In Silico Analysis and Feeding Assays of Some Genes in the Early Steps of Terpenoid Biosynthetic Pathway in Camellia Sinensis
2013-12-13WeiXiangYanXuFuMinWangLiPingGaoMingJunGaoZhengZhuZhangXiaoChunWanShuWei
Wei Xiang,Yan Xu,Fu-Min Wang,Li-Ping Gao,Ming-Jun Gao,Zheng-Zhu Zhang,Xiao-Chun Wan,Shu Wei*
1.Key Laboratory of Tea Biochemistry and Biotechnology,Anhui Agricultural University,130 Changjiang Blvd West,Hefei,Anhui,230036,China.
2.College of Life Sciences,Anhui Agricultural University,130 Changjiang Blvd West,Hefei,Anhui,230036,China.
3.Agriculture and Agri-Food Canada,Saskatoon Research Center,Saskatoon,Saskatchewan,S7N 0X2,Canada.
1.Introduction
Tea(Camellia sinensis)is one of the most popular non-alcoholic beverages worldwide.Terpenoid volatiles,such as linalool and neriodiol,are crucial for the pleasant but distinctive scent in different teas[1,2].Additionally,terpenoid-derived photosynthetic pigments(carotenoids,chlorophylls,and plastoquinone)and regulatory hormones(cytokinins,abscisic acid,and strigolactones)should be involved directly and indirectly in the regulation of tea plant growth and shoot branching as shown in many other plants[3].
Plant terpenoids are all derived from the plastidial methyl-erythritol-phosphate(MEP)pathway and the cytosolic mevalonate(MVA)pathway.The two,often viewed as the early steps of isoprenoid biosynthetic pathways[4],produce common building block of isopentenyl diphosphate(IPP)and its allylic isomer dimethylallyl diphosphate(DMAPP)[5,6].Subcellular compartmentalization of the MEP and MVA pathways leads to distinct sets of isoprenoid products[8],although some metabolic crosstalk between the two pathways exists[9].From the MVA pathway and its downstream reactions, sesquiterpenoids, sterols, and brassinosteroids are produced while gibberellins,monoterpenoids,photosynthetic pigments,abscisic acid and strigolactones are derived from MEP and plastidial terpenoid pathways[8]. The MVA and MEP pathways are comprised of six and eight enzymes,respectively[5].The homologs of these enzymes in the two pathways are found in various plant species[10],suggesting that both MVA and MEP pathways are highly conserved in plants(Fig.1).Thus,their conserved sequences could be used for identifying the tea homologs for dissecting the essential pathways in the plant.
Multiple mechanisms that regulate the MEP and MVA pathways in plantshave been revealed.Genome-wide expression analyses indicate that transcriptional control plays a key role over the entire isoprenoid pathway network[11].Developmental and environmental regulation of transcription of the genes in the MVA and MEP pathways have been extensively studied.Transcription factors such as ORCA3 in Catharanthus roseus,play key role in controlling the gene expression[12].Moreover,metabolite feedback regulation of gene expression was also found in the two pathways in plants.Feeding of the sesquitepenoid farnesol to tobacco cell culture revealed that expression of the gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase(HMGR)in MVA pathway is metabolite mediated.In mycorrhizal maize roots,the expression of genes encoding DXP synthase(DXS)and DXP reductoisomerase(DXR)in MEP pathway is induced by apocarotenoids[13].However,the transcription regulation of the other genes in the two pathways by key metabolites such as the common precursor IPP and starting chemicals acetyl CoA and DXP remains unknown.
In order to identify the main gene components in the MVA and MEP pathways in C.sinensis and learn their possible metabolite-mediated expression,we took advantage of quickly increased transcriptome data in C.sinensis[14]and obtained five complete and three partial transcript sequences of the genes in the MVA and MEP pathways from C.sinensis using the technology of rapid amplification of cDNA ends(RACE).In silico analyses of the genes with complete coding sequences were conducted.Moreover,feeding of precursor metabolites to leaf and cell cultures were also performed in vitro to study metabolite-mediated gene expression in the MEP and MVA pathways in C.sinensis.
2.Materials and methods
2.1.DNA sequence homology search
Transcriptome and incomplete genome sequencing dada were obtained from C.sinensis cv.‘Longjing 43’[14]and‘Tie Guanyin’,respectively.Similarity searches using the preliminarily annotated genes in the terpenoid pathway as query sequences were performed against the database at the National Centre for Biotechnology Information(NCBI)with the BLASTX program with default parameters.Multiple alignments of the protein sequences were conducted using the on-line ClustalW2.Protein subcellular locations were prediction using TagetP

Fig.1.Two distinct biosynthetic pathways for IPP and DMAPP biosynthesis in plants.
2.2.cDNA cloning
Total RNA was extracted from shoot tips of‘Longjing 43’plants using the QIAGEN RNeasy kit.DNase I was used for on-column DNA digestion to minimize genomic DNA contamination.First-strand cDNA was synthesized using a PrimeScript RT reagent kit(Takara)with 1 g total RNA.The full coding sequences of CsAACT,CsHMGS,CsCMK,and CsFPS were obtained by polymerase chain reactions(PCR),and 5'or 3'Rapid Amplification of cDNA Ends(RACE).The First Choice RLM-RACE Kit(Ambion)was used for RACE following the manufacturer's instructions.Briefly,for 5'and 3'RACE,five micrograms of total RNA was ligated to the RNA adapter and a random-primed reverse transcription reaction was performed to synthesize cDNA.A second round of PCR was carried out using a nested adapter primer and gene specific primers.The RACE and regular PCR products were cloned into the pGEM-T Easy vector(Promega)for sequencing.
2.3.Callus,leaf disc cultures,and cell suspensions
The elite tea cultivar‘Nong-Men-Kang’was used in this experiment.For callus induction,seeds were rinsed in 70%(v/v)ethanol for 1 min and then sterilized in 3%(v/v)NaClO for 15 min,followed by five washes with sterile distilled water.After removal of seed coats,seeds were slightly wounded across the surface using a scalpel blade and transferred to B5 solid medium supplemented with 30g/L sucrose,0.5mg/L 2,4-dichlorophenoxyacetic acid(2,4-D)and 0.1mg/L kinetin(KT),pH 5.7.Cultures were incubated at 25 1℃ in darkness,and subcultured every 2 weeks till callus formation.The obtained calli were transferred to liquid B5 medium with the same supplements as for callus induction,and the cell suspension cultures were incubated on a rotary shaker(100 rpm)at 25 1 C.Leaf-disc cultures were prepared using in vitro plantlets of C.sinensis as described by Iwase et al.(2005).
2.4.Metabolite feeding
Chemicals for metabolite feeding,acetyl-CoA,DXP,IPP and DMAPP,were analytical grade and purchased from Sigma.The compounds were fed to 2-week old cell suspension cultures at the linear growth phase(Choi et al.2006).Leaf discs were acclimated for 4 days in a liquid medium under a light intensity of 60 mol.m-2.s-1with a 12-hour photoperiod,and then transferred to the new media supplemented with the chemicals at a final concentration of 1.5 M(Kang et al.2006).
2.5.Real-time quantitative PCR
Cell suspension and leaf-disc samples were collected at 0h,0.5h,2h,4h,and 24h after metabolite feeding.Total RNA extraction,DNA contamination removal,and first-strand cDNA synthesis were conducted as described above for cDNA cloning.Primers used for real-time quantitative PCR(qPCR)assays were designed based on the identified cDNA sequences from C.sinensis using Primer Premier 5 program(Table1).The PCR mixture contained 10 μL of diluted cDNA(25ng),12.5 μL of SYBR Premix Ex Taq II(Takara),and 200 nM of each pair of gene-specific primers in a final volume of 25 μL.the qPCR were performed using a Bio-RAD iQ-5 Detection System under the following conditions:2 min at 50℃,10 min at 95℃,and 40 cycles of 15 s at 95℃ and 1 min at 55℃ in 96-well optical reaction plates.The specificity of amplicons was verified by melting curve analysis(55 to 95℃)after 40 cycles and by agarose gel electrophoresis as well.Glyceraldehyde-3-phosphate dehydrogenase(GAPDH)was used as the endogenous reference gene according to Sun et al.(2010).Transcript levels were normalized and calculated using the 2-ΔΔCtmethod.Three biological replicates for each treatment were used for the analysis.
3.Results
3.1.Identification of genes at the early steps of biosynthetic pathway for terpenoid production
Based on the partial sequences derived from sequencing data as well as the preliminarily gene annotation,a total of seven genes in the early steps of terpenoid biosynthesis were found in C.sinences(Fig.1)(Supplementary Table2).Full coding sequences of CsAACT,CsHMGS,CsCMK,and CsFPS were obtained using PCR and RACE approaches.CsHDS coding sequence was retrieved from the NCBI database(Accession#JQ014629.1).

Table1.Primers for quantitative PCR and expected product sizes.

Table2.Primers used in this study for gene cloning.
The deduced complete protein sequences of the five genes in C.sinences were compared with their functional homologs in other plants.For CsAACT gene,a deduced protein has 408 amino acid residues.Alignment of CsAACT with functional homologs from Arabidopsis and sunflower revealed that CsAACT shares 87% identity(94%similarity)with its homolog AtAACT 2(At5g48230),81%identity(92%)with AtAACT1(At5g47720)in Arabidopsis,and 81%identity(91%)with HaAACT(QC25419)in sunflower.All Arabidopsis and sunflower homologs of CsAACT were characterized as acetyl-CoA C-acetyltransferase[15,16].AtAACT2 can generates the bulk of the acetoacetyl-CoA required forthe cytosol-localized,mevalonate-derived isoprenoid biosynthesisin Arabidopsis[15].TargetP 1.1 program predicted that CsAACT did not contain any signal peptide,suggesting that it is likely localized in cytosol.These data indicate that CsAACT in tea plants should function as an acetyl-CoA C-acetyltransferase.CsAACT does not contain the tripeptide peroxisomal targeting sequence SKL at the C-terminus as found in the sunflower AACT[16].
For CsHMGS,the full-length cDNA encodes a protein containing 464 amino acid residues.CsHMGS shares 86%identity in amino acid with its homolog in Brassica juncea(BjbHMGS1)(AAF69804.1),and 75%identity with Pinus sylvestris(PsHMGS)(CAA65250.1).The CsHMGS protein has the conserved active-site segment corresponding to G107through A125.Moreover,conserved residues C120,H250and N329that are essential for HMGS activity are also present in CsHMGS.
For CsCMK,the full-length gene sequence encodes for a protein of 426 amino acid residues.Although studies on the crystal structures of CMKs in many species of bacteria such as Mycobacterium tuberculosis have clearly revealed the overall and catalytic pocket structure of the enzyme[17],the structure of the plant enzyme is not well characterized so far.In tomato the residues were proposed to represent a putative ATP binding site by Rohdich et al.(2000)[18].Accordingly,in the tea CsCMK the putative ATP binding site was found.
For CsHDS,the deduced protein contains 741 amino acid residues.The plant proteins contain a signal sequence for plastid import,which has been shown to be functional in localizing the Arabidopsis homolog in chloroplast.Additionally,CsHDS and its homologs contain a large and functional insert domain(A*)besides a TIM barrel and a 4Fe4S domain,which are present in bacterial homologs[19].
CsFPS gene encodes a protein consisting of 341 amino acid residues.A multiple alignment indicated that the tea CsFPS had 65.6%and 73.1%identity and 71.6%and 90.0%similarity with AtFPS1L(AAF44787)and AtFPS2(NP_193452)in Arabidopsis,respectively.CsFPS does not contain a NH2-terminal extension of 41 amino acid residues,which is a mitochondrial transit peptide in Arabidopsis AtFPS1L.CsFPS contains residues D93,D96,R101,R102,and D231which are highly conserved and crucial for the plant enzyme activity.
3.2.Expression of the genes in MVA and MEP pathways in response to metabolite feeding

Fig.2.Expression of some of tea genes in the MVA and MEP pathways after feeding with different precursor metabolites.
The relative transcript level of the genes was quantified using quantitative real time PCR.The amplicons for all the genes examined in this study were highly specific,and verified by melting curve analysis and by agarose gel electrophoresis(data not shown).After feeding the tissues with the metabolite,the expression of the genes in both the MVA and MEP pathways were all induced to a different extent(Fig.2).Acetyl-CoA is the starting compound in the MVA pathway.Within half an hour after acetyl-CoA feeding to leaf discs,the expression of CsHMGS was increased by about 3-fold,compared to the non-feeding control(Fig.2A).The CsAACT expression changed slightly over the test period after feeding.In the cell suspension culture,the transcript level of CsAACT,and CsHMGS were increased by >1.5-and >3.0-fold,peaking at 0.5 hr and 4 hr after feeding,respectively(Fig.2B).Acetyl-CoA had the highest induction on the expression of CsHMGS in both leaf disc and cell cultures.
In orderto examine the substrate effecton the gene transcription of the MEP pathway,DXP was fed to the cell and leafdisc cultures.Two hours after feeding,the expression of CsCMK and CsHDS was respectively increased by 1.8-and 1.6-fold(Fig.2C).In cell cultures,the transcript levels of CsCMK and CsHDS were increased>2.2-and 1.5-fold,peaking at 24 hr and 4 hr after feeding,respectively(Fig.2D).These results indicated that the DXP supplementation in leaf and cell cultures induced the expression of CsCMK and CsHDS in the MEP pathway,suggesting that these positive-feedback biochemical steps favor the metabolic flux down to IPP biosynthesis in the MEP pathway.
3.3.Expression of CsFPS in response to the feeding of different precursors
After feeding DXP,IPP or DMAPP to leaf disc cultures,the transcript level of CsFPS was increased by 2.0-,1.7-and 1.8-fold,and peaking at 4 hr,2 hr,and 4 hr,respectively.The transcript level of CsFPS was decreased at 2 hr after acetyl-CoA feeding(Fig.3A).In the cell culture,the transcript level of CsFPS was substantially increased(up to 1.8-fold)2 hr after DXP feeding.But it was decreased at different time after feeding of acetyl-CoA,IPP or DMAPP(Fig.3B).
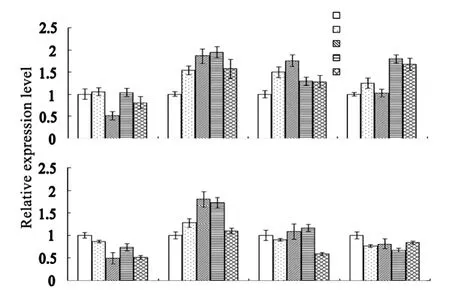
Fig.3.Transcription of CsFPS was affected with the addition of precursor metabolites acetyl-CoA,DXP,IPP,and DMAPP.
4.Discussion
In this study,the full coding sequences of CsAACT,CsHMGS,CsCMK,CsHDS and CsFPS,were obtained based on the transcriptome and genome sequencing data previously obtained from tea plants[14].In silico analyses indicated that all these genes have high identity with their functional homologs in some other plant species and share conserved motifs essential for their function.Additionally,the partial coding sequences of CsHMGR,CsDXR,and CsIDI were also obtained for further characterization.Thus the early steps of terpenoid biosynthesis pathway in C.sinensis began to reveal.However,some other genes in the MVA and MEP pathways such as CsMVK,PMK,CsPMD,and CsCMS were not identified in this study.In addition,it is known that plants often have multiple isoforms in a gene family.For example,Arabidopsis thaliana has three members of DXS family,and Hevea brasiliensis has two members of DXR.Isoforms of some genes identified in this study and other gene components are expected to be found when the sequencing program proceeds to have a better coverage of C.sinensis genome.
In plants metabolite mediators can regulate gene expression[20],due to their capability to interact with some protein factors[21]and riboswitches which are complex folded RNA domains that serve as receptors for specific metabolites.Supplementation of precursor metabolites into in vitro culture media are often employed to study plant terpenoid metabolic flux regulation.In this study,expression of the identified genes in response to precursor metabolites fed to leaf and cell cultures was examined using a quantitative PCR approach.Our results show that for the MVA pathway,the expression of CsHMGS was enhanced due to the addition of the precursor acetyl-CoA in both callus and leaf cultures although it was unclear why the gene expression enhancement occurred hours earlier in leaf disc culture than in cell cultures.The expression of CsAACT was slightly affected by the addition of the precursor(Fig.3)while CsFPS responded unconspicuous to acetyl-CoA feeding,suggesting that different behaviors of the two genes to the addition of the same metabolite.
In the MEP pathway,the expression of CsCMK and CsHDS in leaf disc and cell cultures was enhanced when DXP was added to the culture media(Fig.2C and D).These results suggested that the expression of some MEP genes is highly coordinated,which is firmly supported by the previous findings.FPS catalyzes the reaction where two molecules of IPP and one molecule of DMAPP are condensed to form one intermediate GPP and then the final product FPP.FPS enzyme has been found in cytoplasm,mitochondria,chloroplast,and peroxisome.In this study,CsFPS gene was induced in leaf disc cultures when fed with DXP,IPP and DMAPP.While in the cell culture,CsFPS expression increased in the response to the addition of DXP,but not IPP or DMAPP(Fig.3A and B).Our data suggested that the expression of CsFPS,encoding the cytosolic protein CsFPS,was enhanced by DXP,a metabolite of the plastidal MEP pathway.This is probably because DXP supplementation led to the increased production of IPP in the plastid,which could be further transported to the cytoplasm.This speculation is supported by the findings that metabolic‘crosstalk’between the MVA and MEP pathways exists,particularly from plastids to the cytosol.However,acetyl-CoA feeding did not affect the CsFPS transcript level in both leaf disc and cell cultures at majority of monitoring points.It might be because the amount of acetyl-CoA in tea cells or leaf tissue was not a limiting factor for the metabolic flux in MVA pathway whereas HMGR is usually considered as a limiting enzyme.It was noted that IPP and DMAPP induced transcription of CsFPS only in leaf cultures,not in cell culture(Fig.3B).Further studies will be carried out to find out the mechanisms underlying such a tissue dependent feeding response of CsFPS in C.sinensis.
In this study CsAACT,CsHMGS,CsCMK,CsHDS,and CsFPS at the early steps of terpenoid biosynthetic pathway in C.sinensis have been proposed and their expression was found in the first time being metabolitemediated in different extents.Our results began to unravel the mechanisms that control the low terpenoid biosynthesis in C.sinensis.
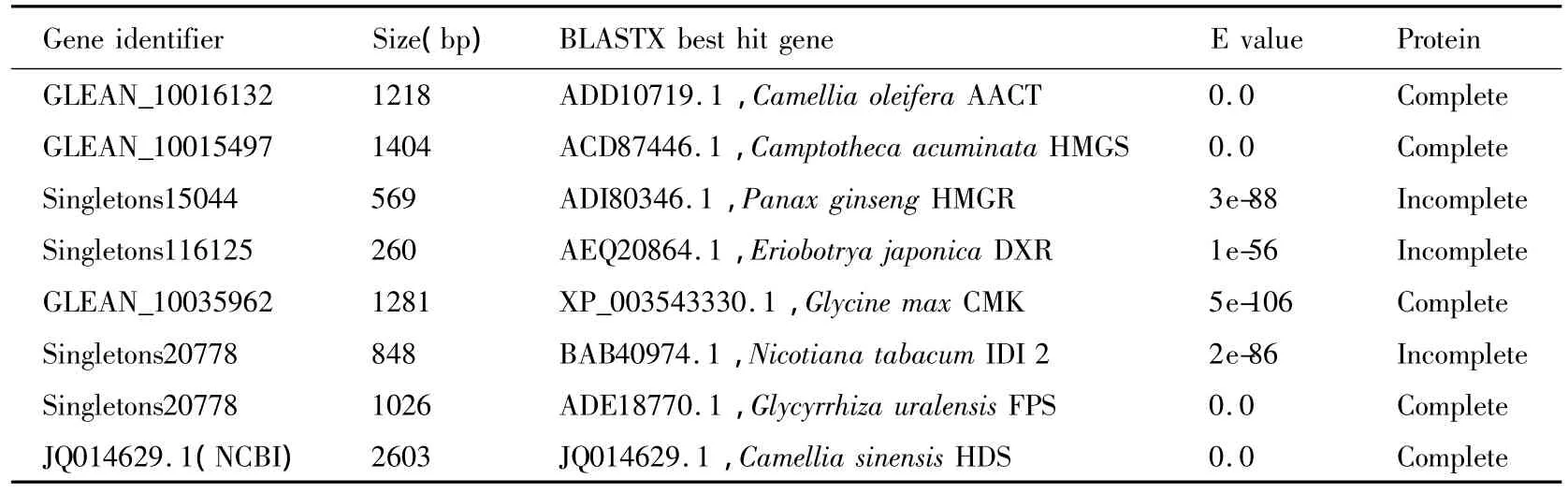
Table3.Genes in terpenoid metabolism in C.sinensis identified in this study.
The authors are grateful to Drs Chaoling Wei and Hua Yang for providing the transcriptome data of C.sinensis.Prof.Rong-Fu Wang and Ms.Juan Li at the Biotechnological Center,Anhui Agricultural University offered their technical help.This work was funded by the National Science Foundation in China(#31070614 to S.Wei),the Research Fund for the Doctoral Program of Higher Education of The ministry of Education(#20123418110002,to S.Wei),the Program for Changjiang Scholars and Innovative Research Team in Universities(IRT1101 to Z.Z.Zhang)the"Twelfth Five-Year"National Key Basic Research and Development Project(973)in China(2012CB722903 to Xiao-Chun Wan).
1.Schuh C,Schieberle P.Characterization of the key aroma compounds in the beverage prepared from Darjeeling black tea quantitative differences between tea leaves and infusion.Journal of Agricultural and Food Chemistry,2006,54:916-924.
2.Yamanishi T.Tea flavor.In:Jain NK(ed)Global Advances in Tea Science.Aravalli Books International:New Delhi,India,1999,pp707-722.
3.Hemmerlin A,Harwood JL,Bach TJ.A raison d'être for two distinct pathways in the early steps of plant isoprenoid biosynthesis.Progress in Lipid Research,2012,51:95-148.
4.Rodríguez-Concepción M.Early steps in isoprenoid biosynthesis:multilevel regulation of the supply of common precursors in plant cells.Phytochemistry Reviews,2006,5(1):1-15.
5.McCaskill D,Croteau R.Isoprenoid synthesis in peppermint(Mentha x piperita):development of a model system for measuring flux of intermediates through the mevalonic acid pathway in plants.Biochemical Society Transactions,1995,23:290.
6.Withers ST,Keasling JD.Biosynthesis and engineering of isoprenoid small molecules.Applied Microbiology and Biotechnology,2007,73:980-990.
7.Vranov E,Coman D,Gruissem W.Structure and dynamics of the isoprenoid pathway network.Molecular Plant,2012,5:318-33.
8.Laule O,F rholz A,Chang HS,Zhu T,Wang X,Heifetz PB,Gruissem W,Lange M.Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana.Proceedings of the National Academy of Sciences of the United States of America,2003,100:6866-8671.
9.Lange BM,Rujan T,Martin W,Croteau R.Isoprenoid biosynthesis:the evolution of two ancient and distinct pathways across genomes.Proceedings of the National Academy of Sciences of the United States of America,2000,97:13172-13177.
10.Botella-Pav a P,Besumbes O,Phillips MA,Carretero-Paulet L,Boronat A,Rodríguez-Concepción M.Regulation of carotenoid biosynthesis in plants:evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors.Plant Journal,2004,40:188-199.
11.van der Fits L,Memelink J.ORCA3,a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism.Science,2000,289:295-297.
12.Walter MH,Fester T,Strack D.Arbuscular mycorrhizal fungi induce the non-mevalonate methylerythritol phosphate synthase pathway of isoprenoid biosynthesis correlated with accumulation of the yellow pigment and other apocarotenoids.Plant Journal,2000,21:571-578.
13.Shi CY,Yang H,Wei CL,Oliver Y,Zhang ZZ,Jiang CJ,Sun J,Li YY,Chen Q,Xia T,Wan XC.Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds.BMC Genomics,2011,12:131.
14.Jin H,Song Z,Nikolau BJ.Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development.Plant Journal,2012,70:1015-1032.
15.Dyer JH,Anthony Maina A,Gomez ID,Cadet M,Oeljeklaus S,Schiedel AC.Cloning,expression and purification of an acetoacetyl CoA thiolase from sunflower cotyledon.International Journal of Biological Science,2009,5:736-744.
16.Shan S,Chen X,Liu T,Zhao H,Rao Z,Lou Z.Crystal structure of 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase(IspE)from Mycobacterium tuberculosis.FASEB Journal,2011,25:1577-1584.
17.Rohdich F,Wungsintaweekul J,Eisenreich W,Richter G,Schuhr CA,Hecht S,Zenk MH,Bacher A.Biosynthesis of terpenoids:4-diphosphocytidyl-2-C-methyl-d-erythritol synthase of Arabidopsis thaliana.Proceedings of the National Academy of Sciences of the United States of America,2000,97:6451-6456.
18.Okada K,Hase T.Cyanobacterial non-mevalonate pathway-(E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase interacts with ferredoxin in Thermosynechococcus elongatus Bp-1.Journal of Biological Chemistry,2005,280:20672-20679.
19.Hannah MA,Caldana C,Steinhauser D,Balbo I,Fernie AR,Willmitzer L.Combined transcript and metabolite profiling of Arabidopsis grown under widely variant growth conditions facilitates the identification of novel metabolite-mediated regulation of gene expression.Plant Physiology,2010,152:2120-2129.
20.Sellick CA,Reece RJ.Eukaryotic transcription factors as direct nutrient sensors.Trends in Biochemical Sciences,2005,30:405-412.
杂志排行
茶叶的其它文章
- Stability of Tea Catechins and Antioxidant Properties of Green Tea Extracts as Affected by Boiling-treatment
- The 30-Day Oral Administration Studies of Liposoluble Tea Polyphenols in Rats
- Oxidative Stability of Green Tea Extract-Enriched Rice Bran Oil During Storage
- Analysis of the Volatile Chemicals of Longjing Tea from Different Production Locations Using Electronic Nose
- Effects of Tannic Acid on Active Aluminum Species Distribution in Various Tea Soils
- Analysis of Volatile Compounds of Jinmudan Oolong Tea by Different Wrapping-Twisting
