Characterization, Expression and Function Analysis of DAX1 Gene of Scallop (Chlamys farreri Jones and Preston 1904) During Its Gametogenesis
2014-05-05LIHailongLIUJianguoHUANGXiaotingWANGDanandZHANGZhifeng
LI Hailong, LIU Jianguo, HUANG Xiaoting, WANG Dan, and ZHANG Zhifeng
Key Laboratory of Marine Genetics and Breeding, Ministry of Education, Ocean University of China, Qingdao 266003, P. R. China
Characterization, Expression and Function Analysis of DAX1 Gene of Scallop (Chlamys farreri Jones and Preston 1904) During Its Gametogenesis
LI Hailong, LIU Jianguo, HUANG Xiaoting, WANG Dan, and ZHANG Zhifeng*
Key Laboratory of Marine Genetics and Breeding, Ministry of Education, Ocean University of China, Qingdao 266003, P. R. China
DAX1, a member of nuclear receptor superfamily, has a function in the sex determination and gonadal differentiation of several vertebrate species. However, little information about DAX1 of invertebrates is available. Here we cloned a homolog of scallop (Chlamys farreriJones and Preston 1904)dax1,Cf-dax1, and determined its expression characteristics at mRNA and protein levels. The cDNA sequence ofCf-dax1was 2093 bp in length, including 1404 bp open reading frame (ORF) encoding 467 amino acids. Unlike those of vertebrates, no conserved LXXLL-related motif was found in the putative DNA binding region of Cf-DAX1. Fluorescencein situhybridization showed thatCf-dax1located on the short arm of a pair of subtelocentric chromosomes. Tissue distribution analysis using semi-quantitative RT-PCR revealed thatCf-dax1expressed widely in adult scallop tissues, with the highest expression level found in adductor muscle, moderate level in mantle, gill and testis, and low level in kidney, ovary and hepatopancreas. The result of quantitative real-time PCR indicated that the expression ofCf-dax1was significantly higher (P<0.05) in testis than in ovary at the same stage, showing a sex-dimorphic expression pattern. Furthermore, immunohistochemical detection found that Cf-DAX1 mainly located in spermatogonia and spermatocytes of testis and in oogonia and oocytes of ovary, implying that DAX1 may involve in gametogenesis of bivalves.
Chlamys farreri;dax1; sex-dimorphic expression; gametogenesis; gonad
1 Introduction
DAX1 (dosage-sensitive sex reversal-adrenal hypoplasia congenital (AHC) critical region on the X chromosome gene 1) is initially identified in human (Homo sapiens) and the mutations of this gene can cause AHC which is characterized by adrenal insufficiency and hypogonadotropic hypogonadism (Zanariaet al., 1994). Furthermore, DAX1 is an unusual nuclear receptor (NR) and classified to 0 subfamily of NR superfamily as it contains only one ligand-binding domain (LBD). Usually, there are two conserved domains, LBD in the C-terminal and DNA-binding domain (DBD) in the N-terminal of NRs (Nuclear Receptors Nomenclature Committee, 1999). In N-terminal of DAX1, although the sequence is not conserved, an LXXLL motif exists commonly in vertebrates, specifically in mammalian with 3.5 repeats of the motif sequence (Guoet al., 1996; Parmaet al., 1997; Zanariaet al., 1994), and implicates in the binding with single-stranded DNA and RNA, as well as the interaction with other proteins (Lalliet al., 2000; Suzukiet al., 2003; Zazopouloset al., 1997). In C-terminal region, the sequence of DAX1 consists of a conserved LBD which is involved in the transcription control of gene (Burriset al., 1996; Guoet al., 1995). In addition, an activation function-2 (AF-2) sequence, which is essential for the transcription regulation of NRs (Moras and Gronemeyer, 1998), is found in the LBD of DAX1.
In vertebrates, the expression and function ofdax1are involved in sex determination and/or differentiation and are different among animals. Expression ofdax1in human, mouse (Mus musculus) and rat (Rattus norvegicus) is mainly restricted to the tissues directly involved in steroid hormone synthesis and reproductive axis, such as adrenal cortex, pituitary gonadotropes, theca and granulosa cells in ovary, Leydig and Sertoli cells in testis, and possibly some other brain cells (Ikedaet al., 1996, 2001; Swainet al., 1996; Tamaiet al., 1996; Zanariaet al., 1994). Bardoniet al. (1994) found that the replication ofdax1in human could lead to male to female sex reversal. After that, more and more studies concentrated on its expression and roles played in sex determination and/or differentiation. In mouse, the transcription ofdax1is firstly detected in the genital ridge at 11.5 d post coitus. After 12 d post coitus, when morphological differences between males and females can be observed in gonad, itsexpression is down-regulated in testis while persists in developing ovary, showing a possible role of DAX1 in ovary development (Swainet al., 1996). However, loss ofdax1does not affect ovarian development or fertility in mouse, but causes the progressive degeneration of testicular germinal epithelium and male sterility, indicating thatdax1plays a critical role in spermatogenesis (Yuet al., 1998). Furthermore, it has been reported that mouse DAX1 has a co-tissue expression site with SF-1 (steroidogenic factor 1) gene (Ikedaet al., 1996, 2001) and mediates independently or cooperatively somatic cell differentiation during testis development (Parket al., 2005). In no-mammalians, researches ondax1gene focus mainly on the expression level. In chicken (Gallus gallus),dax1expresses in embryonic gonads during sex differentiation, and the level increases in gonads with sex differentiation and is higher in ovary than in testis (Smithet al., 2000). In frog (Rana rugos),dax1expresses only in juvenile gonads, and such an expression is stronger in testis than in ovary at stage XXV (just completion of metamorphosis). The expression is maintained only in testis for two months after metamorphosis, implying that it may be involved in testis development of amphibians (Sugitaet al., 2001). Interestingly, the expression ofdax1does not show a sex difference in European sea bass (Dicentrarchus labrax) (Martinset al., 2007) and American alligator (Alligator mississippiensis) (Westernet al., 2000).
Comparing with the extensive research ofdax1gene in vertebrates, poor information is available to date in invertebrates except for in oyster (Crassostrea gigas) in which only the sequence is reported (Zhanget al., 2012a). Shellfish comprises a group of molluscs and their sex variability characteristic involves the interaction between environment and gene.Chlamys farreriis an important commercial shellfish in China and is a kind of good experiment species to identify sex differentiation-related genes due to its relatively stable sex composition (Liaoet al., 1983; Yang and Zhang, 2009). In present study, we cloned a full-length cDNA ofdax1fromC. farreriand determined its expression pattern during the gametogenesis in gonads using quantitative real-time PCR and immunohistochemical method, further presented its location on chromosomes. Our aim was to reveal the expression characteristics ofC. farreridax1(Cf-dax1) during its gametogenesis, identify the expression difference between ovary and testis in reproductive cycle, and provide some basic information for further probing its functional correlation with gametogenesis and sex differentiation.
2 Materials and Methods
2.1 Animals and Sampling
Healthy female and male scallop (mean shell height 6.38 ± 0.33 cm) was purchased from Aquatic Product Market (Qingdao, P. R. China). Tissues (testis, ovary, adductor muscle, gill, mantle, kidney and hepatopancreas) were dissected and immediately frozen in liquid nitrogen, then stored at -80℃ until RNA extraction. Part of gonads was fixed with Bouin’s solution (Picric acid: formaldehyde: acetic acid = 15:5:1) for 24 h, embedded in paraff i n wax, sliced to a thickness of 5 μm and stained with haematoxylin-eosin (H & E) for histologically determining gonadal developmental stages. Another part of gonads was fixed in 4 % paraformaldehyde (pH 7.4) at 4℃ for 24 h and then dehydrated with a methanol series (25%, 50%, 75%, and 100%) and stored in 100% methanol at -20℃for immunohistochemistry analysis.
According to the morphologic characteristics described by Liaoet al. (1983), the gonads (ovaries and testes) were grouped into three stages based on histological structure and the gonadosomatic index
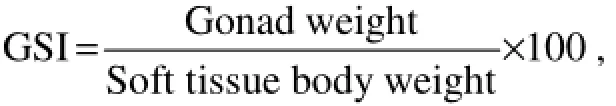
as proliferative stage (GSI=3.18 ± 0.008 for ovary and GSI= 3.89 ± 0.008 for testis), growing stage (GSI= 4.20 ± 0.013 for ovary and GSI= 3.93 ± 0.012 for testis), and mature stage (GSI= 4.41 ± 0.004 for ovary and GSI= 4.53 ± 0.009 for testis).
2.2 3’ and 5’ –RACE Analysis
Total RNA was isolated from ovary at proliferative stage with Trizol reagents (Takara Bio Inc., Otsu, Japan) according to the manufacturer’s instructions. The RNA was then treated with DNase (Takara Bio Inc., Otsu, Japan) and purified using RNeasy mini kit (TransGen Bio Inc., Beijing, China). RACE-Ready First-strand cDNA was synthesized using SMARTTM-RACE cDNA Amplification Kit (Invitrogen, Carlsbad, CA, USA). 5’ RACEPCR and 3’ RACE-PCR were conducted using the SMARTTM-RACE cDNA Amplification Kit (Invitrogen, Carlsbad, CA, USA) and gene specific primers (5’-CT TTCAGTTCGCTGTCACCCCGACACAC-3’and 5’-GG CAGGAAGCAGCAACAACAAGCGTGAG-3’) were designed according to a 692 bp expressed sequence tag (EST) (GenBank accession no. DT716557) obtained by BLAST search fromC. farreriEST collection (http://www.ncbi. nlm.nih.gov/nucest/115428840). The RACE cDNAs were denatured at 94℃ for 5 min, followed by 35 cycles at 94℃ 30 s, 68℃ 30 s, and 72℃ 3 min, ended with 5 min of extension at 72℃. PCR products were separated by an agarose gel (1.2%) and purified with a gel purification kit (Takara Bio Inc., Otsu, Japan). Then, the purified 5’ and 3’ RACE products were subcloned into PMD-18T vector (Takara Bio Inc., Otsu, Japan) and sequenced.
2.3 Bioinformatics Analysis
The 5’ and 3’ RACE fragments were assembled by the software DNAstar to get the whole length ofCf-dax1cDNA. Sequence identity and similarity of Cf-DAX1 with those of other species were analyzed using the online BLAST suite of programs at National Center for Biotechnology Information (NCBI). Multiple sequence alignment was performed using the CLUSTAL X algorithm and GeneDoc software. Phylogenetic analysis was conducted using the MEGA 4 software by neighbor-joining method.
2.4 Semi-Quantitative Reverse Transcription (RT) PCR Analysis
Tissue distribution ofCf-dax1inC. farreriat the proliferative stage was analyzed using semi-quantitative RTPCR (sqRT-PCR) method. Total RNA was isolated for various tissues according to the method described above, and was transcribed to cDNA using the PrimeScript RT reagent Kit (Takara Bio Inc., Otsu, Japan) as the initial template for sqRT-PCR. A 331 bp fragment of target gene was amplified using specific primers (forward primer: 5’-GGCAGGAAGCAGCAACAACAAGCGTGAG-3’, reverse primer: 5’-CTTTCAGTTCGCTGTCACCCCGAC ACAC-3’). Theβ-actinwas selected as the reference following Zhanget al. (2012b), and a 594 bp fragment ofC. farreriβ-actin(AY335441) was amplified using primers (forward primer: 5’-ATGCCCTCCCTCACGCTAT-3’ and reverse primer: 5’-GCCAGACTCGTCGTATTCCT-3’), respectively. The thermocycles were optimized to obtain a single product within the linear range of PCR amplification. The PCR condition was 5 min at 95℃, followed by 30 cycles (95℃ 30 s, 68℃ 30 s, 72℃ 30 s) and 5 min at 72℃ forCf-dax1, and forβ-actin, that was 5 min at 94℃, followed by 25 cycles (94℃ 30 s, 60℃ 30 s, 72℃ 45 s) and 5 min at 72℃. All the PCR products were separated in 1.5% agarose gel and visualized with ethidium bromide staining. The PCR was repeated twice.
2.5 Quantitative Real-Time PCR Analysis
Quantitative real-time PCR (qRT-PCR) for analyzing expression level ofCf-dax1in gonads during the reproductive cycle was conducted according to the method described previously (Zhouet al., 2010). Specific primers for amplifying a 190 bp fragment were designed according to the no-conservative domain ofC. farreri dax1. The forward primer was 5’-TCTTCCTCGCCTCATTGTCG-3’and the reverse primer was 5’-CGTCGGTATTGGAGCC TTTG-3’. A 129 bp fragment ofC. farreri β-actin(AY 335441) was amplified as reference using the forward primer 5’-TTCTTGGGAATGGAATCTGC-3’ and the reverse primer 5’-ATTGTGCTACCACCGGAAAG-3’. Primer specificity was verified by a single distinct peak obtained in the melting curve analysis of qRT-PCR system. Quantification of target and reference genes was conducted simultaneously by ABI 7500 detection system (Applied Biosystems) with SYBR Green Master Mix (Takara Bio Inc., Otsu, Japan). The qRT-PCR reaction consisted of 5 min at 94℃, followed by 40 cycles of 94℃15 s, 60℃ 1 min. Gonadal RNA from five individuals at the same stage and duplicate assays for each gonad sample were detected. Data were analyzed using the ABI 7500 system SDS software version 1.4 (Applied Biosystems) with automatically set baseline and cycle threshold values. The 2-ΔΔctmethod was employed to analyze the relative expression level ofCf-dax1(Kenneth and Thomas, 2001).
All data were presented as mean ± SEM (n=5). Difference was tested using one-way ANOVA followed by the least significant difference test (SPSS software version 18.0; SPSS Inc., Chicago, IL, USA), and a significance level was set atP<0.05.
2.6 Immunohistochemistry
The complete ORF fragment ofCf-dax1was amplified using the forward primer 5’-GGATCCA TGGACCTCG GACACGATAAAG-3’ (BamHI site underlined) and the reverse primer 5’-CTCGAGCAAATTTTGTATCATCTC CTTC-3’ (XhoI site underlined). After the fragment was subcloned into PMD-18T vector (Takara Bio Inc., Otsu, Japan), the recombinant plasmid was extracted and then digested byBamHI andXhoI. The digested fragment was then inserted into theBamHI /XhoI site of bacterial expression vector pet28a (Invitrogen, Carlsbad, CA, USA) and verified by sequencing. The recombinant plasmid was then transformed intoE. coliBL21 (DE3) (TransGen Bio inc, Beijing, China). Glutathione-S-transferase (GST)-DAX1 fusion protein was expressed and affinity purified on a anti-GST Ni-NTA His Bind Resins (Invitrogen, Carlsbad, CA, USA), and was injected into New Zealand white rabbits for the production of polyclonal antibodies. For immunohistochemical analysis, gonad tissues were dehydrated with ethanol, embedded in paraffin, and 5 µm sections were cut using a Histostart 820 Rotary microtome (Reichert, Co. Ltd., USA). Sections were deparaffinized in xylenes and descending ethanol, followed by antigen retrieval in citrate buffer (0.01 mol L-1, pH 6.0). In order to assess the specificity of immuoreaction, testis and ovary tissues from different stages were used as a negative control. Sections were blocked in BSA (3%) for 1 h and incubated with preimmune serum (for the negative control) and anti-DAX1 primary antibody (1:200 dilutions) respectively for 1 h at room temperature. Speci fi c goat-anti-rabbit IgG (conjugated to horseradish peroxidase, 1:1000 dilutions) was used as secondary antibody and incubated for 1 h at room temperature. After washing with PBS (phosphate buffer saline) +0.1% Tween-20, sections were then color development with DAB (3, 3’-diaminobenzidine) and counterstained with hematoxylin. Tissue sections were then observed and photographed using a Nikon E80i microscope.
2.7 Fluorescencein situHybridization (FISH)
The qRT-PCR primers ofCf-dax1described previously were used to screen a bacterial artificial chromosome (BAC) library ofC. farrericonstructed by Cheng (2010). The selected BAC clone was then labeled with biotin-11-dUTP using a Nick Translation Kit (Roche) following the manufacturer’s instructions. Chromosomal preparation for FISH was performed as described by Huanget al. (2006). Probe hybridization and detection were performed according to Zhanget al. (2008). Biotinylated probes were detected with avidin-FITC (Vector). Chromosomes were counterstained with Propidium iodide (1.5 μg mL-1, Vector) at room temperature for 10 min. Examination of slides was carried out with a Nikon epifluorescence mi-croscope (Eclipse E-600) and analyzed with Lucia-FISH Image System.
3 Results
3.1 Sequencing and Characterization ofCf-dax1
The 3’-and 5’ -RACE fragments of target cDNA were obtained fromC. farreriovary at the proliferative stage, 902 bp and 1525 bp in length, respectively. The full-length cDNA was 2093 bp (JQ071986), consisting of an ORF of 1404 bp, encoding a deduced protein of 467 amino acids with an estimated molecular mass of 51.8 kDa and an isoelectric point of 6.39. The sequence contained a 181 bp 5’ untranslated region (UTR) and a 508 bp 3’UTR.
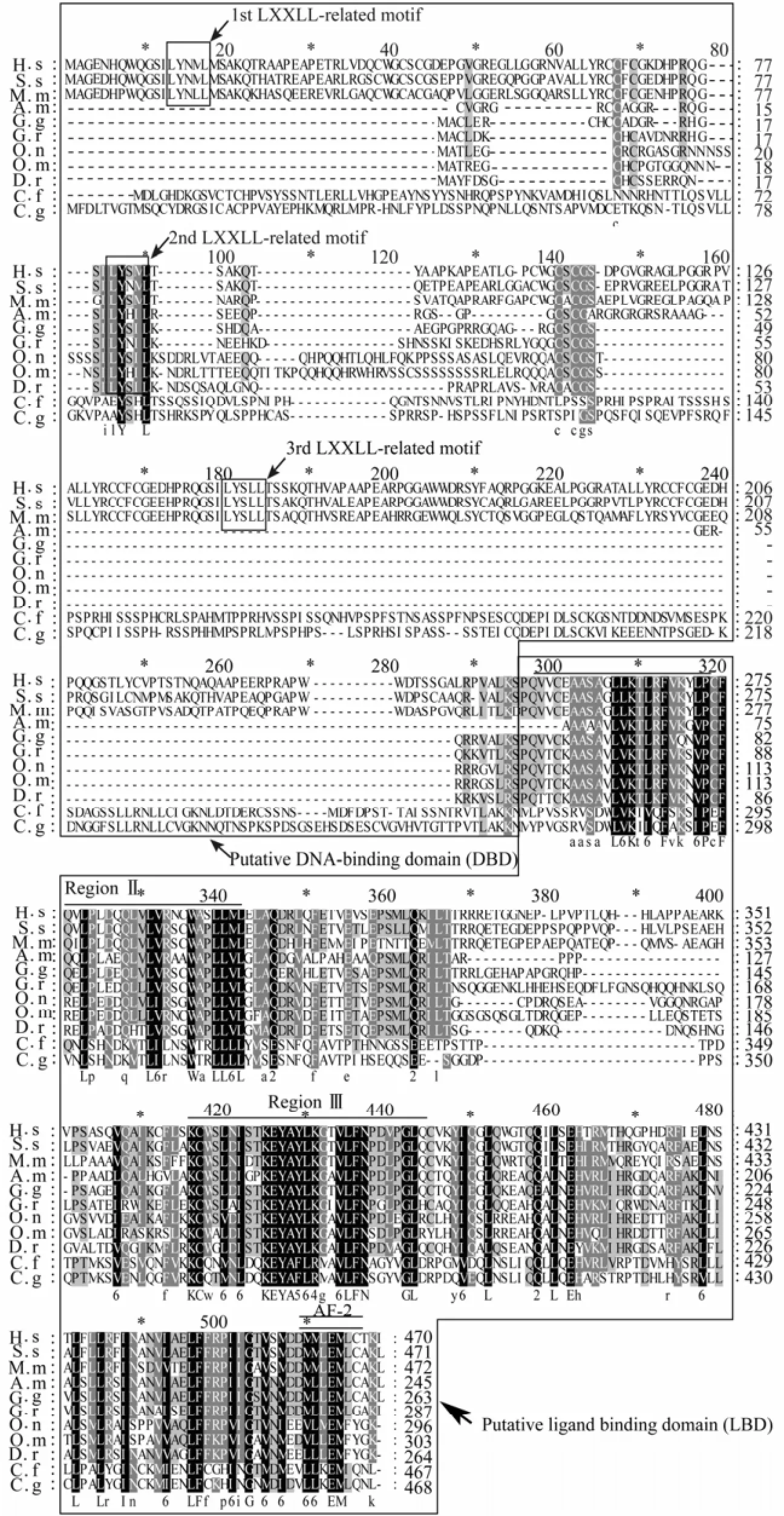
Fig.1 Alignment of the amino acid sequence of DAX1.
Cf-DAX1 consisted of two putative domains of NR superfamily, a C-terminal LBD and an N-terminal DBD (Fig.1). The LBD shared the identities of 80%, 32%, 31%, 28%, 28% and 27% with that ofC. gigas,A.mississippiensis,Oreochromis niloticus,H.sapiens,G.gallusandXenopus laevis, respectively. Two regions (II and III) that are important for the formation of a ligand-binding pocket were relatively more conservative than other regions in LBD. Regions II showed an identity of 93% with that ofC. gigasDAX1 homolog. The region III shared a high identity with those of other species and the identity score ranged from 61% to 91%, with the highest identity of 91% with that ofC. gigas. Also, an AF-2 sequence was found in LBD (Fig.1). However, the putative N-terminal DBD region of Cf-DAX1 was less conservative than LBD, and shared no significant sequence homology with other proteins except for that ofC.gigas, which shared a 38% identity with those deposited in NCBI. Moreover, LXXLL-related motif which is conserved in vertebrates DAX1s did not exist in the putative DNA binding region of Cf-DAX1 alignment (Fig.1).
Phylogenetic analysis indicated that most DAX1s clustered firstly within the same class, such as amphibians (G.rugosaandX.laevis), mammals (H.sapiensandM.musculus), and fishes (Danio rerioandO.niloticus). Also, Cf-DAX1 clustered primarily with those of other shellfish and finally grouped with the branch formed by vertebrates (Fig.2).
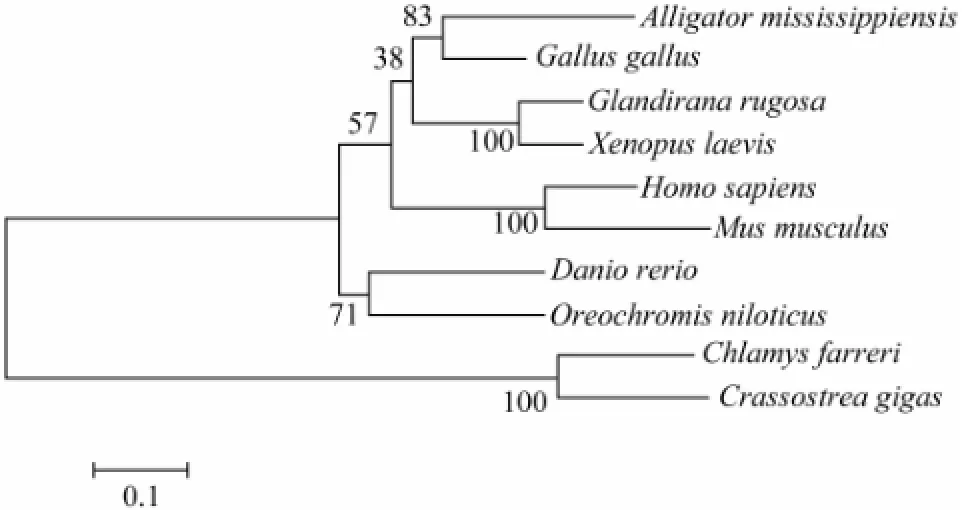
Fig.2 Phylogenetic relationship of DAX1 constructed using MEGA 4 based on amino acid sequences aligned using CLUSTAL X.
The putative DNA-binding domain (DBD) and ligand-binding domain (LBD) are boxed, respectively. The LXXLL-related motifs are boxed inside DBD box. Region II and III in LBD are marked with single overlines, and the activation function-2 (AF-2) core is marked with a double overline. Beneath the alignment, identical amino acids are indicated by capital letters, similar amino acids are indicated by lowercase letters. The numbers indicate the scoring features in scoring table of GeneDoc, which means the replacement of these amino acids is more frequent than the expected at random during evolution. Accession numbers: A. m:A.mississippiensis(AAD 55095); C. f:C.farreri(JQ071986); C. g:C.gigas(EKC 26839); D. r:D.rerio(NP_001076416); G. g:G.gallus(NP_989924); G. r:G.rugosa(BAB85864); H. S:H.sapiens(NP_000466); M. m:M.musculus(NP_031456); O. m:Oncorhynchus mykiss(ACM80361); O. n:O.niloticus(ABB88833); S. s:Sus scrofa(AAB81101).
Numbers in the branches represent bootstrap values (percentage) with 1000 replicates. The scale bar indicates an evolutionary distance of 0.1 amino acid substitution per site. Accession numbers:A.mississippiensis(AAD 55095);C.farreri(JQ071986);C.gigas(EKC26839);D.rerio(NP_001076416);G.gallus(NP_989924);G.rugosa(BAB85864);H.sapiens(NP_000466);M.musculus(NP_ 031456);O.niloticus(ABB88833);X.laevis(BAF52037).
3.2 Spatial Expression ofCf-dax1
Using sqRT-PCR,Cf-dax1was detected in all the examined tissues (Fig.3). However, the expression level ofCf-dax1was different among tissues, being high in adductor muscle, moderate in mantle, gill and testis, and low in kidney, ovary and hepatopancreas.
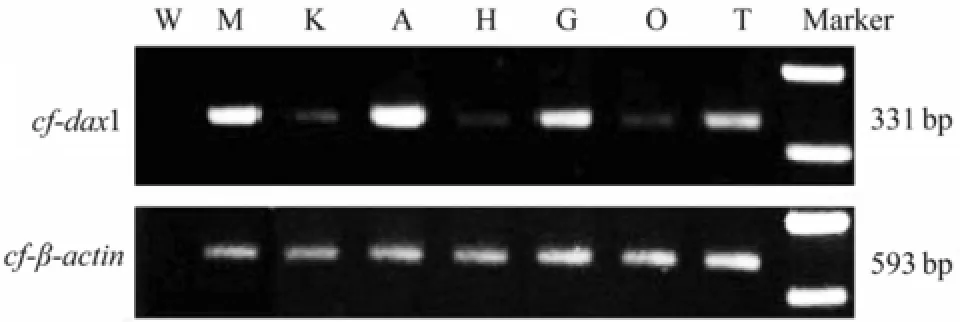
Fig.3 Expression of Cf-dax1 in C. farreri tissues analyzed by sqRT-PCR. β-actin was used as reference. W, water; M, mantle; K, kidney; A, adductor muscle; H, hepatopancreas; G, gill; O, ovary; T, testis.
3.3 Quantitative Expression ofCf-dax1in Gonads During Reproductive Cycle
The qRT-PCR results indicated thatCf-dax1expressed in both testis and ovary ofC. farreri(Fig.4). However, expression level was different in gonad at different development stage. The highest level was found in testis of growing stage which was significantly higher (P<0.05) than that at other stages. While in ovaries,dax1gene remained nearly the same expression level between proliferative and growing stages, and decreased sharply atmature stage. Furthermore, a significantly (P<0.05) higher expression was found in testis than in ovary at the same stage during reproductive cycle. That was about 1 time higher in testis than in ovary at proliferative stage, 1.5 times at growing stage and 2 times at mature stage.
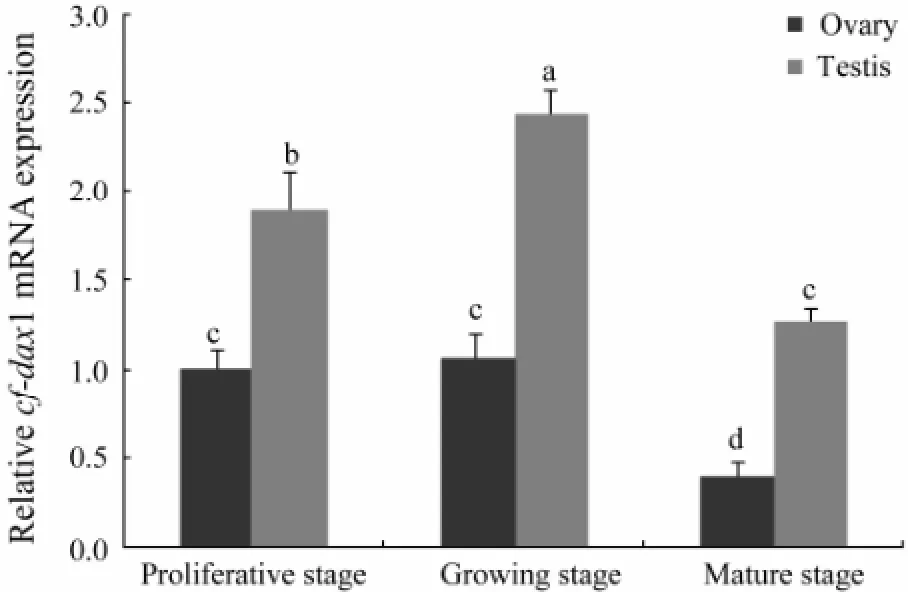
Fig.4 Expression of Cf-dax1 in C. farreri gonads detected by qRT-PCR. The expression level in proliferative ovary was set as 1.00 to calibrate the relative level in gonads at various stages. Values are mean ± SEM (n=5); different letters (a–d) indicate significant differences (P <0.05).
3.4 Cyto-Location of Cf-DAX1 in Gonads During Gametogenesis
In testis, a strong immunoreaction was observed in spermatogonia at three developmental stages, and in spermatocytes at growing stage. In ovary, DAX1 was detected in oogonia and oocytes, and the immunostaining intensities decreased along with the gametogenesis and only a faint staining was observed in oocytes at mature stage (Fig.5). No visible expression was found in somatic cells of both testis and ovary during the whole reproductive cycle.
3.5 Location ofCf-dax1on Chromosomes
One BAC clone containingCf-dax1gene was successfully isolated from the BAC library ofC. farreri. After FISH, theCf-dax1gene was mapped on the short arm of a pair of subtelocentric chromosomes (Fig.6).
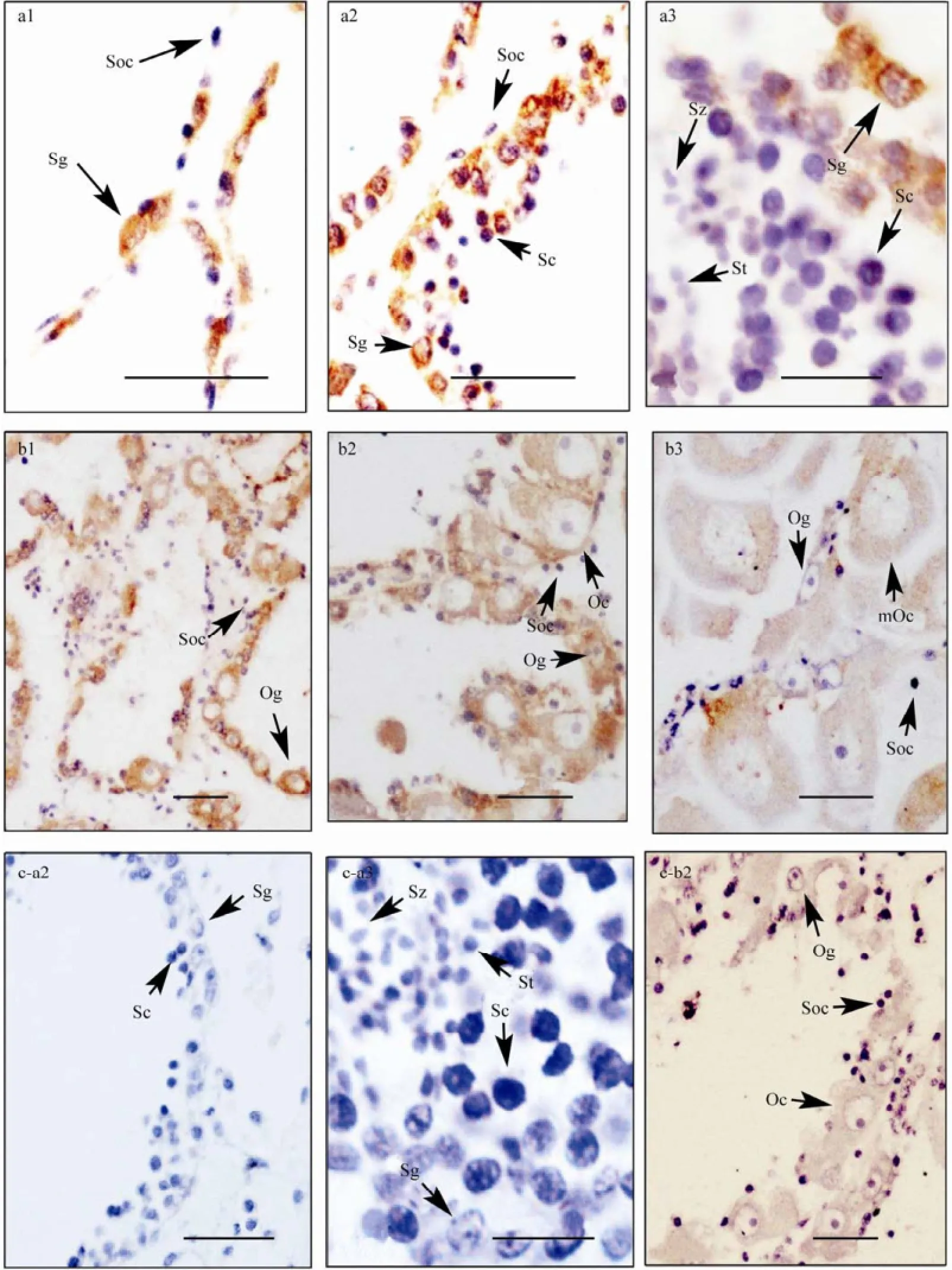
Fig.5 Cf-DAX1 protein expression in C. farreri adult gonads detected by immunohistochemistry. a, testis; b, ovary; c, negative control; 1, proliferative stage; 2, growing stage; 3, mature stage; Og, oogonium; Oc, oocyte; mOc, mature oocyte; Sg, spermatogonium; Sc, spermatocyte; St, spermatid; Sz, spermatozoon; Soc, somatic cells; Scale bars, a3 and c-a3 are 10 µm, others are 30 µm.

Fig.6 Hybridization of Cf-dax1 gene to metaphase chromosomes. Arrows indicate the location of Cf-dax1. Chromosomes are counterstained with Propidium iodide. Scale bar, 5 µm.
4 Discussion
4.1 Structure Characteristics of Cf-DAX1
In this study, we have identified a 2093 bp full length cDNA sequence in scallop, which encodes a 467 amino acids sequence containing a putative LBD and a putative DBD of NR superfamily. To be specific, the putative LBD is well-conserved and shared the identity of 80% with that of oyster DAX1. While the putative N-terminal DBD region shared no sequence homology to those of other proteins deposited in NCBI, except for an identity of 38% with that ofC.gigasDAX1. The characteristics of no-conserved DBD region were congruent with that of the other members of subfamily 0 of NR superfamily (Niakan and McCabe, 2005). These features indicated that the cloned full length cDNA sequence wasC. farreri dax1, which was named asCf-dax1.
In vertebrate DAX1s, an LXXLL motif exists commonly in the DBD region and there are 3.5 repeats of this motif in mammals (Guoet al., 1996; Parmaet al., 1997; Zanariaet al., 1994). Furthermore, in mammalians, it has been reported that the motifs function in NR binding by interacting with other NRs, such as SF-1, androgen receptor, estrogen receptor and progestin receptor (Agoulniket al., 2003; Holteret al., 2002; Itoet al., 1997; Zhanget al., 2000). In this study, multisequence alignments conducted by CLUSTAL X showed that no conserved LXXLL-related motif existed in the putative DBD of Cf-DAX1 (Fig.1). We suggest that this difference betweenC. farreriand vertebrates maybe due to the difference of species, and Cf-DAX1 may need the assistances of a mediate molecule to bind to DNA and RNA. Whatever, it merits further studies.
The C-terminal region of DAX1s shows a high degree of conservation among species reported so far (Martinset al., 2007; Smithet al., 2000; Sugitaet al., 2001). To date, all mutations in AHC are localized in the C-terminal part, suggesting that this domain plays crucial roles in DAX1 (Achermannet al., 2001). Also, this region is shown to have the transcriptional silencing activity in mammals (Lalliet al., 1997). Considering the conservative property of Cf-DAX1 in this region, we speculate that Cf-DAX1 might have the activity of a transcriptional silencing.
4.2 Wide Spatial Expression Characteristics ofCf-dax1
Cf-dax1had a broad tissue expression, which is similar to most vertebrates. In European sea bass,dax1mRNA is detected in gut, heart, gill, muscle and kidney (Martinset al., 2007), also a similar expression is observed in frog, mouse and Nile talipia (Baeet al., 1996; Sugitaet al., 2001; Wanget al., 2002), indicating an evolutionary conservatism of the gene expression. However,dax1expression is not visible in the adult gonads ofR. rugos(Sugitaet al., 2001) while it can be detected in the gonad ofC. farreriand the tissues of brain-pituitary-gonadal axis in other vertebrates, implying thatdax1may have a function in spermatogenesis, gonadal development and function maintenance in frog, mouse, chicken and scallop. Meanwhile, we noted that the transcriptional level ofCf-dax1was different among tissues, with the highest found in adductor muscle, high levels in testis, mantle and gill, and low level in ovary, hepatopancreas and kidney (Fig.3). The different expression levels among the tissues have also been reported for frogdax1, which was detected at higher level in liver and pancreas, lower level in brain and heart, none in lung, spleen, kidney/adrenal, testis, ovary and muscle (Sugitaet al., 2001). Obviously, this broad tissue expression and difference of expression level among tissues imply thatdax1gene may have different functions in different biological processes.
4.3 DAX1 May be Involved in Gametogenesis of Bivalves
In most species,dax1presents sexually dimorphic expression pattern in gonad, while this sex-related expression is different among species. In embryo gonads of mouse and chicken,dax1expresses at higher level in ovary than in testis (Swainet al., 1996; Smithet al., 2000). On the contrary, in juvenile frog and rainbow trout during early gametogenesis,dax1is predominantly transcribed in testis (Baronet al., 2005; Sugitaet al., 2001). In present study,Cf-dax1also showed a sexually dimorphic expression, with a significantly higher expression level in testis than in ovary (Fig.4). Immunohistochemical results with strong staining in testis while a relative low signal in ovary further proved its sex dimorphic expression at protein level (Fig.5). The expression ofdax1predominantly in testis is similar to that of frog, in whichdax1mRNA abundance is higher in testis than in ovary at stage XXV (just completion of metamorphosis), and only in testis in two months after metamorphosis (Sugitaet al., 2001). Sugitaet al. (2001) suggested that the expression ofdax1in frog may be involved in testicular development.Considering the similarity ofdax1expressions between scallop and frog, we hypothesized thatCf-dax1takes part in the testis development ofC.farreri. Furthermore, thedax1gene may function in early ovarian development of adult scallop as well, since its expression level is relatively high in ovaries at proliferative stage.
In gonads of most vertebrates such as mouse, rat and medaka, DAX1 gene is only expressed in somatic cells (Nakamotoet al., 2007; Swainet al., 1996; Tamaiet al., 1996). In present study, the Cf-DAX1 in gonad was mainly located in germ cells, in spermatogonia and spermatocytes of testis, and in oogonia and oocytes of ovary (Fig.5). We could not explain the differences of cellular location between Cf-DAX1 and most vertebrates DAX1s, while the mainly expression pattern in germ cells suggests that Cf-DAX1 may be involved in the development of germ cells inC. farrei.
4.4 Equivalent Copy ofCf-dax1on Chromosomes
The chromosome locations ofdax1are different among species, some are located on autosomes and some are on sex chromosomes. As it is well known that humandax1is located on X chromosome (Zanariaet al., 1994). While in chicken and marsupial, thedax1is autosomal, and lies on chromosome 1 and chromosome 5, respectively (Pasket al., 1997; Smithet al., 2000). So it is necessary to identify the location ofCf-dax1on chromosomes. In this study, the FISH results showed thatCf-dax1located on the short arm of a pair of subtelocentric chromosome (Fig.6), which indicated that both male and female scallop embryos might have equivalent copy of the gene, and the differences in expression are therefore due to the different regulation between sexes. To date, there are no reports about the systematic numbering and the existence of sex chromosomes inC. farreri, so more studies are necessary to revealCf-dax1location.
5 Conclusions
We cloned a full length cDNA ofdax1from scallop, and a LXXLL motif of vertebrate type was not found in Cf-DAX1. This is the first report on the expression characteristics and chromosomal localization ofdax1in invertebrates. In adult scallop,Cf-dax1expression was examined in various tissues with different levels. The mRNA level ofCf-dax1showed a sex dimorphic feature in favor of testis, which is similar to many vertebrates. Cf-DAX1 was located in the germ cells and may take part in the gametogenesis of bivalves.
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (863 Program) (2012AA10A402), the Natural Science Foundation of Qingdao (11-2-4-1(10)-jch) and the Key Natural Science Foundation of Shandong Province (Z2008D02).
Achermann, J. C., Meeks, J. J., and Jameson, J. L., 2001. Phenotypic spectrum of mutations in DAX-1 and SF-1. Molecular and Cellular Endocrinology, 185 (1-2): 17-25.
Agoulnik, I. U., Krause, W. C., Bingman III, W. E., Rahman, H. T., Amrikachi, M., Ayala, G. E., and Weigel, N. L., 2003. Repressors of androgen and progesterone receptor action. Journal of Biological Chemistry, 278: 31136-31148.
Bae, D. S., Schaefer, M. L., Partan, B. W., and Muglia, L., 1996. Characterization of the mouse DAX-1 gene reveals evolutionary conservation of a unique amino-terminal motif and widespread expression in mouse tissue. Endocrinology, 137: 3921-3927.
Bardoni, B., Zanaria, E., Guioli, S., Floridia, G., Worley, K. C., and Tonini, G., 1994. A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nature Genetics, 7: 497-501.
Baron, D., Houlgatte, R., Fostier, A., and Guiguen, Y., 2005. Large-scale temporal gene expression prof i ling during gonadal differentiation and early gametogenesis in rainbow trout. Biology of Reproduction, 73: 959-966.
Burris, T. P., Guo, W., and McCabe, E. R. B., 1996. The gene responsible for adrenal hypoplasia congenita, DAX1, encodes a nuclear hormone receptor that defines a new class within the superfamily. Recent Progress in Hormone Research, 51: 241-260.
Cheng, J., 2010. Construction and application of large insert libraries for Zhikong Scallop (Chlamys farreri). PhD thesis, Ocean University of China (in Chinese).
Guo, W., Mason, J. S., Stone, C. G., Morgan, S. A., Madu, S. I., Baldini, A., Lindsay, E. A., Biesecker, L. G., Copeland, K. C., Horlick, M. N. B., Pettigrew, A. L., Zanaria, E., and McCabe, E. R. B., 1995. Diagnosis of X-linked adrenal hypoplasia congenita by mutation analysis of the DAX1 gene. Journal of the American Medical Association, 74: 324-330.
Guo, W. W., Rhonda, S. L., Zhang, Y. H., Huang, B. L., Thomas, P. B., William, J. C., and McCabe, E. R. B., 1996. Ahch, the mouse homologue of DAX1 cloning, characterization and synteny with GyK, the glycerol kinase locus. Gene, 178: 31-34.
Holter, E., Kotaja, N., Makela, S., Strauss, L., Keitz, S., Janne, O. A., Gustafsson, J. A., Palvimo, J. J., and Treuter, E., 2002. Inhibition of androgen receptor (AR) function by the reproductive orphan nuclear receptor DAX1. Molecular Endocrinology, 16: 515-528.
Huang, X. T., Bao, Z. M., Bi, K., Hu, J. J., Zhang, C., Zhang, Q. Q., and Hu, X. L., 2006. Chromosomal localization of the major ribosomal RNA genes in scallop Chlamys farreri. Acta Oceanologica Sinica, 25 (3): 108-115.
Ikeda, Y., Swain, A., Weber, T. J., Hentges, K. E., Zanaria, E., Lalli, E., Tamai, K. T., Sassone-Corsi, P., Lovell-Badge, R., Camerino, G., and Parker, K. L., 1996. Steroidogenic factor 1 and Dax-1 colocalize in multiple cell lineages: potential links in endocrine development. Molecular Endocrinology, 10: 1261-1272.
Ikeda, Y., Takeda, Y., Shikayama, T., Mukai, T., Hisano, S., and Morohashi, K., 2001. Comparative localization of Dax-1 and Ad4BP/SF-1 during development of the hypothalamic-pituitary-gonadal axis suggests their closely related and distinct functions. Developmental Dynamics, 220: 363-376.
Ito, M., Yu, R., and Jameson, J. L., 1997. DAX-1 inhibits SF-1 mediated transactivation via a carboxy-terminal domain thatis deleted in adrenal hypoplasia congenita. Molecular and Cellular Biology, 17: 1476-1483.
Kenneth, J. L., and Thomas, D. S., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△ctmethod. Methods, 25: 402-408.
Lalli, E., Bardoni, B., Zazopoulos, E., Wurtz, J. M., Strom, T. M., Moras, D., and Sassone-Corsi, P., 1997. A transcriptional silencing domain in DAX-1 whose mutation causes adrenal hypoplasia congenita. Molecular Endocrinology, 11 (13): 1950-1960.
Lalli, E., Ohe, K., Hindelang, C., and Sassone-Corsi, P., 2000. Orphan receptor DAX-1 is a shuttling RNA binding protein associated with polyribosomes via mRNA. Molecular and Cellular Biology, 20: 4910-4921.
Liao, C. Y., Xu, Y. F., and Wang, Y. L., 1983. Reproductive cycles of scallop Chlamys farreri (Jones and Preston 1904) at Qingdao. Journal of Fisheries of China, 1: 1-13 (in Chinese).
Martins, R. S., Deloffre, L. A., Mylonas, C. C., Power, D. M., and Canario, A. V., 2007. Developmental expression of DAX1 in the European sea bass, Dicentrarchus labrax: Lack of evidence for sexual dimorphism during sex differentiation. Reproductive Biology and Endocrinology, 5 (19): 1-13.
Moras, D., and Gronemeyer, H., 1998. The nuclear receptor ligand-binding domain: Structure and function. Current Opinion in Cell Biology, 10 (3): 384-391.
Nakamoto, M., Wang, D. S., Suzuki, A., Matsuda, M., Nagahama, Y., and Shibata, N., 2007. Dax1 suppresses P450arom expression in medaka ovarian follicles. Molecular Reproduction and Development, 74: 1239-1246.
Niakan, K. K., and McCabe, E. R. B., 2005. DAX1 origin, function, and novel role. Molecular Genetics and Metabolism, 86: 70-83.
Nuclear Receptors Nomenclature Committee, 1999. A unified nomenclature system for the nuclear receptor superfamily. Cell, 97: l6l-l63.
Park, S. Y., Joshua, J. M., Gerald, R., Liza, E. P., Weiss, J., Hammer, G. D., and Jameson, J. L., 2005. Nuclear receptors Sf1 and Dax1 function cooperatively to mediate somatic cell differentiation during testis development. Development, 132: 2415-2423.
Parma, P., Pailhoux, E., Puissant, C., and Cotinot, C., 1997. Porcine Dax-1 gene: Isolation and expression during gonadal development. Molecular and Cellular Endocrinology, 135: 49-58.
Pask, A., Toder, R., Wilcox, S. A., Camerino, G., and Graves, J. A. M., 1997. The candidate sex-reversing DAX1 gene is autosomal in marsupials: Implications for the evolution of sex determination in mammals. Genomics, 41: 422-426.
Smith, C. A., Clifford, V., Western, P. S., Wilcox, S. A., Bell, K. S., and Sinclair, A. H., 2000. Cloning and expression of a DAX1 homologue in the chicken embryo. Journal of Molecular Endocrinology, 24 (1): 23-32.
Sugita, J., Takase, M., and Nakamura, M., 2001. Expression of Dax-1 during gonadal development of the frog. Gene, 280 (1-2): 67-74.
Suzuki, T., Kasahara, M., Yoshioka, H., Morohashi, K., and Umesono, K., 2003. LXXLL-related motifs in Dax-1 have target specif i city for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Molecular and Cellular Biology, 23: 238-249.
Swain, A., Zanaria, E., Hacker, A., Lovell-Badge, R., and Camerino, G., 1996. Mouse Dax1 expression is consistent with a role in sex determination as well as in adrenal and hypothalamus function. Nature Genetics, 12: 404-409.
Tamai, K. T., Monaco, L., Alastalo, T. P., Lalli, E., Parvinen, M., and Sassone-Corsi, P., 1996. Hormonal and developmental regulation of DAX-1 expression in Sertoli cells. Molecular Endocrinology, 10: 1561-1569.
Wang, D. S., Kobayashi, T., Senthilkumaran, B., Sakai, F., Sudhakumari, C. C., Suzuki, T., Yoshikuni, M., Matsuda, M., Morohashi, K., and Nagahama, Y., 2002. Molecular cloning of DAX1 and SHP cDNAs and their expression patterns in the Nile tilapia, Oreochromis niloticus. Biochemical and Biophysical Research Communications, 297 (3): 632-640.
Western, P. S., Harry, J. L., Graves, J. A. M., and Sinclair, A. H., 2000. Temperature-dependent sex determination in the American alligator: Expression of SF1, WT1 and DAX1 during gonadogenesis. Gene, 241 (2): 223-232.
Yang, F. Y., and Zhang, B., 2009. Comparative nutrition components in various populations of Chlamys farreri. Journal of Anhui Agricultural Science, 37 (9): 4073-4075 (in Chinese).
Yu, R. N., Ito, M., Saunders, T. L., Camper, S. A., and Jameson, J. L., 1998. Role of Ahch in gonadal development and gametogenesis. Nature Genetics, 20: 353-357.
Zanaria, E., Muscatelli, F., Bardoni, B., Strom, T. M., Guioli, S., Guo, W., Lalli, E., Moser, C., Walker, A. P., McCabe, E. R. B., Meitinger, T., Monaco, A. P., Sassone-Corsi, P., and Camerino, G., 1994. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature, 372: 635-641.
Zazopoulos, E., Lalli, E., Stocco, D. M., and Sassone-Corsi, P., 1997. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature, 390: 311-315.
Zhang, H., Thomsen, J. S., Johansson, L., Gustafsson, J. A., and Treuter, E., 2000. DAX1 functions as an LXXLL-containing corepressor for activated estrogen receptors. Journal of Biological Chemistry, 275: 39855-39859.
Zhang, L. L., Bao, Z. M., Wang, S., Hu, X. L., and Hu, J. J., 2008. FISH mapping and identification of Zhikong scallop (Chlamys farreri) chromosomes. Marine Biotechnology, 10: 151-157.
Zhang, G., Fang, X., and Guo, X., 2012a. The oyster genome reveals stress adaptation and complexity of shell formation. Nature, 490: 49-54.
Zhang, H., Pan, L., and Zhang, L., 2012b. Molecular cloning and characterization of estrogen receptor gene in the scallop Chlamys farreri: Expression prof i les in response to endocrine disrupting chemicals. Comparative Biochemistry and Physiology Part C, 156: 51-57.
Zhou, Q. R., Shao, M. Y., Qin, Z. K., Kang, K. H, and Zhang, Z. F., 2010. Cloning, characterization, and expression analysis of the DEAD-box family genes, Fc-vasa and Fc-PL10a, in Chinese shrimp (Fenneropenaeus chinensis). Chinese Journal of Oceanology and Limnology, 28: 37-45.
(Edited by Qiu Yantao)
(Received February 12, 2013; revised April 24, 2013; accepted June 16, 2014)
© Ocean University of China, Science Press and Spring-Verlag Berlin Heidelberg 2014
* Corresponding author. Tel: 0086-532-82031647 E-mail: zzfp107@ouc.edu.cn
杂志排行
Journal of Ocean University of China的其它文章
- Construction of Inorganic Elemental Fingerprint and Multivariate Statistical Analysis of Marine Traditional Chinese Medicine Meretricis concha from Rushan Bay
- A Comparative Study of Intensive Litopenaeus vannamei Culture on Four Bottom Substrates Without Water Change
- Secondary Metabolites of a Deep Sea Derived Fungus Aspergillus versicolor CXCTD-06-6a and Their Bioactivity
- Preparation, Characterization and Pharmacokinetics of Fluorescence Labeled Propylene Glycol Alginate Sodium Sulfate
- Early Development of Silvetia babingtonii (Fucales, Phaeophyceae)
- A Comparison of Different Gracilariopsis lemaneiformis (Rhodophyta) Parts in Biochemical Characteristics, Protoplast Formation and Regeneration
