幽门螺旋杆菌尿素酶A亚基抗原表位肽与霍乱毒素B亚基融合蛋白的生物信息学设计及表达优化研究
2013-06-07刘昆梅李小康
郭 乐, 刘昆梅, 李小康, 汤 锋, 奚 涛
1.宁夏医科大学检验学院,银川750021;
2.中国药科大学生命科学与技术学院,南京210009
幽门螺旋杆菌尿素酶A亚基抗原表位肽与霍乱毒素B亚基融合蛋白的生物信息学设计及表达优化研究
郭 乐1,2*, 刘昆梅1,2, 李小康2, 汤 锋2, 奚 涛2
1.宁夏医科大学检验学院,银川750021;
2.中国药科大学生命科学与技术学院,南京210009
幽门螺旋杆菌(Hp)是慢性胃炎、消化性溃疡和胃癌的重要致病因子。研发基于Hp尿素酶的表位疫苗是一种很有前景的防治Hp感染的策略。本研究主要通过生物信息学软件对Hp尿素酶表位肽U21和霍乱毒素B亚基(CTB)的连接顺序和间隔序列加以分析,设计出一种由U21和CTB构成的Hp表位疫苗CTB-UA。通过基因克隆技术构建含有融合基因CTB-UA的重组表达载体pETCUA及其重组菌株;重组菌株经经蛋白表达和优化后,利用Ni-NTP镍离子亲和层析和DEAE Sepharose FF阴离子交换层析纯化融合蛋白CTB-UA,获得高纯度(94.8%)的融合蛋白CTB-UA。并进一步通过腹腔注射免疫BALB/c小鼠,鉴定Hp表位疫苗CTB-UA的免疫学活性,经间接ELISA鉴定小鼠能够产生针对CTB和表位肽U21的高滴度特异性抗体。结果证明,Hp表位疫苗CTB-UA具有科学合理的结构,能在大肠杆菌表达系统中获得较高水平的表达,且具有较高的免疫学特异性,为研发防治Hp感染的表位疫苗奠定一定的实验基础。
Helicobacter pylori(Hp)has been recognized as themajor aetiological determinant of various gastro-duodenal diseases,such as chronic gastritis,peptic ulcers and gastric cancer[1~3].Current therapies,based on triple drug therapy containing three or four different antibiotics together with a proton-pump inhibitor,entail problems such as patient compliance,increasing antibiotic resistance,re-infection and high cost.Therefore,the development of an effective prophylactic or therapeutic vaccine against Hp will be of enormous scientific value and economic benefits.Epitope vaccine with unique design ideas is a new technology of vaccine development.
Urease is considered important colonization factor and pathogenic factor[4,5].It consists of a polymeric structure that comprises two subunits,UreA and UreB.The design of H vaccinesmainly paid attention to UreB in previous study because UreB is the catalytic subunit of Hp urease.However,more and more experimental evidences have proved that UreA is also an ideal candidate for vaccine design,especially somemonoclonal antibodies(mAb)against UreA.A mousemonoclonal antibody(mAbs)HpU-2 showed the strongest inhibitory effect on Hp urease activity.Further research indicated that HpU-2mainly recognized UreA butweakly UreB and the epitope sequence recognized by HpU-2 was a stretch of UreA-derived 21 amino acid residues(U21)[6].
In this study,an epitope vaccine named CTB-UA was designed by bioinformatics software.After gene cloning,expression optimization and purification,the fusion protein CTB-UA with high purity was obtained.The activity of epitope vaccine CTB-UA was investigated by intraperitoneal immunization experiments in BALB/c mice.
1 Materials and M ethods
1.1 M aterials
The plasmid pET22b,pUC57-T simple vector,and E.coli DH5αand E.coli BL21(DE3)were preserved in our laboratory.The recombinant plasmid pETCtUBE containing CTB and the epitope F8 gene from UreB was kindly provided by professor Wu Wutong.TIANpure Mini Plasmid Kit and TIANgel Midi Purification Kitwere purchased from Titan Biotech Ltd.The restriction enzymes NcoⅠ,XhoⅠ,Hin dⅢ,KpnⅠand T4 DNA ligase were purchased from TAKARA BIO INC.Ni2+-charged column chromatography was purchased from Bio Basic Inc.DEAE Sepharose FF anion-exchange chromatography was purchased from Amersham Pharmacia Biotech.
1.2 Design of the epitope vaccine CTB-UA
The theoretically optimal combination of the intramolecule adjuvant CTB,the linkers and the epitope U21 from UreA was established on the basis ofmodeling and prediction using RANKPEP,DNAstar software and molecular operating environment(MOE).
1.3 Construction of the recombinant p lasm ids
1.3.1 Construction of the recombinant plasmid pETC
The CTB gene was cloned by PCR amplification from pETCtUBE,using the forward primer P1-CTB(5′-CATGCCATGGGCACACCTCAAAATATTACTGATTTGTGTGC-3′),containing a NcoⅠrestriction site and the reverse primer P2-CTB(5′-CCCAAGCTTCGGTACCCGCGGATCATTTGCCATACTAATTGCGGCAATC-3′),containing KpnⅠ and Hin dⅢ restriction sites.The CTB gene was cloned into pET22b through NcoⅠ and Hin dⅢrestriction sites,generating the plasmid pETC.
1.3.2 Construction of the recombinantplasmid pETCUA
Two complementary single stranded DNA sequences coding U21 gene were synthesized:P1-U21(5′-CGAGCAGCAGCGTGGAACTGATTGATATTGGCGGCAACCGCCGCATTTTTGGCTTTAACGCGCTGGTGGATC-3′),containing a Kpn I restriction site and P2-U21(5′-TCGAGATCCACCAGCGCGTTAAAGCCAAAAATGCGGCGGTTGCCGCCAATATCAATCAGTTCCACGCTGCTGCTCGGTAC-3′),containing a XhoⅠ restriction site.These two sequences were mixed together,annealed and inserted subsequently into the plasmid pETC through KpnⅠ and XhoⅠ restriction sites,generating the recombinant plasmid pETCUA.
1.3.3 Screening and identification of the recombinants
Experimental methods are outlined below:①Recombinant plasmids were prepared from recombinant strains.The fusion geneswere identified by PCR amplification using the primersmentioned above.②The recombinant plasmids was identified by restriction endonuclease and detected with a 1%agarose gel electrophoresis.③After successful identification by PCR and restriction endonuclease,the recombinant plasmids were sent to Nanjing Jinsirui Biotechnology Company and further identified by gene sequencing.
1.4 Exp ression and optim ization of the recombinant strain
The recombinant plasmids pETCUA were transformed into E.coli BL21(DE3)for expression of the recombinant protein CTB-UA.The successful transformants were selected by resistance to ampicillin and verified by PCR,plasmid restriction digestand sequencing.The positive recombinantswere subcultured and used to express recombinant protein CTB-UA.
1.5 Purification of the fusion protein CTB-UA
The recombinant protein CTB-UA was purified by Ni2+-charged column chromatography according to the recommendation of the manufacturer.After the adsorption of CTB-UA protein to Ni-NTA SefinoseTMResin(Bio Basic Inc.),washes with 10 volumes of binding buffer containing 10,20,and 50 mmol/L imidazole were performed.The column was eluted with 5 volumes of the same solution containing 500 mmol/L imidazole.The obtained fusion proteins were dialyzed in 2 L of 30 mmol/L Tris-HCl buffer(pH 8.0)in one step sufficiently.Finally,the recombinant proteins were then purified by anion-exchange chromatography using DEAE Sepharose FF in binding buffer(30 mmol/L Tris-HCl,pH 8.0)and eluted with elution buffer(30 mmol/L Tris-HCl,1 mol/L NaCl,pH 8.0).After purification,the purity of the fusion peptide CTB-UA was analyzed by 12%SDS-PAGE and computer scan.The samples were dialyzed in 2 L of PBS(20 mmol/L sodium phosphate,pH 7.4)and finally concentrated and stored at-70℃.
1.6 Activity identification of the epitope vaccine CTB-UA
An experimental procedure to identify the activity of the epitope vaccine CTB-UA was performed as previously described with some modifications[7].BALB/c mice were randomized into three groups(6 mice in each group)and were respectively immunized with 100μg of the purified CTB-UA,rCTB and PBS by intraperitoneal immunization in presence of aluminum hydroxide adjuvant for three times at 1 week intervals.Themice were also immunized with rCTB or PBSusing the samemethod as control.Antisera were separated on the fifth day after the final immunization.
The epitope peptides U21 were synthesized commercially in Shanghai TASH Biotechnology Company by the Fmoc solid-phasemethod.The synthesized peptides were purified and analyzed by reverse-phase HPLC,and then,the purified peptides were identified by use of a mass spectrometer.The purity of the epitope peptide was 95.27%.The epitope peptides or CTB were coated overnight on an ELISA plate at 4℃ (10μg/mL,100 μL/well)and the subsequent steps of the assay were performed as described above for the indirect ELISA.All assays were performed in triplicate.
2 Result
2.1 Design on the coup ling sequence of CTB andU21
The fusion proteins CTB-UA and UA-CTB had good antigenicity,hydrophobicity and tertiary structure on the basis of modeling and prediction using RANKPEP,DNAstar software and molecular operating environment(MOE).Experimental studies have reported that the C-terminal fusion to CTB showed most efficient pentamerization and significant GM1-binding.Therefore,U21 was coupled to the C-terminal of CTB in design on the coupling sequence of CTB and U21.
2.2 Design on linker between CTB and U21
In order to decrease the interaction between CTB and U21,a seven-amino-acid,proline-containing segment(DPRVPSS)was used as a spacer at linkage sites between them.CTB and U21 werewell separated by the linker(NPRVPSS)because it was easy to form turn andβ-sheet exceptα-helix.Besides,the linker was helpful for antigen-presentation of the epitope peptideU21 because the linker was easy to form flexible construction.Therefore,we still chose the linker(DPRVPSS)to decrease the interaction between CTB and U21 in constructing the CTB-UA epitope vaccine(Fig.1).
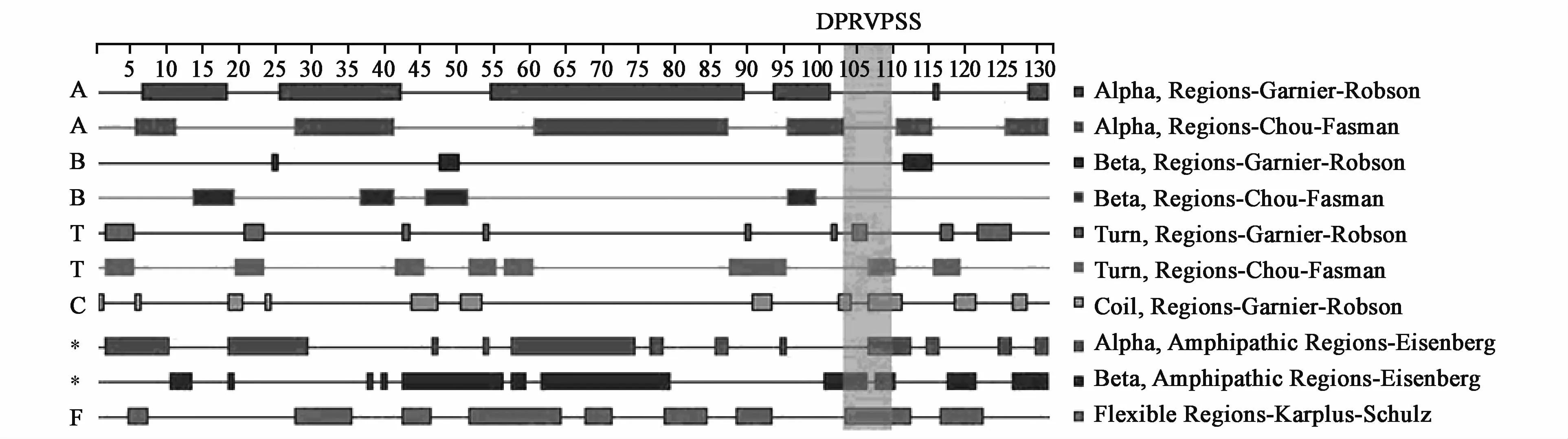
Fig.1 Analysis of the linker(DPRVPSS)in epitope vaccine CTB-UA w ith DNAstar software.The linker(DPRVPSS)ismarked in shadowed region.
2.3 Assessm ent on three dim ensional structure of
t
he epitope vaccine CTB-UA
The three dimensional(3D)structure of the epitope vaccine CTB-UA was predicted by Geno3D and MOE software(Fig.2).The epitope peptide U21 was in the external of CTB-UA,and the structure of CTB-UA was helpful for antigen-presentation of the epitope peptide U21.

Fig.2 3D Ribbon cartoon structure prediction of epitope vaccine CTB-UA by Geno3D and MOE software.
2.4 Identification of the recombinant p lasm id pETC
The CTB gene was amplified from pET-CtUBE using the primer P1-CTB and P1-CTB.The amplified DNA fragment was 342 bp by analysis of 1%agarose gel electrophoresis,and was consistent with the expected size of CTB gene(Fig.3a).After enzyme digestion by NcoⅠand XhoⅠ,the digested DNA fragment from the recombinant plasmids pETC was 342 bp,and was consistent with the expected size of CTB gene(Fig.3b).After successful identification by PCR and restriction endonuclease,the recombinant plasmids pETC were sent to Nanjing Jinsirui Biotechnology Company and further identified by gene sequencing.The result of gene sequencing was exactly consistent with nucleotide sequence of CTB gene.
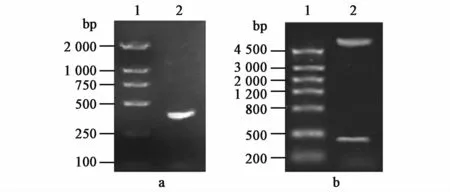
Fig.3 Identification of the recombinant plasm id pETC.(a)Identification of PCR product of CTB gene.1:DNA Marker DL2000;2:PCR product of CTB gene.(b)The double restriction enzyme digestion map of recombinant expression vector pETC.1:DNA MarkerⅢ;2:pETC digested by NcoⅠand XhoⅠ.
2.5 Identification of the recom binant p lasm id pETCUA
The U21 gene was synthesized by PCR using the primer P1-U21 and P1-U21.The synthesized U21 gene was 80 bp by analysis of 1%agarose gel electrophoresis,and was consistent with the expected size of U21 gene(Fig.4a).After enzyme digestion by NcoⅠ and XhoⅠ,the digested DNA fragment from the recombinant plasmids pETCUA was405 bp,and was consistent with the expected size of fusion gene CTB-UA(Fig.4b).After successful identification by PCR and restriction endonuclease,the recombinant plasmids pETCUA were sent to Nanjing Jinsirui BiotechnologyCompany and further identified by gene sequencing.The result of gene sequencing was exactly consistent with nucleotide sequence of CTB-UA fusion gene.
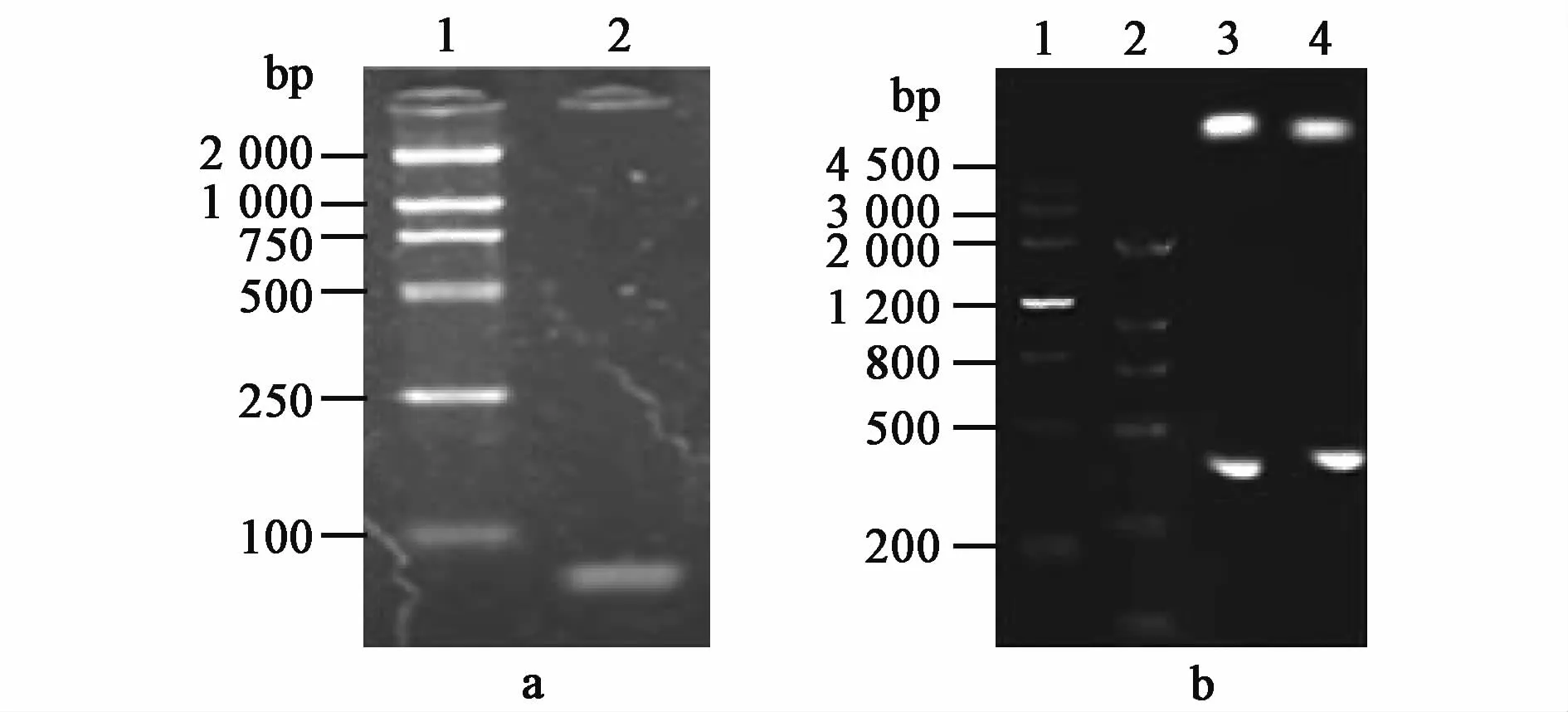
Fig.4 Identification of the recombinant p lasm id pETCUA.
2.6 Expression and optim ization of the fusion protein CTB-UA
E.coli BL21(DE3)bearing the recombinant plasmid pETCUA was cultured in 5 mL Luria-Bertani(LB)containing 100μg/mL ampicillin at37℃ for overnight.The overnight culture was used to inoculate a fresh 100 mL LB culture in a 500 mL flask and allowed to continue to grow at 37℃ with shaking(200 r/min).When the OD600value of the culture reached 0.6,the culture was divided into the groups under IPTG with different concentration(0.2,0.4,0.6,0.8,1.0,1.5 and 2.0 mmol/L),different temperature(28℃,32℃ and 37℃),or different induction time(2 h,3 h,4 h,5 h,6 h,7 h and 8 h).After identification by SDS-PAGE,the optimum induction conditionswere as follows:IPTG concentration of 1.0 mmol/L,induction temperature of 37℃ and induction time of 6 h(Fig.5).

Fig.5 Expression optim ization of fusion protein CTB-UA.
2.7 The fusion protein CTB-UA purified by Ni-NTA affinity chrom atography
The fusion protein CTB-UA was purified by Ni2+-charged column chromatography according to the recommendation of the manufacturer.After the adsorption of CTB-UA protein to Ni-NTA SefinoseTMResin,washes with 10 volumes of wash buffer containing 10 mmol/L imidazole were performed.The column was eluted with 5 volumes of the same solution containing 50 mmol/L or 500 mmol/L imidazole.The obtained fusion proteins were dialyzed in 2 L of 30 mmol/L Tris-HCl buffer in one step sufficiently.After purification by Ni2+-charged column chromatography,the purity of the fusion protein CTB-UA,analyzed by 12%SDS-PAGE(Fig.6)and computer scan,was 82.6%.

Fig.6 SDS-PAGE analysis of the protein peak(CTB-UA)purified by Ni-NTA affinity chromatography.
2.8 The fusion protein CTB-UA purified by anion-exchange chromatography
The fusion proteinswere then purified by anion-exchange chromatography using DEAE Sepharose FF in binding buffer and eluted with elution buffer.After purification by anion-exchange chromatography,the purity of the fusion protein CTB-UA,analyzed by 12%SDSPAGE(Fig.7)and computer scan,was 94.8%.

Fig.7 SDS-PAGE analysis of the protein peak(CTB-UA)purified by anion exchange chromatography.
2.9 Activity identification of the epitope vaccine CTB-UA
The activity of the epitope vaccine CTB-UA was evaluated by ELISA.A modest antibody level was observed in sera from mice immunized with the fusion protein CTB-UA.Compared with intraperitoneal immunization with PBS,immunization with CTB-UA or rCTB significantly increased the levels of specific IgG(P<0.001)against the CTB protein.Besides,compared with intraperitoneal immunization with PBS,immunization with CTB-UA significantly increased the levels of specific IgG(P<0.001)against the peptides U21,butmice immunized with rCTB could not produce specific antibodies against the epitope peptides U21(Fig.8).
3 Discussion
Helicobacter pylori(Hp)is a gram-negative spiral bacterium that infects greater than 50% of the world population and can cause a variety of diseases,including chronic gastritis,peptic ulcers and gastric cancer.Vaccination would be a cost-effective means to control this public health problem faced by one half of the world’s population.In this study,we designed an epitope vaccine CTB-UA with a scientific and reasonable structure composed of cholera toxin B subunit(CTB)and an epitope peptide named U21 from urease A subunit by analyzing the coupling sequence and linker between CTB and U21 using bioinformatics software.
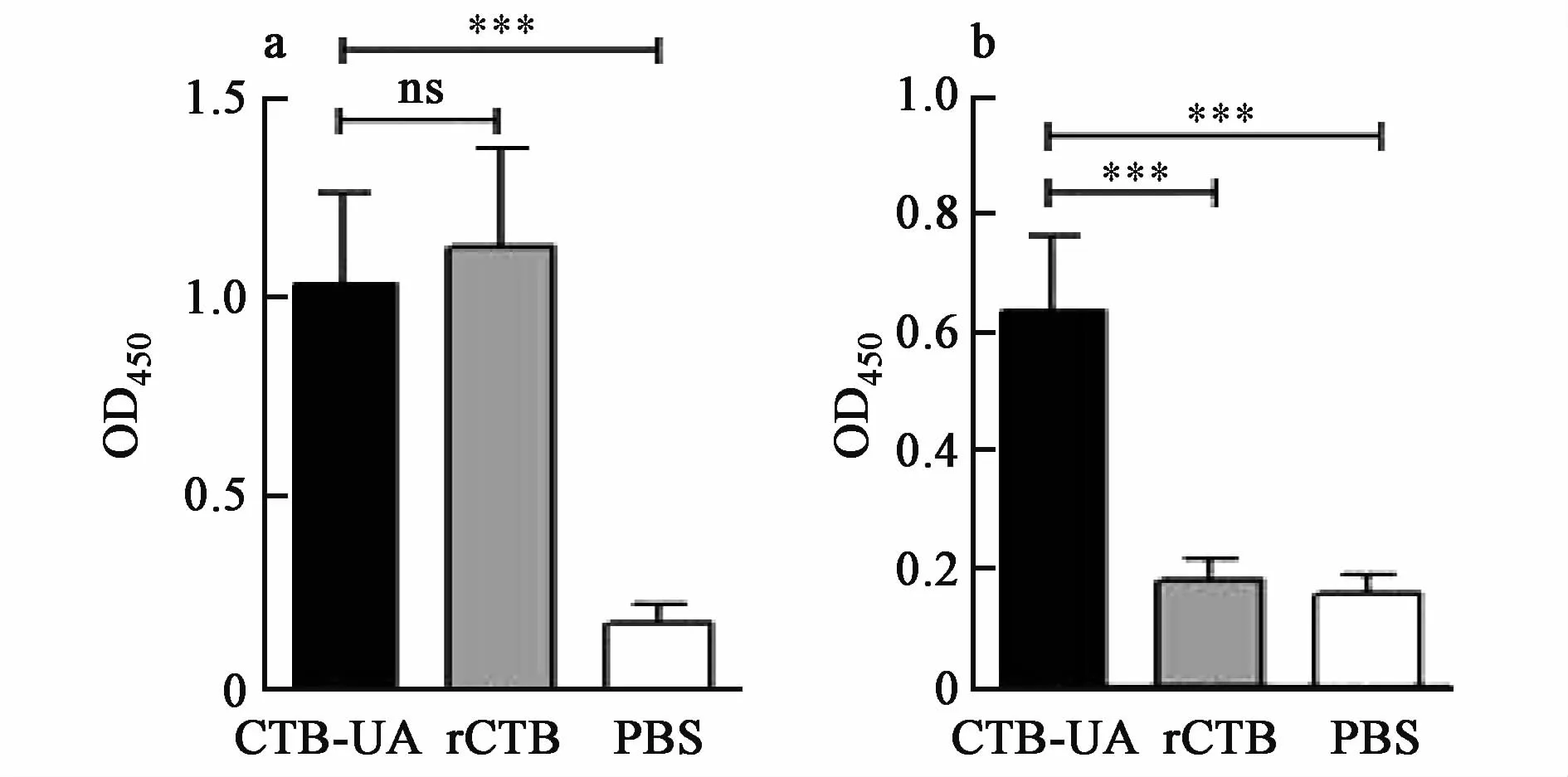
Fig.8 Detection of specific antibodies after immunization.
The design of an epitope vaccine is very important,and a number of factors have been shown to influence its overall success at inducing an immune response against the desired peptide sequence[8,9].The epitope peptides U21 have very low immunogenicity for their low molecularweight.CTB is a potentmucosal adjuvant and chemical conjugations with CTB have been performed usingmany different heterologous antigens.The pentameric form of CTB is responsible for the binding to the GM1 ganglioside,which is present on all nucleated mammalian cells and abundant on intestinal epithelial cells.Themucosal carrier and associated immunological properties of CTB are thought to be critically dependent on its pentameric structure and its ability to bind to GM1 receptors.Experimental studies have shown that the C-terminal fusion to CTB showed more efficient pentamerization than the N-terminal fusion to CTB[10].Therefore,U21 was coupled to the C-terminal of CTB in design on the coupling sequence of CTB and U21.Besides,in order to decrease the interaction between CTBand U21,the linker(DPRVPSS)was used as a spacer at linkage sites between them.It has been proved that the linker(DPRVPSS)could retain the antigenicity of E.coli heat stable enterotoxin(ST)in the LTB-ST fusion protein.DNAstar software predicted that the linker(DPRVPSS)was helpful for antigen-presentation of the epitope peptide U21 because itwas easy to form flexible construction.
More and more experimental evidences have proved that Hp urease-specific polyclonal antibodies generated by immunization with purified Hp urease protein did not inhibit its enzymatic activity at all[11,12].In addition,it has recently been reported that poor response to the ureasemay favor persistence of H.pylori infection and promote the progress of stomach pathology.Amousemonoclonal antibody(mAb),named HpU-2,showed the strongest inhibitory effect on the enzymatic activity of the urease suppressing the urease activity by 82%in twenty-six urease monoclonal antibodies.Further research indicated that the HpU-2 mAbs mainly recognized UreA but weakly UreB and the epitope sequence recognized by HpU-2 mAb was a stretch of UreA-derived 21 amino acid residues(U21).In this study,the epitope vaccine CTB-UA was composed of cholera toxin B subunit(CTB)and the epitope U21.The results indicated thatmice immunized with CTB-UA using aluminum hydroxide adjuvant could induce high level of antibodies specific for both CTB and U21 by ELISA.
In conclusion,we have designed an epitope vaccine named CTB-UA with scientific and reasonable structure.After gene cloning,expression optimization and purification,the fusion protein CTB-UA with high purity(94.8%)was obtained.The epitope vaccine CTB-UA using aluminum hydroxide adjuvant could induce high level of antibodies specific for both CTB and U21.This study will provide much experimental evidences for the development of epitope vaccines against Hp for human use.
[1] Hirai Y,Hayashi S,Shimomura H,et al..Association of Helicobacter pylori with gastroduodenal diseases[J].Jpn.J.Infect.Dis.,1999,52:183-197.
[2] Khan M K,Bemana M.Association of Helicobacter pylori infection and gastric carcinoma[J].Mymensingh.Med.J.,2012,21:80-84.
[3] Tsimmerman Ia S.Gastroduodenal diseases and Helicobacter pylori infection:overview[J].Klin.Med.(Mosk).,2009,87:9-15.
[4] Eaton K A,Brooks C L,Morgan D R,et al..Essential role of urease in pathogenesisof gastritis induced by Helicobacter pylori in gnotobiotic piglets[J].Infect.Immun.,1991,59:2470 -2475.
[5] Marshall B J,Barrett L J,Prakash C,et al..Urea protects Helicobacter(Campylobacter)pylori from the bactericidal effect of acid[J].Gastroenterology,1990,99:697-702.
[6] HifumiE,Yamada Y,Uda T.A catalytic antibody heavy chain HpU-2 degrading its epitope peptide and H.pylori urease[J].Immunol.Lett.,2006,103:68-74.
[7] Guo L,Li X,Tang F,et al..Immunological features and the ability of inhibitory effects on enzymatic activity of an epitope vaccine composed of cholera toxin B subunitand B cell epitope from Helicobacter pylori urease A subunit[J].Appl.Microbiol.Biotechnol.,2012,93:1937-1945.
[8] Liu W,Peng Z,Liu Z,et al..High epitope density in a single recombinant protein molecule of the extracellular domain of influenza A virus M2 protein significantly enhances protective immunity[J].Vaccine,2004,23:366-371.
[9] Livingston B,Crimi C,Newman M,et al..A rational strategy to design multiepitope immunogens based on multiple Th lymphocyte epitopes[J].J.Immunol.,2002,168:5499-5506.
[10] Liljeqvist S,Stahl S,AndreoniC,et al..Fusions to the cholera toxin B subunit:influence on pentamerization and GM1 binding[J].J.Immunol.Methods,1997,210:125-135.
[11] Hirota K,Nagata K,Norose Y,et al..Identification of an antigenic epitope in Helicobacter pylori urease that induces neutralizing antibody production[J].Infect.Immun.,2001,69:6597-6603.
[12] Nagata K,Mizuta T,Tonokatu Y,et al..Monoclonal antibodies against the native urease of Helicobacter pylori:synergistic inhibition of urease activity by monoclonal antibody combinations[J].Infect.Immun.,1992,60:4826-4831.
Design based on Bioinformatics and Expression Optim ization of Fusion Protein w ith Cholera Toxin B Subunit and an Epitope Peptide from Helicobacter pylori Urease A Subunit
GUO Le1,2,LIU Kun-mei1,2,LIXiao-kang2,TANG Feng2,XU Guang-xian1,XITao2*
1.School of Laboratory Medicine,Ningxia Medical University,Yinchuan 750004,China;
2.School of Life Science and Technology,China Pharmaceutical University,Nanjing 210009,China
Helicobacter pylori(Hp)is associated with the development of chronic gastritis,peptic ulcer and gastric cancer.Epitope vaccine based on the enzyme urease of Hp is a promising option for prophylactic and therapeutic vaccination against Hp infection.An epitope vaccine CTB-UA composed of cholera toxin B subunit(CTB)and an epitope peptide named U21 from urease A subunit was constructed by analyzing the coupling sequence and linker between CTB and U21 using bioinformatics software.The recombinant expression vector pETCUA containing the fusion gene CTB-UA and its recombinant strain were constructed by gene cloning technology.After protein expression and optimization,the recombinant protein CTB-UA was purified by Ni2+-charged column chromatography and anion-exchange chromatography using DEAE sepharose FF,about 51 mg of pure target protein was obtained from 1 L of fermentation broth and the purity of CTB-UA was 94.8%.The activity of epitope vaccine CTB-UA was investigated by intraperitoneal immunization experiments in BALB/c mice.Mice immunized with CTB-UA using aluminum hydroxide adjuvant could induce high levelofantibodies specific for both CTB and U21 by ELISA.The epitope vaccine CTB-UA with a scientific and reasonable structure was expressed at a medium level in E.coli and had good immunological specificity.This study will provide much experimental evidences for the development of epitope vaccines against Hp for human use.
Helicobacter pylori;urease;epitope
2012-10-22;Accepted:2012-10-29
s:Supported by the Science Foundation of China Pharmaceutical University(JKY2009023),Postgraduate Innovation Projectof Jiangsu Province(CXZZ11_0817)and National Major Special Program of New Drug Research and Development(2012ZX09103-301-008).
10.3969/j.issn.2095-2341.2013.01.08
Author:Guo Le,lecturer,majored in microbial and biochemical pharmacology.E-mail:guoletian1982@163.com.*Correspondance:Xi Tao,professor,majored in molecular pharmacology.E-mail:nihaoandly@qq.com
关键词:幽门螺旋杆菌;尿素酶;表位
