Expression of CD44, CD24 and ESA in pancreatic adenocarcinoma cell lines varies with local microenvironment
2011-07-05HongJiWeiTaoYinZhuZhuPengFeiShiYuanTianandChunYouWang
Hong-Ji Wei, Tao Yin, Zhu Zhu, Peng-Fei Shi, Yuan Tian and Chun-You Wang
Wuhan, China
Expression of CD44, CD24 and ESA in pancreatic adenocarcinoma cell lines varies with local microenvironment
Hong-Ji Wei, Tao Yin, Zhu Zhu, Peng-Fei Shi, Yuan Tian and Chun-You Wang
Wuhan, China
BACKGROUND:Emerging evidence suggests that pancreatic adenocarcinoma is hierarchically organized and sustained by pancreatic cancer stem cells. Furthermore, elimination of these cells is possible and therapeutically relevant. This study aimed to investigate the expression patterns of pancreatic cancer stem cell surface markers CD44, CD24 and ESA in pancreatic adenocarcinoma cell lines and explore the influence of their local microenvironment.
METHODS:Flow cytometry was used to analyze the expression patterns of CD44, CD24 and ESA in five pancreatic adenocarcinoma cell lines (PANC-1, PC-2, MIA-Paca-2, AsPC-1 and BxPC-3). In addition, the capacity for sphereformation in serum-free medium of four cell lines (PANC-1, PC-2, MIA-Paca-2 and BxPC-3) was assessed. Then, the same assays were performed when tumor cell spheres were developed. The role of sonic hedgehog (SHH) in cell spheres from PANC-1 and MIA-Paca-2 were also assessed by RT-PCR.
RESULTS:CD44 and CD24 were detected in PANC-1. Only CD44 expression was detected in PC-2, MIA-Paca-2 and AsPC-1. CD44, CD24 and ESA were all detected in BxPC-3. Tumor cell spheres developed in PANC-1 and MIA-Paca-2 in serumfree medium. This was accompanied by an increase in CD24 expression and a decrease in CD44 expression in PANC-1. Interestingly, the expression of CD44 and CD24 returned to initial levels once the medium was changed back from serumfree to serum-containing medium. No significant change in the expression of CD44 was detected in MIA-Paca-2. Furthermore, the relative quantification of SHH mRNA in PANC-1 cell spheres was significantly higher than that in cells cultured in the serum-containing medium.
CONCLUSION:The expression patterns of the pancreatic cancer stem cell surface markers CD44, CD24 and ESA were diverse in different pancreatic adenocarcinoma cell lines and changed with their local microenvironment.
(Hepatobiliary Pancreat Dis Int 2011; 10: 428-434)
cell surface antigen; sonic hedgehog; microenvironment; tumor stem cells; pancreatic neoplasms
Introduction
Although the incidence of pancreatic cancer currently ranks the tenth among all cancers and only makes up 3% of all annual new cancer cases in both men and women, it is the fourth leading cause of cancer death in the United States.[1,2]Pancreatic cancer is a fatal disease, with a median survival time of only about 6 months and a 5-year relative survival lower than 5%. Even in patients who undergo a pathologically negative margin resection, the median survival remains at about 2 years, and for those with metastatic disease, the median survival is less than 6 months.[3,4]Over the past decades, the prognosis of pancreatic cancer has remained dismal despite the advances in diagnosis and treatment, mainly because most patients already have local or distant metastasis when diagnosed. In addition, pancreatic cancer is resistant to nearly all conventional therapies. It is imperative to understand the unique biological characteristics of pancreatic cancer and the specific mechanisms for its obstinate malignancy.
The cancer stem cell hypothesis is important to better understand the intrinsic biological characteristics of pancreatic cancer. This suggests that tumor progression is initiated and driven by a small subset of undifferentiated cells with the ability of self-renewal and differentiation into different integrated and heterogeneous tumor populations.[5]The first evidence of cancer stem cells wasfound about a half a century ago[6]and their existence was first verified in acute myelogenous leukemia.[7,8]They were subsequently demonstrated in several solid tumors, including those of the breast,[9]brain,[10]prostate,[11]liver,[12]lung,[13]and colon.[14]The existence of pancreatic cancer stem cells has recently been shown by Li[15]and Hermann.[16]Through analysis of the expression of the distinct cell surface markers CD44, CD24, ESA and CD133, by fluorescence-activated cell sorting (FACS), magnetic-activated cell sorting (MACS), and xenograft tumorigenicity assays, researchers isolated and identified a small subset of pancreatic cancer cells with the capacity for self-renewal, differentiation, and the ability to recapitulate the phenotype of the original tumor. However, these two populations are not identical but only overlap. To better explain this inconsistency, we speculated that the expression patterns of pancreatic cancer stem cell surface markers differ among individuals. Furthermore, the phenotypes of parental cells can change with local microenvironment or niche. To test our hypothesis, we used flow cytometry to detect the expression of the surface markers CD44, CD24 and ESA in five pancreatic adenocarcinoma cell lines (PANC-1, PC-2, MIA-Paca-2, AsPC-1 and BxPC-3). Then, the same assays were performed when the local microenvironment was changed using sphere-formation culture. In addition, the role of sonic hedgehog (SHH) in tumor cell spheres was explored by RT-PCR.
Methods
Experimental materials
Epidermal growth factor and fibroblast growth factorbasic were from PeproTech, USA. The insulin-transferrinselenium liquid media supplement (100×) was from Sigma-Aldrich, Shanghai, China. Bovine serum albumin (BSA) and trypsin were from Amresco, USA, while fetal bovine serum was from Sijiqing, Hangzhou, China. Dulbecco's modified Eagle's medium F12 (DMEM-F12) was from Hyclone, Wuhan, China. Anti-CD44 allophycocyanin (APC), anti-CD24 phycoerythrin (PE), anti-ESA fluorescein isothiocyanate (FITC), and their respective isotype control antibodies were all from Biolegend, USA. Primers for SHH and β-actin amplification were synthesized by Invitrogen.
Cell lines and cultures
The human pancreatic adenocarcinoma cell lines PANC-1, MIA-Paca-2, AsPC-1 and BxPC-3 were from the American Type Culture Collection, Manassas, Virginia, while PC-2 was from the Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China. All five cell lines were grown as monolayer cultures in serum-containing medium (SCM), which was composed of DMEM-F12 and 10% fetal bovine serum, mixed with penicillin (100 U/mL) and streptomycin (100 μg/mL). All cells were cultured in an incubator at 37℃with 5% CO2.
Sphere formation culture
Four pancreatic adenocarcinoma cell lines (PANC-1, PC-2, MIA-Paca-2 and BxPC-3) were chosen for evaluation of sphere-forming capacity using a previously reported method.[17,18]Initially, all cells were cultured in SCM, which was then removed and the cells were cultured in DMEM-F12 containing 0.4% BSA for 24 hours and dissociated with trypsin-EDTA solution (trypsin, 0.25%; EDTA, 0.02%). The enzymatically dissociated single cells were counted and diluted to a density of 5000 cells/mL with serum-free medium (SFM) in culture flasks. The SFM consisted of DMEM-F12 supplemented with 10 ng/mL fibroblast growth factorbasic, 20 ng/mL epidermal growth factor, 5 μg/mL insulin, 2.75 μg/mL transferrin, 2.5 ng/mL sodium selenite, and 0.4% BSA. SFM cultures yielded semi-adherent spherical clusters of cells, usually called spheres. The cells were observed daily under an Olympus CKX41 microscope, and images were captured by an Olympus C5050Z camera as needed. Spheres were enzymatically dissociated every 8 to 14 days with trypsin-EDTA solution for 2 minutes at 37℃. Dissociated single cells were re-plated in SFM at a density of 5000 cells/mL to form more spheres; they were also seeded in SCM when necessary.
Flow cytometry
Dissociated single cells were transferred to a 5-mL tube, washed twice with phosphate buffer solution (PBS), counted, and re-suspended in PBS at 1×106cells/100 μL. Then, the antibodies APC anti-human CD44, PE antihuman CD24, and FITC anti-human ESA (each at a dilution of 1∶40) were added and incubated for 20 minutes on ice in the dark. The respective isotype control antibodies were used at the same concentrations according to the manufacturer's instructions. After washing twice with PBS, samples were re-suspended in 500 μL PBS and analyzed on a flow cytometer (BD LSR II, USA). Side-scatter and forward-scatter profiles were used to eliminate cell doublets. Data were analyzed with BD FACS Diva software.
Quantitative real-time RT-PCR
To assess the level of SHH mRNA in PANC-1 andMIA-Paca-2, three samples of cells from tumor cell spheres and those cultured in SCM were analyzed. Singlecell suspensions of samples were prepared and total RNA was extracted. cDNA was synthesized using a firststrand cDNA synthesis kit (Fermentas) in accordance with the manufacturer's instructions. A reaction mixture with a total volume of 25 μL was used for real-time PCR, containing 12.5 μL SYBR Green PCR Master mix, 100 nmol sense and antisense primer, and 1 μL cDNA. β-actin was used as control. The primers for real-time RT-PCR were: SHH forward, 5'-GAG ATG TCT GCT GCT AGT CC-3'; SHH reverse, 5'-GGA GAT CTT CCC TTC ATA CC-3'; β-actin forward, 5'-CAT TAA GGA GAA GCT GTG CT-3'; and β-actin reverse, 5'-GTT GAA GGT AGT TTC GTG GA-3'. The reaction was as follows: 60 minutes at 37℃; 5 minutes at 95℃ for enzyme inactivation; 40 cycles of denaturation at 94℃ for 20 seconds, annealing at 55℃ for 20 seconds, and extension at 72℃ for 30 seconds. Fluorometric PCR was carried out using the FTC-2000 system. Each reaction was done in triplicate. Quantification of SHH mRNA in samples was determined relative to β-actin using the comparative Ct method.[19]
Statistical analysis
All analyses were performed using SPSS 15.0. Data were expressed as mean±standard deviation. Statistically significant differences were determined by Student's t test, andPvalues less than 0.05 were considered statistically significant.
Results
Presence of CD44,CD24 and ESA on cell surface of pancreatic carcinoma cell lines
Flow cytometric analysis was used to determine the presence of CD44, CD24 and ESA on the cell surface of the pancreatic adenocarcinoma cell lines PANC-1, PC-2, MIA-Paca-2, AsPC-1 and BxPC-3. In PANC-1, 91%-100% (98.88±2.38%) of cells expressed the cell surface marker CD44, and 0.8%-2.2% (1.22±0.57%) expressed CD24; while 0.7%-2% (1.43±0.59%) of cells were CD44+CD24+. All samples were confirmed as ESA negative. Only CD44 expression was detected in PC-2, MIA-Paca-2, and AsPC-1, in 24.3%-49.7% (37.48± 13.70%), 99.1%-99.8% (99.43±0.30%) and 0.5%-2.7% (1.38±0.94%), respectively. In BxPC-3, all three markers were detected, with 99%-99.9% (99.44±0.43%) CD44+, 87.6%-97.3% (93.50±4.82%) CD24+, and 43.9%-98.6% (68.88±24.02%) ESA+. In addition, 42.8%-88.3% (68.30± 19.08%) of cells were CD44+CD24+ESA+. Typical samples are shown in Fig. 1.
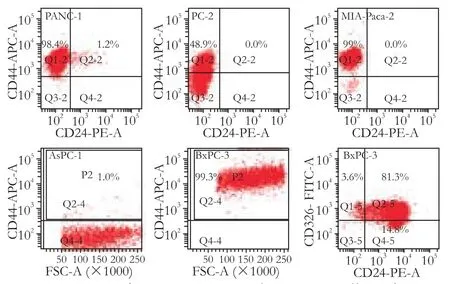
Fig. 1. Presence of CD44, CD24 and ESA on cell surface in pancreatic carcinoma cell lines. Flow cytometric analysis was used to determine the presence of CD44, CD24, and ESA on the cell surface of five pancreatic adenocarcinoma cell lines. PANC-1: CD44+99.6%, CD24+1.2%, CD44+CD24+1.2%; PC-2: CD44+48.9%, CD24-; MIA-Paca-2: CD44+99.0%, CD24-; AsPC-1: CD44+1.0%; BxPC-3: CD44+99.3%, CD24+95.4%, ESA+84.3%, CD44+CD24+ESA+80.7%.
Sphere formationin vitro
A recent study demonstrated that pancreatic cancer stem cells form tumor cell spheres from both primary pancreatic adenocarcinoma specimens[20]and the pancreatic adenocarcinoma cell line PANC-1.[17]To assess the capacity for sphere formation, four pancreatic adenocarcinoma cell lines (PANC-1, PC-2, MIA-Paca-2, and BxPC-3) were cultured in SFM. Results showed that only PANC-1 and MIA-Paca-2 formed tumor cell spheres. Furthermore, sphere formation occurred later in MIA-Paca-2 than in PANC-1. The amount of spheres developed was dramatically less in MIA-Paca-2 than in PANC-1. Images of cells cultured in SFM or SCM are presented in Fig. 2.
Changeable percentage expression of cell surface markers in PANC-1 and MIA-Paca-2
The expression patterns of the cell surface markers CD44, CD24 and ESA were analyzed in PANC-1 and MIA-Paca-2 after the formation of cell spheres in SFM. In PANC-1, CD24 expression increased to 6.4%-23.6%, CD44 expression decreased to 72.5%-95.4%, CD24+CD44+expression increased to 6.6%-21% (Table), and ESA remained negative. In MIA-Paca-2, CD44+cells decreased to 82.6%-98.2% (93.76±6.36% vs 99.43±0.30%, P=0.123), while CD24 and ESA remained negative. Interestingly, the expression of the markers immediately returned to initial levels once the medium was shifted back to SCM, specifically CD44+to 87.5%-99.4% (94.80± 4.77% vs 98.88±2.38%, P=0.093). CD24+to 0.7%-2.4%(1.57±0.68% vs 1.22±0.57%, P=0.388), and CD44+CD24+to 0.7%-2.4% (1.55±0.75% vs 1.43±0.59%, P=0.802) in PANC-1 (Table), and CD44+to 99%-99.8% (99.4±0.37% vs 99.43±0.30%, P=0.919) in MIA-Paca-2. Two typical samples are shown in Fig. 3.
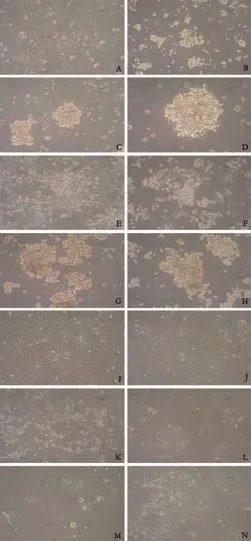
Fig. 2. Sphere-formation ability of pancreatic adenocarcinoma cell lines. To assess the capacity of sphere-formation, four pancreatic adenocarcinoma cell lines (PANC-1, PC-2, MIA-Paca-2 and BxPC-3) were cultured in serum-free medium (SFM). Images of cells were captured at days 7 and 14, as were the images of corresponding cells cultured in serum-containing medium (SCM) as control. A-D: PANC-1 (A: SCM ×100; B: SFM 7 days ×100; C: SFM 14 days ×100; D: SFM 14 days ×200) E-H: MIA-Paca-2 (E: SCM ×100; F: SFM 7 days ×100; G: SFM 14 days ×100; H: SFM 14 days ×200) I-K: PC-2 (I: SCM ×100; J: SFM 7 days ×100; K: SFM 14 days ×100) L-N: BxPC-3 (L: SCM ×100; M: SFM 7 days ×100; N: SFM 14 days ×100).
Expression of SHH mRNA up-regulated in PANC-1 cell spheres
The SHH pathway is critical for normal mammalian development and the pathogenesis of cancer.[21-24]Furthermore, SHH is up-regulated in some types of cancer stem cells, such as those in glioma cancer,[25]breast cancer,[26]small-cell lung cancer,[27]and pancreatic cancer.[15]To determine whether SHH expression was increased in cell spheres of PANC-1 and MIA-Paca-2, realtime quantitative RT-PCR was done using three samples of cell spheres developed in SFM and three samples of cells cultured in SCM. The 2-ΔΔΔCTmethod was used in the analysis of mRNA expression. Results showed that themean level of SHH mRNA in PANC-1 cell spheres was 4.23±0.85, and was higher than that in cells cultured in SCM (1.52±0.60) (P<0.01, Fig. 4). However, the mean level of SHH mRNA in MIA-Paca-2 showed an insignificant elevation (7.88±1.15 vs 6.63±1.05, P>0.05, Fig. 4). This shows that SHH expression was up-regulated in PANC-1 cell spheres compared with cells cultured in SCM.
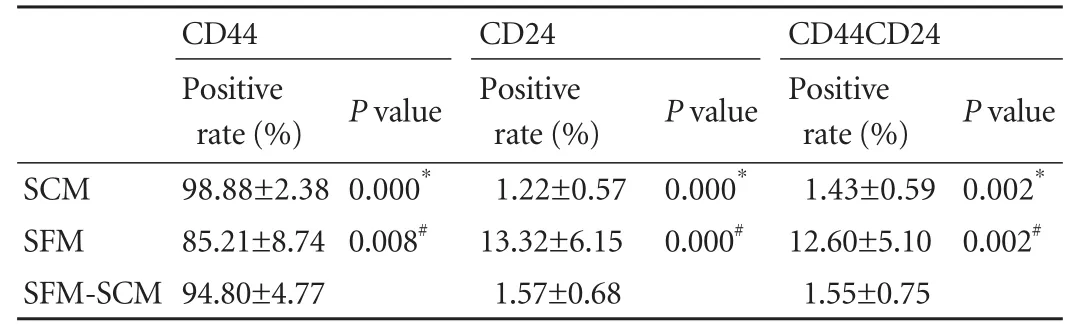
Table. Expression of CD44 and CD24 in PANC-1
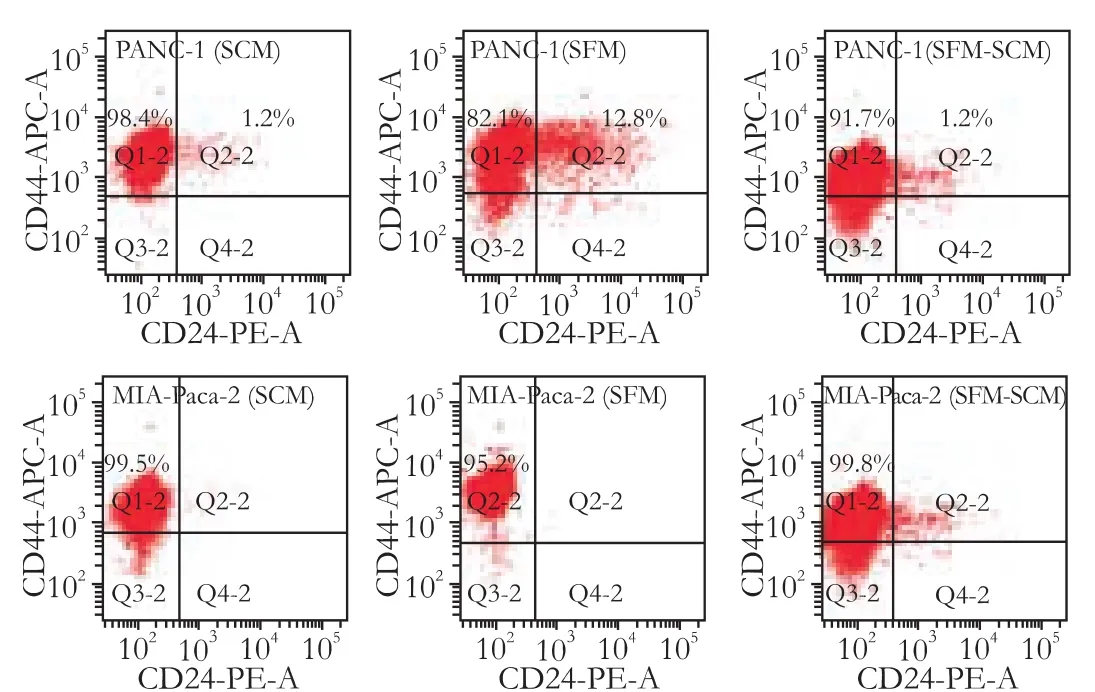
Fig. 3. Changeable percentage expression of CD44 and CD24 in PANC-1 and MIA-Paca-2. PANC-1: SCM CD44+99.6%, CD24+1.2%, CD44+CD24+1.2%; SFM CD44+94.9%, CD24+13.3%, CD44+CD24+12.8%; SFM-SCM CD44+92.9%, CD24+1.3%, CD44+CD24+1.2%; MIA-Paca-2: SCM CD44+99.5%, SFM CD44+95.2%; SFM-SCM CD44+99.8%. SCM: serum-containing medium; SFM: serum-free medium; SFM-SCM: from serum-free medium to serum-containing medium.
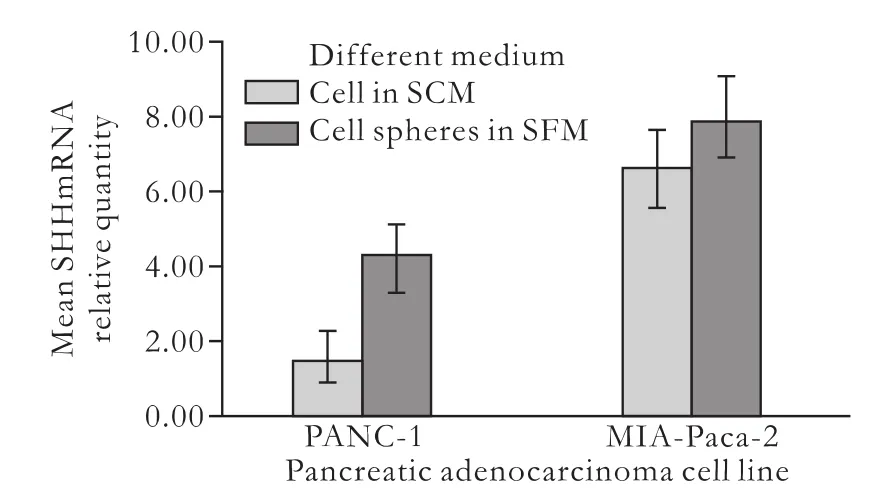
Fig. 4. Expression of SHH mRNA in PANC-1 and MIA-Paca-2. Real-time quantitative RT-PCR was done to assess the levels of SHH mRNA. In PANC-1, the expression of SHH mRNA was upregulated in cell spheres (4.23±0.85 vs 1.52±0.60, P=0.005). In MIA-Paca-2, the mean level of SHH mRNA in cell spheres was slightly but not significantly elevated, compared with that in cells cultured in SCM (7.88±1.15 vs 6.63±1.05, P=0.236).
Discussion
Emerging evidence suggests that solid tumors, including pancreatic adenocarcinoma, are hierarchically organized and sustained by a distinct subpopulation of cancer stem cells, which is responsible for tumor growth, metastasis, and resistance to therapy. Furthermore, elimination of these cells is possible and therapeutically relevant. Using the FACS method and a xenograft model, Li and colleagues[15]identified a subpopulation of highly tumorigenic cancer cells within human pancreatic adenocarcinomas. These highly tumorigenic CD44+CD24+ESA+cancer cells display the ability to selfrenew, generate differentiated progeny, and recapitulate the phenotype of the tumor from which they were derived. As few as 100 CD44+CD24+ESA+pancreatic cancer cells form tumors that are histologically indistinguishable from the original tumors when injected into NOD/SCID mice.[15]Hermann et al[16]demonstrated that CD133+cells in human pancreatic cancer tissue are exclusively tumorigenic and highly resistant to standard chemotherapy using a xenograft model in which primary human pancreatic adenocarcinoma cells were implanted in NMRI-nu/nu mice and investigated by the MACS method. In addition, they showed that CD133+CXCR4+cancer cells are essential for tumor metastasis. However, these two reports leave several questions to be addressed. First, obtaining adequate amounts of cells from patients for direct sorting is difficult in practice. An alternative is to allow proliferation of established xenografts in immunodeficient mice, but this procedure is complicated and time-consuming, usually requiring 3-4 months. Second, the expression of cell surface markers depends on the tumor of origin and its ability for self-renewal and differentiation. Besides, results of flow cytometry may vary because of the method of cell preparation, gate setting, and the specific antibody used. Finally, potential contamination of sorted cells should also be addressed. For example, ESA-cells probably include non-epithelial inflammatory, stromal, and vascular cells,[28]and CD133-may represent differentiated hematopoietic and endothelial cells.
Meanwhile, Gou et al[17]used the clonal colonyforming assay to verify that cell spheres of PANC-1 developed in SFM have the cancer stem cells properties of self-renewal and production of differentiated progeny. Dembinski and colleagues[29]also identified a subpopulation of slow-cycling DiI+tumor cells in BxPC-3 and Panc03.27 using a label-retention technique. The identified sub-population had enhanced tumorigenicity and morphological characteristics, resembling cells that have undergone an epithelial-to-mesenchymal transition. At present, direct evidence for the existence of cancer stem cells in human pancreatic cancer in various studies has not been consistent. To isolate and identify pancreatic cancer stem cells more efficiently, these methods should be further investigated.
In this study, we found that the expression patterns of CD44, CD24 and ESA in pancreatic adenocarcinoma cell lines were dramatically different. In PANC-1, almost all cells expressed CD44, while few expressed CD24, or were CD44+CD24+. All samples were confirmed to be ESA negative. In PC-2, MIA-Paca-2 and AsPC-1, only the expression of CD44 was detected. In contrast, all three markers were detected in BxPC-3. Moreover, a change in the local microenvironment of cells, as exemplified by culture in SFM, led to an increase in CD24 expression and a decrease in CD44 expression in PANC-1. Once the medium was shifted back to SCM and the local microenvironment was restored, the expression of CD44 and CD24 quickly returned to the initial levels even during the first passage. However, significant changes in CD44 expression were not detected in MIA-Paca-2 despite similar alteration of the local microenvironment. These findings indicate that the expression patterns of the stem cell surface markers CD44, CD24 and ESA in pancreatic adenocarcinoma cell lines are diverse. Furthermore, the expression of CD44 and CD24 changed with variations of the local microenvironment,and this change was reversible. However, these expression changes cannot be definitely attributed to the variable expression of cell surface proteins, because the distinct propagation capacity of subpopulations with different expression patterns in SFM can also lead to changes in the percentage expression. Our results suggest that using one or a few fixed surface markers to isolate pancreatic cancer stem cells from all pancreatic adenocarcinoma specimens is not feasible.
In addition, we found that not all pancreatic adenocarcinoma cell lines develop tumor cell spheres in SFM. This result is in agreement with a newly published report in which Gaviraghi et al[30]showed that PANC-1, PSN-1, and CFPAC-1 are capable of forming spheres. Moreover, elevated expression of SHH was not detected in all tumor cell spheres. SHH mRNA expression was up-regulated in PANC-1 spheres, further verifying that these are cancer stem cells. This also implies that sphere formation culture is a potentially simple and convenient method to obtain more pancreatic cancer stem cells from certain individuals. All these conclusions should be further validated in primary human pancreatic adenocarcinoma cells.
In summary, this study demonstrated that the expression patterns of the pancreatic cancer stem cell surface markers CD44, CD24 and ESA in pancreatic adenocarcinoma cell lines are clearly different. Their expression varies with local microenvironment or niche, although the exact mechanism of these changes and their correlation with the SHH signaling pathway need further investigation.
Acknowledgement
We thank the Research Laboratory of General Surgery, Union Hospital, Wuhan, for technical assistance.
Funding:This study was supported by a grant from the National Natural Science Foundation of China (30801100).
Ethical approval:Not needed.
Contributors:YT and WCY designed the research. WHJ and SPF performed the majority of experiments. TY provided analytical tools. WHJ and ZZ analyzed the data. WHJ and YT wrote the paper. All authors contributed to the design and interpretation of the study and to further drafts. WCY is the guarantor.
Competing interest:No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
1 Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225-249.
2 Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300.
3 Philip PA, Mooney M, Jaffe D, Eckhardt G, Moore M, Meropol N, et al. Consensus report of the national cancer institute clinical trials planning meeting on pancreas cancer treatment. J Clin Oncol 2009;27:5660-5669.
4 Zhang J, Dhakal I, Yan H, Phillips M, Kesteloot H; SEER Cancer Registries. Trends in pancreatic cancer incidence in nine SEER Cancer Registries, 1973-2002. Ann Oncol 2007;18: 1268-1279.
5 Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature 2001;414:105-111.
6 Bruce WR, Van Der Gaag H. A quantitative assay for the number of murine lymphoma cells capable of proliferation in vivo. Nature 1963;199:79-80.
7 Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730-737.
8 Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994;367:645-648.
9 Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100:3983-3988.
10 Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature 2004;432:396-401.
11 Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005;65:10946-10951.
12 Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007;132: 2542-2556.
13 Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 2005;121:823-835.
14 Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007;445:111-115.
15 Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res 2007;67:1030-1037.
16 Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007;1:313-323.
17 Gou S, Liu T, Wang C, Yin T, Li K, Yang M, et al. Establishment of clonal colony-forming assay for propagation of pancreatic cancer cells with stem cell properties. Pancreas 2007;34:429-435.
18 Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005;65:5506-5511.
19 Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-408.
20 Simeone DM. Pancreatic cancer stem cells: implications for the treatment of pancreatic cancer. Clin Cancer Res 2008;14:5646-5648.
21 Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001;15: 3059-3087.
22 Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature 2003;425:846-851.
23 Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature 2004;431:707-712.
24 Lau J, Kawahira H, Hebrok M. Hedgehog signaling in pancreas development and disease. Cell Mol Life Sci 2006;63: 642-652.
25 Clement V, Sanchez P, de Tribolet N, Radovanovic I, RuiziAltaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol 2007;17:165-172.
26 Lu ZH, Jia J, Ren J, Ma B, DI LJ, Song GH. Detection of breast cancer stem cells and the expression of key molecules in Hedgehog signaling pathway. Beijing Da Xue Xue Bao 2008; 40:480-485.
27 Watkins DN, Berman DM, Baylin SB. Hedgehog signaling: progenitor phenotype in small-cell lung cancer. Cell Cycle 2003;2:196-198.
28 Lonardo E, Hermann PC, Heeschen C. Pancreatic cancer stem cells-update and future perspectives. Mol Oncol 2010;4: 431-442.
29 Dembinski JL, Krauss S. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin Exp Metastasis 2009;26:611-623.
30 Gaviraghi M, Tunici P, Valensin S, Rossi M, Giordano C, Magnoni L, et al. Pancreatic cancer spheres are more than just aggregates of stem marker-positive cells. Biosci Rep 2010; 31:45-55.
Received March 28, 2011
Accepted after revision May 26, 2011
The horizon of life is broadened chiefly by the enlargement of the heart.
—Black
Author Affiliations: Department of Pancreatic Surgery (Wei HJ, Yin T, Zhu Z, Shi PF and Wang CY) and Laboratory of General Surgery (Tian Y), Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China
Chun-You Wang, MD, Department of Pancreatic Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Ave, Wuhan 430022, China (Tel: 86-27-85351621; Fax: 86-27-85351669; Email: chunyouwang52@126.com)
© 2011, Hepatobiliary Pancreat Dis Int. All rights reserved.
杂志排行
Hepatobiliary & Pancreatic Diseases International的其它文章
- Prognostic role of diabetes mellitus in hepatocellular carcinoma patients after curative treatments: a meta-analysis
- Risk factors of severe ischemic biliary complications after liver transplantation
- Surgical treatment of Budd-Chiari syndrome: analysis of 221 cases
- Efficacy of liver transplantation for acute hepatic failure: a single-center experience
- Combined invagination and duct-to-mucosa techniques with modifications: a new method of pancreaticojejunal anastomosis
- Large regenerative nodules in a patient with Budd-Chiari syndrome after TIPS positioning while on the liver transplantation list diagnosed by Gd-EOB-DTPA MRI
