the crystal water molecules and the coordinated water molecules.The dehydration substance shows great stability before 339.3°C and then experiences a mass loss.Following that,the intermediates of 1-3 are slowly decomposed,and do not form stable compounds until 1000°C.
3.3 Photoluminescent property
The solid-state photoluminescent properties of ligand and their polymeric compounds 1-3 were investigated at room temperature.The excitation bands(λex)of the three compounds and the ligand are at about 362nm for 1,336nm for 2,373nm for 3,359 nm for the ligand,respectively(Fig.S2(see Supporting Information)and Table 2).As indicated in Fig.5,in the solid state,all compounds and the ligand in this study exhibit strong emission in high energy region.There are two intense fluorescent emission bands(λem)at 421,463 nm for 1,419,457 nm for 2,421,481 nm for 3,respectively(Table 2).The emissions bands of 1-3 and the ligand at about 421 nm show that the nature of ligand plays an important role in the photoluminescence of coordination compounds.The maximum emission of compounds 1-3 excited at discrepancy wavelengths would be attributed to dual effects of the terminal coordination water molecules and the coordination environment around Cu(II).The luminescence of compounds 1-3 may be ascribed to an intraligand phosphorescent emission and ligand-to-metal charge transfer(LMCT).36
3.4 UV-Vis spectra of the ligand and the compounds

Fig.1 (a)Coordination unit of compound 1 and(b)1D Z-shaped double-chain

Fig.2 (a,b)Central atom coordination configurations of 2 and 3,(c)Z-shaped double-chains in 2,and(d,e)hydrogen bond systems in 2 and the 3D framework
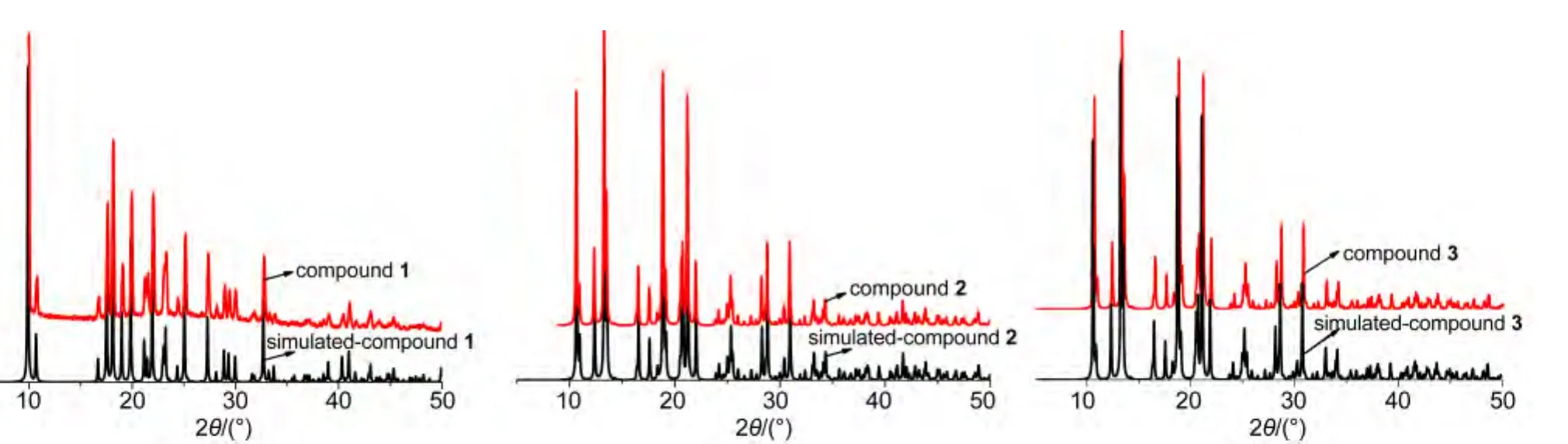
Fig.3 X-ray powder diffraction patterns of 1-3
UV-Vis spectra of the ligand and the compounds could be reliably recorded over the full range.Compared to UV-Vis spectrum of the ligand,little changes in the wavelengths of absorption maxima were observed in those of the corresponding compounds(Fig.S3(see Supporting Information)and Table 3),indicating that the compounds in solvated molecule state occur in dimethyl sulfoxide(DMSO).
3.5 Solubility and molar conductivity of the compounds
All of the three compounds are insoluble in water and methanol,carbon tetrachloride,chloroform,and acetonitrile,while moderately soluble(50 mg/100 mL solvent)in dimethylformamide(DMF)and DMSO.The solid state compounds are fairly stable in air so as to allow physical measurements.Molar conductivities on those compounds fall in the expected range for nonelectrolytes,37,38as shown in Table 3.
3.6 Antifungal activities of compounds
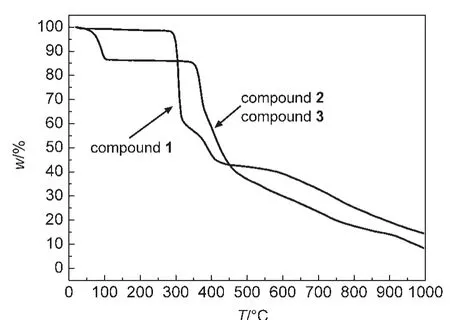
Fig.4 TG curves of compounds 1-3

Table 2 Photoluminescent properties of the ligand and compounds 1-3 in the solid state
According to the results and analyses,the three compounds show a degree of antifungal efficacy(Fig.6 and Table S2(see Supporting Information)).Compound 1 presents the best antifungal capacity among the three compounds.It is worth mentioning that at 120 µmol·L-1antifungal percentage of compound 1 is 73.5%against Fusarium graminearum,whereas compounds 2 and 3 are only 52.2%and 50.0%,repectively(Table S3(see Supporting Information)).Through our calculation,compound 1 always shows a higher antifungal percentage than the other two compounds on all the five fungi.It means that compound 1 has a decent antifungal activity.

Fig.5 Emission spectra of the ligand and compounds in the solid state at ambient temperature

Table 3 Molar conductivities and UV-Vis data for the ligand and compounds in dimethyl sulfoxide(DMSO)solvent at room temperature
The five closely agriculture related fungi show the different sensitivity to the three titled compounds treatment.Vasa mali and Alternaria alternate are much more sensitive than the other three fungi.Data reveal that at the concentration of 160 µmol·L-1the antifungal percentages are almost high to 96.9%on Vasa mali and 94.6%on Alternaria alternate(Table S3(see Supporting Information)).
3.7 DNA cleavage of compounds
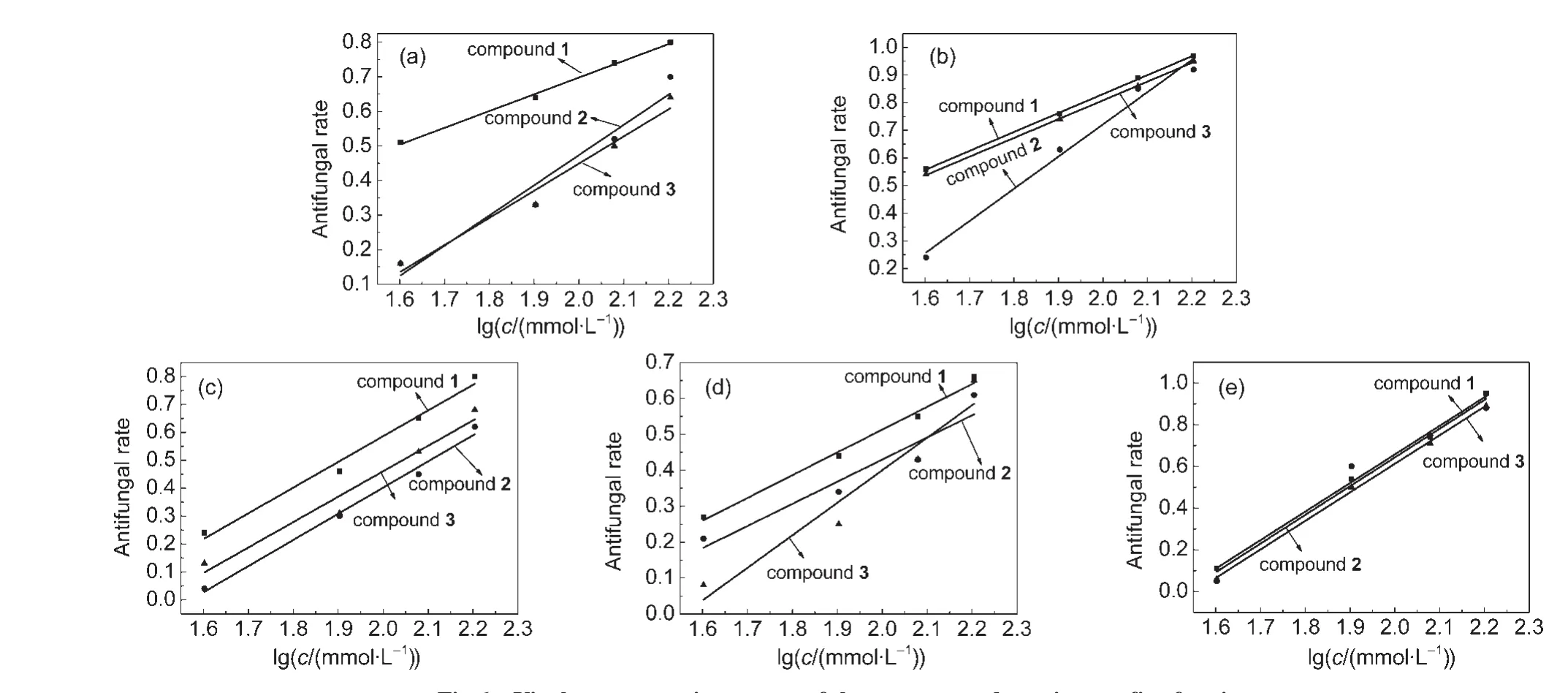
Fig.6 Virulence regression curves of three compounds against on five fungi
Different DNAshapes lead to different moving rates in AGE.39For instance,the intact supercoil form(Form I)will exhibit relatively fast migration when circular plasmid DNA is subjected to AGE.40If cleavage occurs on one strand,supercoil form DNAwill relax to produce a slower-migrating open circular form(Form II).If both strands are cleaved,a linear form DNA(Form III)will be generated and its migration is between the intact supercoil form and open circular form.41Since the plasmid DNA migration pattern under AGE condition is:supercoil>linear>open circular,42that is,Form I>Form III>Form II.Hence,the cleavages obtained by the titled compounds could be carried out and analyzed byAGE.43
As the images after AGE of solutions in different combinations show,the compounds could degrade plasmid DNA(pUC 18).With increasing concentration of compounds,the amount of supercoil(Form I)DNA diminishes gradually.In Fig.7(a),Form I vanishes gradually with the increasing concentration of compound 1,whereas the amount of Form II and Form III begins to increase.At 0.06 µmol·L-1(Lane 4),Form I is cut into Form II and Form III thoroughly and Form III has a significant increase compared to Lanes 2 and 3.Compound 1 promotes complete degradation of plasmid DNAat the concentration of 0.08 µmol·L-1(Lane 5).
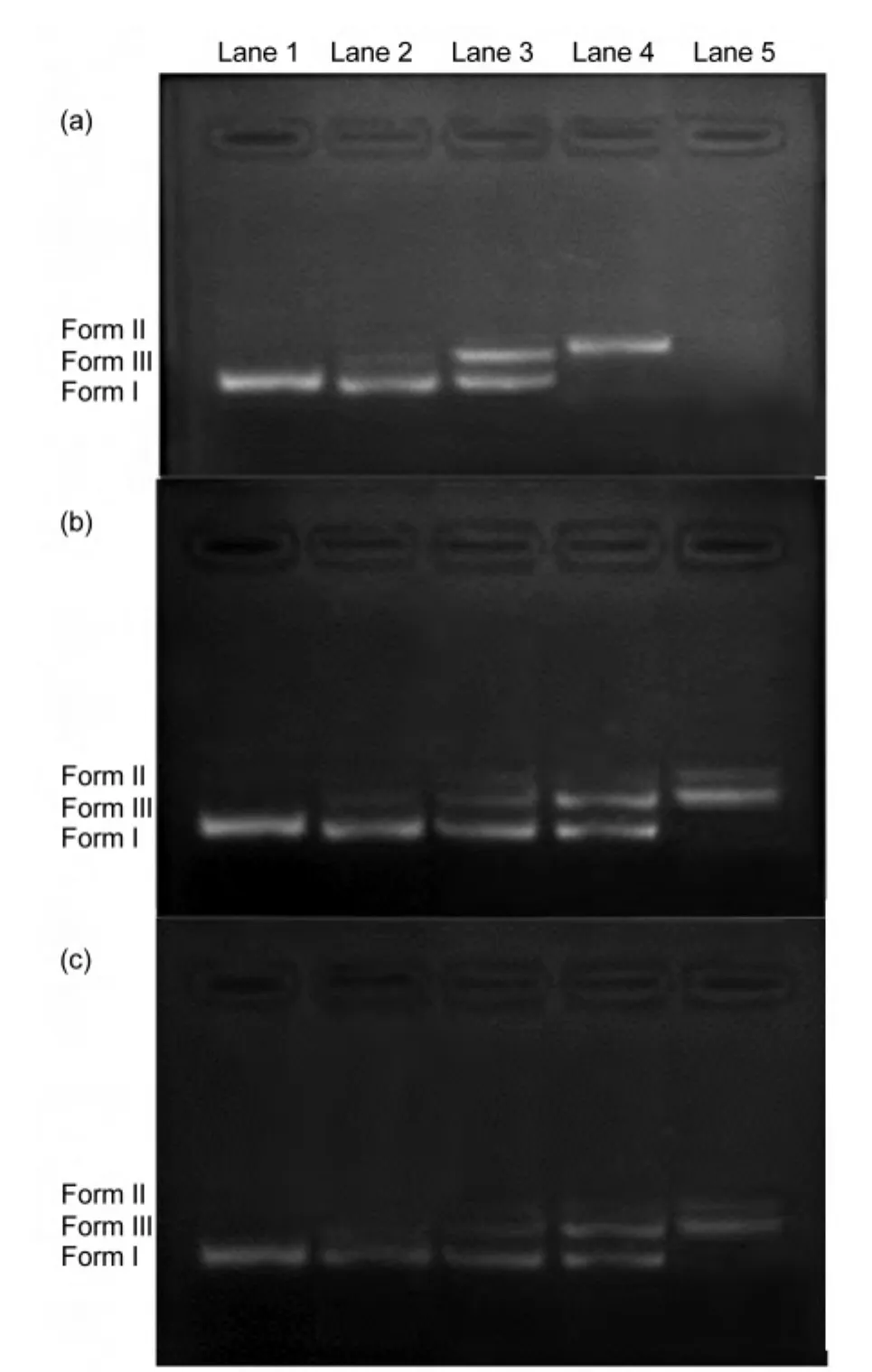
Fig.7 Agarose gel electrophoresis patterns for the cleavage of plasmid DNAby compounds
As for compound 2 at 0.08 µmol·L-1,there still are open circular and linear forms but the supercoil isabsent(Fig.7(b)).All the forms are present at 0.06 µmol·L-1,whereas compound 1 is not.There is no significant difference between compounds 2 and 3 on proportions of plasmid DNA forms.Compounds 2 and 3 almost present an identical pattern on DNA cleavage.With increasing concentration of compounds 2 and 3(Fig.7(b)and Fig.7(c)),the amount of Form I of plasmid DNA diminishes gradually.At 0.08µmol·L-1,both compounds can change Form I into Form II and III completely.
3.8 Absorption peak of DNA displacement
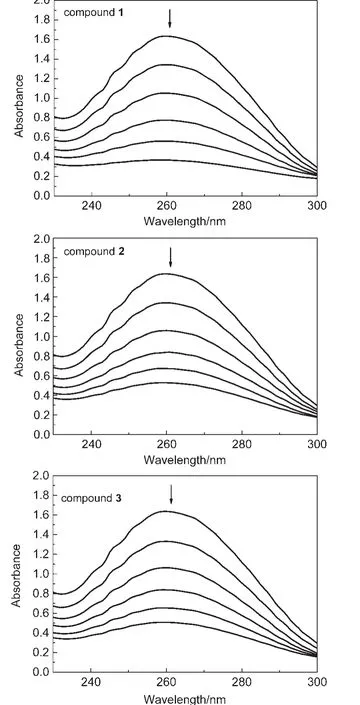
Fig.8 Absorption spectra of the compounds upon the plasmid DNA
To explore the interaction pattern between the titled compounds and plasmid DNA,the absorption peak of DNAdisplacement tests are also performed.The intrinsic absorption peak of DNAis at 260 nm.If the compounds intercalate into DNA,the unique structure of DNA will be changed,and the absorption peak will move.In our tests,as shown in Fig.8,the absorbance of DNA at 260 nm shows a remarkable decreasing tendency with increasing the concentration of the compounds.In test concentration,the endpoint of compound 1 is 0.33 at 260 nm while those of the other compounds are around 0.47.The decreased absorbance indicates that the compounds change DNA double-helix structure,which may be elucidated as the intercalation of the compounds between the base pairs of DNA.44,45Among the three compounds,compound 1 presents the lowest absorbance at 260 nm,showing that the more base pairs are intercalated by 1 compared to the others.46
The antifungal activities of compounds exhibit strong antifungal efficacy,following the order of 1>3≥2.Significantly,further investigations on DNA cleavage experiments reveal that such compounds show different intercalation activities.Notably,1 had an obviously higher inhibitory rate than other compounds,which is related with its DNAcleavage activities.Compared with compounds 2 and 3,compound 1 demonstrates an infinite 1D double-chain without interchain interaction.The better cleavage properties of the compound 1 may be attributed to the simple structure,which means that supramolecular interaction decreases the ability of the compounds to intercalate into DNA,and further influences the DNA cleavage activities.Although 2 and 3 show the isomorphous structure,their DNA cleavage activities are not exactly the same.The difference reveals that metal ion also affects the intercalation ability.As noticed,Cu(II)compounds have been the theme aimed at establishing the presumed synergy between the Cu(II)ion and the drug.47-50In conclusion,the cooperative effect of the supramolecular interaction and metal ions results in DNA cleavage activities.
4 Conclusions
In the present work,three transition metal compounds 1-3 with 4-(1H-1,2,4-triazol-1-ylmethyl)benzoic acid have been hydrothermally synthesized.Single-crystal X-ray diffraction analysis revealed that compound 1 features a 1D chain,while 2 and 3 exhibit 3D network structure.The detailed optical property investigations reveal that:1-3 exhibit remarkable luminescence emissions,which may be ascribed to the cooperative effects of intraligand emission and ligand-to-metal charge transfer(LMCT).Antifungal activities analyses show that compound 1 has the greatest antifungal efficacy compared to 2 and 3 on the five fungi.Photographs taken after agarose gel electrophoresis demonstrate the three titled compounds could cause plasmid DNAcleavage.In order to evaluate the effect of compounds concentration increase and research the pattern of cleavage,we studied different combination of the plasmid DNA(pUC 18)and compounds.The addition of increasing amount of compounds causes a significant increase in the DNA breakage.It is understandable that the three compounds can beak the DNAof fungi even cause its degradation.Although the three compounds can break the DNA,their breakage situations are not same.Among our presupposed concentrations compounds 2 and 3 can promote the conversion of DNA from Form I to Forms II and III,whereas compound 1 can fully degrade DNAat 0.08 µmol·L-1.This means that compound 1 has the highefficacy capacity on DNA breakage and that is the reason for compound 1 to present such high antifungal efficacy.
Supporting Information: available free of charge via the internet at http://www.whxb.pku.edu.cn.
(1)Yaghi,O.M.;Davis,C.E.;Li,G.M.;Li,H.L.J.Am.Chem.Soc.1997,119,2861.doi:10.1021/ja9639473
(2) Seo,J.S.;Whang,D.;Lee,H.;Jun,S.I.;Oh,J.;Jeon,Y.J.;Kim,K.Nature 2000,404,982.doi:10.1038/35010088
(3) Datta,A.;Karan,N.K.;Mitra,S.;Gramlich,V.J.J.Chem.Cryst.2003,33,579.doi:10.1023/A:1024299005045
(4) Gillon,B.;Mathoniere,C.;Ruiz,E.;Alvarez,S.;Cousson,A.;Kahn,T.M.J.Am.Chem.Soc.2002,124,14433.doi:10.1021/ja020188h
(5) Kozlov,I.A.;Kubareva,E.A.;Ivanovskaya,M.G.Antisense Nucleic A 1997,7,279.doi:10.1089/oli.1.1997.7.279
(6) Purmal,A.A.;Shabarv,T.V.;Gumport,R.I.Nucleic Acids Res.1998,20,3713.
(7)Trawick,B.N.;Daniher,A.T.;Bashkin,J.K.Chem.Rev.1998,98,939.doi:10.1021/cr960422k
(8) Budrìa,J.G.;Raugei,S.;Cavallo,L.J.Inorg.Chem.2006,22,1732.
(9) Liu,J.;Mei,W.J.;Xu,A.W.;Shi,S.;Tan,C.P.;Ji,L.N.Antivir.Res.2004,62,65.doi:10.1016/j.antiviral.2003.12.004
(10) Chabner,B.A.;Roberts,T.G.Nat.Rev.Cancer 2005,5,65.
(11) Chen,L.M.;Liu,J.;Chen,J.C.;Shi,S.J.Mol.Struct.2008,881,156.doi:10.1016/j.molstruc.2007.09.010
(12)Li,V.S.;Choi,D.;Wang,Z.;Jimenez,L.S.;Tang,M.S.;Kohn,H.J.Am.Chem.Soc.1996,18,2326.
(13)Zuber,G.;James,C.Q.;Hecht,S.M.J.Am.Chem.Soc.1998,120,9368.doi:10.1021/ja981937r
(14) Hecht,S.M.J.Nat.Prod.2000,63,158.doi:10.1021/np990549f
(15) Liu,J.G.;Ye,B.H.;Li,H.;Zhen,Q.X.;Ji,L.N.;Fu,Y.H.J.Inorg.Biochem.1999,76,265.doi:10.1016/S0162-0134(99)00154-3
(16) Strekowski,L.;Wilson,B.Mutat.Res.2007,623,3.doi:10.1016/j.mrfmmm.2007.03.008
(17) Zhou,C.Y.;Zhao,J.;Wu,Y.B.;Yin,C.X.;Yang,P.J.Inorg.Biochem.2007,101,10.doi:10.1016/j.jinorgbio.2006.07.011
(18) Gao,F.;Chao,H.;Zhou,F.;Yuan,Y.X.;Peng,B.;Ji,L.N.J.Inorg.Biochem.2006,100,1487.doi:10.1016/j.jinorgbio.2006.04.008
(19) Cater,D.C.;Ho,J.X.Adv.Protein Chem.1994,45,153.doi:10.1016/S0065-3233(08)60640-3
(20) He,X.M.;Cater,D.C.Nature 1992,358,209.doi:10.1038/358209a0
(21) Curry,S.;Brick,P.;Frank,N.P.Biochim.Biophys.A 1999,1141,131.
(22)Shen,X.C.;Yuan,Q.;Liang,H.Sci.Chin.Ser.B-Chem.2003,46,387.doi:10.1360/02yb0062
(23)Wang,Y.M.;Song,Y.;Kong,D.L.Chin.Sci.Bull.2005,50,1839.doi:10.1360/982004-405
(24) Mehra,R.K.;Tran,K.;Scott,G.W.;Mulchandani,P.;Saini,S.S.J.Inorg.Biochem.1996,61,125.doi:10.1016/0162-0134(95)00046-1
(25) Koh,L.L.;Ranford,J.O.;Robinson,W.T.;Swensson,J.O.;Tan,A.L.;Wu,D.Inorg.Chem.1996,35,6466.doi:10.1021/ic9606441
(26)Zhang,P.Z.;Fu,Q.Y.;Chi,R.X.;Yang,C.X.;Xu,J.G.J.Zhejiang Univ.Sci.Technol.2003,15,143.
(27) Xiong,P.P.;Li,J.;Bu,H.Y.;Wei,Q.;Zhang,R.L.;Chen,S.P.J.Solid State Chem.2014,215,292.doi:org/10.1016/j.jssc.2014.04.012
(28)Zhao,X.X.;Ma,J.P.;Dong,Y.B.;Huang,R.Q.Cryst.Growth Des.2007,7,1058.doi:10.1021/cg060583+
(29)Qin,J.;Ma,J.P.;Liu,L.L.;Huang,R.Q.;Dong,Y.B.Acta Cryst.2009,65,66.
(30) Sheldrick,G.M.SHELXS-97,Program for Solution of Crystal Structures;University of Göttingen:Göttingen,Germany,1990.
(31) Sheldrick,G.M.SHELXK-97,Program for Refinement of Crystal Structures;University of Göttingen:Göttingen,Germany,1997.
(32) Mann,A.;Banso,A.;Clifford,L.C.;Tan.J.Health.Res.2008,10,34.
(33)Li,Y.T.;Liu,Z.Q.;Wu,Z.Y.J.Inorg.Biochem.2008,102,1790.doi:org/10.1016/j.jinorgbio.2008.05.011
(34)Zhang,J.P.;Chen,X.M.Chem.Commun.2006,1689.
(35) Ouellette,W.;Hudson,B.S.;Zubieta,J.Inorg.Chem.2007,46,4887.doi:10.1021/ic062269a
(36) Zheng,L.L.;Li,H.X.;Leng,J.D.;Wang,J.;Tong,M.L.Eur.J.Inorg.Chem.2008,213.
(37)AbouEl,E.S.;El,S.A.;Emam,S.M.;Ell,S.M.A.Spectrochim.Acta Part A 2008,71,421.doi:10.1016/j.saa.2007.12.031
(38) Valent,A.;Melnik,M.;Hudecova,D.;Dudova,B.;Kivekas,R.;Sundberg,M.R.Inorg.Chim.Acta 2002,340,15.doi:10.1016/S0020-1693(02)01062-9
(39)Song,Y.M.;Wu,Q.;Yang,P.J.;Luan,N.N.J.Inorg.Biochem.2006,100,1685.doi:10.1016/j.jinorgbio.2006.06.001
(40) Zavitsanos,K.;Nunes,A.M.;Malandrinos,G.;Hadjiliadis,N.J.Inorg.Biochem.2011,105,1329.doi:10.1016/j.jinorgbio.2011.07.014
(41) Barton,J.K.;Raphael,A.L.J.Am.Chem.Soc.1984,106,2466.doi:10.1021/ja00320a058
(42) Scheppler,J.A.;Cassin,P.E.;Gambier,R.M.Biotechnology Explorations:Applying the Fundamentals;ASM Press:Washington DC,2000.
(43) Xi,P.X.;Xu,Z.H.;Chen,F.J.;Zeng,Z.Z.J.Inorg.Biochem.2009,103,210.doi:10.1016/j.jinorgbio.2008.10.010
(44) Li,Y.T.;Liu,Z.Q.;Wu,Z.Y.J.Inorg.Biochem.2008,102,1790.doi:10.1016/j.jinorgbio.2008.05.011
(45) Rajendiran,V.;Karthik,R.;Palaniandavar,M.;Evans,S.H.;Periasamy,V.S.;Akbarsha,M.A.;Srinag,B.S.;Krishnamurthy,H.Inorg.Chem.2007,46,8208.doi:10.1021/ic700755p
(46)Khoramdareh,Z.K.;Yazdi,S.A.;Spingler,H.B.;Khandar,A.A.Inorg.Chim.Acta 2014,415.7.
(47)Jiang,J.;Tang,X.L.;Dou,W.;Zhang,H.H.;Liu,W.S.;Wang,C.X.;Zheng,J.R.J.Inorg.Biochem.2010,104,583.doi:10.1016/j.jinorgbio.2010.01.011
(48)Melnik,M.Coord.Chem.Rev.1982,42,259.doi:10.1016/S0010-8545(00)80537-8
(49) Kato,M.;Muto,Y.Coord.Chem.Rev.1988,92,45.doi:10.1016/0010-8545(88)85005-7
(50)Weder,J.E.;Dillon,C.T.;Hambley,T.W.;Kennedy,B.J.;Lay,P.A.;Biffin,J.R.;Regtop,H.L.;Davies,N.M.Coord.Chem.Rev.2002,232,95.doi:10.1016/S0010-8545(02)00086-3
